Abstract
Autosomal dominant polycystic kidney disease (ADPKD) is the most common renal genetic disease, with most patients carrying mutations in PKD1. The main feature is the formation of bilateral renal cysts, leading to end stage renal failure in a significant proportion of those affected. Despite recent advances made in understanding ADPKD, there are currently no effective curative therapies. The emergence of human induced pluripotent stem cell (hiPSC)-derived kidney disease models has led to renewed hope that more physiological systems will allow for the development of novel treatments. hiPSC-derived organoid models have been used to recapitulate ADPKD, however they present numerous limitations which remain to be addressed. In the present study, we report an efficient method for generating organoids containing a network of polarised and ciliated epithelial tubules. PKD1 null (PKD1−/−) organoids spontaneously develop dilated tubules, recapitulating early ADPKD cystogenesis. Furthermore, PKD1−/− tubules present primary cilia defects when dilated. Our model could therefore serve as a valuable tool to study early ADPKD cystogenesis and to develop novel therapies.
Similar content being viewed by others
Introduction
Autosomal dominant polycystic kidney disease (ADPKD) is the most common kidney genetic disorder, with a point prevalence of clinically diagnosed cases in Europe of approximately 3 to 4 per 10,0001. According to registry data, around 10% of patients undergoing renal replacement therapy have ADPKD2. The main feature of ADPKD is massive growth and enlargement of fluid-filled cysts in both kidneys, slowly impairing renal function over time and causing end-stage renal disease (ESRD) in most patients by the age of 603,4. This process appears to begin in renal tubular epithelia, which initially become dilated, increasing in lumen size, followed by the separation of the cystic tissue from the original tubule5. This condition is caused by mutations in the genes PKD1, which encodes polycystin-1 (PC1), and PKD2, which encodes polycystin-2 (PC2) and are responsible for approximately 85% and 15% of cases, respectively3. PC1 is a large transmembrane protein with a large extracellular domain, while PC2 may function as a Ca2+ channel. It has been shown that the two proteins form a heterodimer located mainly at the primary cilium5. Although the function of this complex remains controversial, the dominant hypothesis is that PC1-PC2 heterodimers act as mechanosensors that signal via Ca2+ entry into the cell, as disruption of either protein leads to impaired Ca2+ signaling6,7. In ADPKD, impaired Ca2+ inflow through the tubular cell membrane results in reduced intracellular Ca2+ concentration and secondary adenyl cyclase over-activation with increased cyclic adenosine monophosphate (cAMP) levels8. Medications, such as the vasopressin type 2 receptor antagonist Tolvaptan or the somatostatin analogue Octreotide-LAR, that reduce intracellular cAMP, have been tested in clinical trials in ADPKD patients9,10,11,12. Tolvaptan, currently the only FDA-approved treatment for rapidly progressive ADPKD13, has a modest ability to slow disease progression and its use is limited by major side effects such as hepatotoxicity9,10. In recent trials, 3-year Octreotide-LAR treatment slowed total kidney and cyst volume growth, reduced chronic decline of renal function11, and, in patients with stage 3b-4 chronic kidney disease (CKD), the progression to ESKD12. Moreover, Octreotide-LAR therapy reduced total liver volume in ADPKD patients with associated polycystic livers14 and improved left ventricular function in these patients15. Notably, combined therapy with Octreotide-LAR and Tolvaptan has additional beneficial effects, at least in the early phase of ADPKD16. Octreotide-LAR studies have been performed in relatively small cohorts of ADPKD patients and larger series of patients with longer exposure to treatment would be needed to investigate these signals further. Therefore, given that so far the available therapies are encouraging but not yet fully curative for ADPKD patients, there is a pressing need of robust in vitro models that recapitulate essential features of ADPKD to test novel candidates in light of their potential use17,18,19.
Many current models are based on culturing single renal epithelial cells suspended in an extracellular matrix, where cystogenesis is generally induced with cAMP-stimulating agents, such as forskolin or arginine vasopressin18,20. cAMP signalling has been identified as one of the central pathways that is altered following PKD1 and PKD2 dysfunction, however it does not appear to be the sole mechanism leading to cyst formation5,21. Therefore, an ideal in vitro model should recapitulate the tubular architecture and exhibit cystogenesis in a genotype-dependent manner, without the need for cAMP activation, as this may bias studies towards identifying cAMP modulators as drug candidates. Recently we demonstrated that it is possible to engineer tubular structures to model ADPKD using primary cyst-derived cells isolated from a patient22. These structures can be used for preclinical drug discovery and testing, although the culture of primary cells presents limitations, and their isolation constitutes a risk for patients.
The discovery that adult somatic cells can be reprogrammed into human induced pluripotent stem cells (hiPSCs)23 has completely revolutionised the field24. As for kidney disease, various differentiation protocols have been established that can generate both specific renal cell types25,26, as well as nephron27,28,29 and ureteric bud (UB)/collecting duct (CD) organoids30,31,32,33,34, containing multiple cell types. Such organoid models have demonstrated their ability to mimic ADPKD cystogenesis35,36,37, but most require chemical agents to induce cysts and do not replicate early processes, such as tubular dilation, an initial step leading to cystogenesis. Nishinakamura and colleagues showed cystogenesis in hiPSC-derived UB organoids, but the cysts occurred at a low rate, required cAMP stimulation and mostly appeared to originate from nontubular regions37. Freedman and colleagues have demonstrated that in nephron organoids the removal of extracellular matrix leads to spontaneous cystogenesis in PKD1 and PKD2 mutant organoids38. During the preparation of this manuscript, Osafune and colleagues reported a methodology for long term expansion of UB progenitors and the generation of cortical CD cells with enhanced maturity that allowed for spontaneous cystogenesis from PKD1 null organoids34.
In the present report, taking advantage of the many advances that have been made in the studies described above, we established a highly robust methodology to generate ciliated tubular epithelial organoids that express key specific markers. Furthermore, we demonstrated that PKD1 homozygous mutant organoids spontaneously develop dilated tubules, recapitulating an early process in ADPKD cystogenesis.
Results
UB spheroid differentiation from hiPSCs
We started out using a differentiation protocol by Xia and colleagues22,39 that generated UB progenitors. Experiments to optimise the protocol were carried out using the hiPSC line IRFMNi003-A, generated previously by our group25. The 4-day procedure involves seeding hiPSCs as clusters and culture in monolayer. As expected, after 4 days of differentiation, the cells most closely resembled the anterior intermediate mesoderm stage, as many early genes were upregulated, but RET, an essential receptor for ureteric bud branching40, was not induced (Fig. S1). Consequently, we decided to prolong the maturation protocol. Moreover, due to the high variability in cluster formation affecting the reproducibility of the protocol, we decided to culture the cells as spheroids on low attachment plates.
We applied the 4-day protocol to hiPSC spheroids, which were cultured in the presence of BMP4 and FGF2 for the first 2 days to derive primitive streak (Fig. 1A, Step 1), then anterior intermediate mesoderm was specified using Activin A, BMP2 and Retinoic acid for 2 additional days (Fig. 1A, Step 2). In order to promote maturation towards the UB, we investigated factors involved in embryonic development. Wnt and fibroblast growth factor (FGF) signals are essential in renal development31 as is RET, whose expression is maintained by retinoic acid (RA) signaling41,42 and inhibited by BMP ligands40,43. We therefore cultured day 4 differentiated spheroids for additional 2 days in medium containing CHIR99021, a Wnt agonist, FGF9, RA and LDN193189 Tetrahydrochloride (LDN), a strong BMP antagonist (Fig. 1A, Step 3). Over the course of the 6 days, spheroids grew substantially (Fig. 1B), while acquiring the expression of typical early UB lineage markers, including PAX2, GATA3, HOXB7 and LHX1 (Fig. 1C).
UB spheroid differentiation from hiPSCs. (A) Overview of UB spheroid differentiation protocol. (B) Time course brightfield images of UB spheroid differentiation. Scale bar, 100 μm. (C) Gene expression analysis showing the specification of UB spheroids over 10 days. Expression levels are relative to HPRT1, n = 3 independent experiments. Data represented as mean \(\:\pm\:\) SD. (D) Wholemount confocal immunofluorescence images of day 10 UB spheroids showing expression of the ureteric bud markers HOXB7, AQP2, PAX2, CDH1 and GATA3. Scale bar, 50 μm.
We then dissociated the day 6 spheroids and seeded the cells on low attachment plates in medium containing GDNF to promote epithelialisation of the spheroids (Fig. 1A, Step 4). Spheroids were then matured for additional 2 days in modified medium (Fig. 1A, Step 5). The gene expression profile at day 10 showed the expression of UB markers WNT9B and AQP2, and upregulation of RET (Fig. 1C). Immunofluorescence analysis revealed that the UB spheroids contained epithelialised regions and expressed the lineage markers HOXB7, AQP2, E-cadherin (CDH1), PAX2, and GATA3 (Fig. 1D). The fact that not all cells become epithelialised is in line with similar UB specification protocols, where UB cells are enriched with fluorescent sorting31 or by manual procedures33,34.
To test the robustness of the protocol we then differentiated the hiPSC lines IRFMNi001-B, and Gibco Episomal hiPSCs and determined that they differentiated into UB spheroids with similar efficiency to our reference line, as shown by gene expression (Fig. S2A) and immunofluorescence analysis (Fig. S2B). As expected, cells with a homozygous deletion of PKD1 (PKD1−/−) generated by our group44 could also be efficiently differentiated (Fig. S2).
UB spheroids can generate tubular organoids with tight epithelial barriers
We then assessed whether the UB spheroids could generate branching tubular structures resembling the CD. GDNF-RET signalling is implicated in UB growth40, and numerous other signalling pathways contribute to specifying the correct lineage. FGF ligands play key roles, such as FGF1, which promotes branching and patterning of the stalk, and FGF7, which increases proliferation of the UB45, while Wnt signalling controls an important gene hierarchy46 and is activated in the UB in developing human kidneys47. Day 10 UB spheroids were cultured in branching medium (Fig. 2A), without the addition of basement membrane extracts, to promote tubule formation and branching for 14 days. This resulted in the formation of numerous tubules with a clear lumen inside the spheroids, evolving into a complex tubular network (Fig. 2B,C). The tubules showed expression of UB markers AQP2, Calbindin (CALB1), Cytokeratin 8 (KRT8), SOX9 and HOXB7 (Fig. 2D). Following branching culture, we observed that AQP2 expression was maintained, while cells acquired the expression of ELF5, a transcription factor specifically expressed in principal cells of the CD48, and AVPR1A, an arginine vasopressin receptor expressed early during human kidney development37 (Fig. 2E). AVPR2, a mature marker involved in CD cyst formation49, was not detected similarly to the previously mentioned studies aimed at generating CD organoids30,32,33,37 (data not shown). These organoids expressed numerous UB markers, however since certain key markers, including RET in UB tip cells, were not detected we termed them tubular epithelial organoids.
Generation of tubular epithelial organoids. (A) Outline of the culture conditions used to generate tubular epithelial organoids. (B) Brightfield image showing a day 24 organoid. Scale bar, 200 μm. (C) Histological PAS staining of a day 24 organoid. Scale bar, 200 μm. (D) Wholemount confocal immunofluorescence images of day 24 organoids showing expression of key ureteric bud markers CALB1, AQP2, KRT8, SOX9, and HOXB7. Scale bar, 50 μm. (E) Gene expression analysis showing that day 24 tubular organoids maintain expression of AQP2, while acquiring expression of genes characteristic of embryonic ureteric markers ELF5 and AVPR1A. Expression levels are relative to HPRT1, n = 3 independent experiments. Data represented as mean \(\:\pm\:\) SD. (F,G) Heatmaps showing differentially expressed typical UB (F) and urothelial (G) markers between day 24 and day 10 evaluated with RNA sequencing. Each column indicates a single replicate at day 24 compared to average day 10 expression.
In order to better characterise the cells, we then performed transcriptomic analysis with RNA sequencing on the spheroids and organoids. This confirmed the expression of some UB lineage markers (Fig. 2F). Due to the observation of multilayered tubules in our organoids and the common origin of the renal CD and the ureter, we investigated the expression of common urothelial markers in our samples. Indeed, upregulation of the urothelial markers UPK1A, UPK1B, TP63, UPK3A, KRT20, SHH and FOXA2 was found between day 10 and day 24 (Fig. 2G).
We then generated tubular epithelial organoids from hiPSC lines IRFMNi001-B and Gibco Episomal hiPSCs and observed similar marker expression compared to our reference cell line (Fig. S3), confirming the robustness of the protocol.
PKD1 −/− tubular organoids reproduce early ADPKD processes
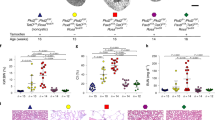
Next, we verified the potential of the generated tubular organoids to mimic ADPKD. As expected, PKD1−/− hiPSCs (IRFMNi003-A-4) were similarly differentiated, as shown by lineage marker expression (Fig. S3), however as tubules emerged, we observed that, unlike in isogenic control organoids (IRFMNi003-A), they became highly dilated and the luminal space continuously expanded (Fig. 3A). The tubular lumen surface was significantly higher in PKD1−/− organoids than in controls (p < 0.0001, Fig. 3B, left panel) and the luminal space occupied a significantly higher proportion of the total organoid area (p < 0.0001, Fig. 3B, right panel). PAS staining of the organoids (Fig. 3C) and serial confocal z-stack images (Fig. 3D) further revealed the cystic dilations in PKD1−/− organoids. Additionally, we determined that PKD1−/− organoids recapitulate cystogenesis in multiple culture formats, enabling the upscaling of their culture for prospective drug screening efforts (Fig. S5).
PKD1−/− tubular organoids develop dilated tubules resembling early cystogenesis. (A) Brightfield images of Control and PKD1−/− day 24 organoids showing the tubular dilations in PKD1−/− organoids. Scale bar, 200 μm. (B) Scatter plots showing the total lumen surface (left panel) and the percentage occupied by the lumen (right panel) in control and PKD1−/− organoids. Graphs represent data from n = 60 control and n = 61 PKD1−/− organoids from 3 independent experiments. Red asterisk indicates p value < 0.0001. Unpaired t-test with Welch’s correction was used. (C) Histological PAS staining of control and PKD1−/− tubular organoids indicating tubular dilations in PKD1−/− organoids. Scale bar, 200 μm. (D) Wholemount confocal immunofluorescence staining of KRT8 in control and PKD1−/− tubular organoids displaying the tubular structure and varying depths. Scale bar, 100 μm. (E) Time course brightfield images of control and PKD1−/− organoids upon forskolin treatment. Forskolin was added at a concentration of 25 μm from day 16 to day 18. Scale bar, 200 μm. (F) Immunohistochemical analysis of control and PKD1−/− tubular organoids. Ki67, left panels, cleaved caspase 3 (Casp3), right panels. Scale bars, 200 μm. (G) Quantification of Ki67+ cells in tubular epithelial cells in control and PKD1−/− organoids, n = 3 organoids in 3 independent experiments.
We then investigated whether the organoids could respond to cAMP stimulation with forskolin, a known inducer of cystogenesis50. Organoids were treated with forskolin for 2 days, from day 16 to day 18, when tubules had already developed. Treatment with forskolin for 2 days led to gross expansion of the luminal space of tubules, with no difference between control and PKD1−/− organoids (Fig. 3E). After the withdrawal of forskolin, the tubules of PKD1−/− organoids enlarged continuously and by day 24 they had a drastically larger luminal space than control organoids (Fig. 3E).
To better understand the mechanisms of tubular lumen expansion, proliferation and apoptosis were investigated in day 24 organoids. Tubular epithelial cells in PKD1−/− organoids were significantly more proliferative than controls (p < 0.005, n = 3 organoids in 3 independent experiments), as shown by Ki67 staining (Fig. 3F–G). Conversely, apoptosis, determined by cleaved caspase 3 staining, was exclusively observed in nontubular cells concentrated in the center of the organoids (Fig. 3F), likely due to the size of the organoids that prevented nutrient access to these cells and the lack of a vascular endothelial network.
Ultrastructural analysis of PKD1 −/− organoids reveals loss of cilia
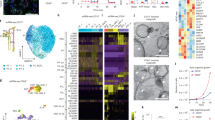
Finally, we sought to better characterise the morphology of the organoids. We therefore performed ultrastructural analysis using scanning electron microscopy (SEM) on the organoids. The outer appearance of control and PKD1−/− organoids was highly similar, and high magnification images showed that they were coated with a fiber-like extracellular matrix (ECM) (Fig. 4A). As the organoids are cultured in the absence of Matrigel or any other ECM component, it is likely that the ECM observed in the tubular organoids was produced by the epithelial cells. Organoids were then fractured to allow observation of the inner structures. Analysis of control organoids revealed tubules with a small lumen (Fig. 4B, left panel, white arrows) and an apical membrane characterised by numerous microvilli, as well as primary cilia extending towards the lumen (Fig. 4B, right panels, red arrows and inset). In PKD1−/− organoids, on the other hand, the tubules appeared severely dilated (Fig. 4B, left panel, white asterisks). The apical side of such tubules was mostly devoid of microvilli or primary cilia (Fig. 4B, right panels and inset), while it appeared that tubules with smaller lumens did possess microvilli and cilia (Fig. 4C, red arrows and inset). In some cystic tubules, there appeared to be small areas with microvilli and cilia, followed by a transition zone of abrupt switch to the smooth phenotype seen in large cysts (Fig. 4D, black arrows), possibly hinting at a dynamic transition from normal to cystic tubules, which is accompanied by the loss of microvilli and cilia. Immunostaining of ARL13B, a primary cilium marker51, confirmed the finding that cystic tubules in PKD1−/− organoids mostly lack primary cilia (Fig. 4E, middle panel), while some areas did possess cilia (Fig. 4E, right panel). By contrast, ARL13B staining could be found on the apical side of all tubules in control organoids (Fig. 4E, left panel).
PKD1−/− tubular organoids show a dynamic loss of cilia in dilated tubules. (A) Scanning electron micrographs showing the outer structure of control and PKD1−/− organoids. Scale bars, 100 μm for whole organoids, 10 μm for magnified images. (B) Scanning electron micrographs, with the corresponding high magnification insets, showing the inner structure of control and PKD1−/− organoids. White arrows indicate tubules in control organoids while red arrows indicate microvilli and cilia in control organoids. Cystic tubules in PKD1−/− organoids are marked with an asterisk. Scale bar, 100 μm, left panels, 10 μm in centre and right panels. (C) Scanning electron micrograph, with the corresponding high magnification insets, showing a PKD1−/− tubule exhibiting normal microvilli and primary cilia (red arrows, inset), scale bar, 2 μm and (D) a cystic PKD1−/− tubule showing the transition zone from ciliated to non-ciliated phenotype (black arrows, inset), scale bar, 10 μm. (E) Wholemount confocal immunofluorescence images of control and PKD1−/− tubular organoids showing loss of cilia (ARL13B) in dilated PKD1−/− tubules. Scale bar, 25 μm.
Discussion
In ADPKD, large and severe cysts are found predominantly in the renal CD52,53,54 hinting that there may be cell-autonomous processes that drive disease progression. Numerous studies have reported on methodologies to generate UB/CD tissues from hiPSCs30,31,32,33 and they have demonstrated their ability to model ADPKD cystogenesis34,37. Despite these remarkable advances, such procedures remain technically challenging and time intensive, which may limit their feasibility.
In the present study we generated tubular epithelial organoids from hiPSCs. Starting from a previously reported method to generate UB progenitors, we further differentiated the cells into ciliated tubular epithelial organoids expressing key lineage markers. This methodology is highly reproducible between experimental batches and does not necessitate optimisation of culture conditions for different cell lines. Importantly, the culture of the spheroids, and subsequently organoids, does not require complex techniques or cell sorting at any stage, ensuring that this methodology can be applied by other research groups. Organoids generated with our protocol appear to have multilayered epithelial tubules, which in mouse development is only seen during the initial stages of UB branching. During human renal development, however, it has been observed that the UB is multilayered at various stages, ranging from the initial branching event, up to at least 10 weeks of gestation55. Notably, the ureter shares a common origin with the UB lineage and it is known to be a multilayered epithelium56. Consistent with this, analysis of common urothelial markers revealed significant upregulation of some of these genes, raising the possibility that this cell fate was also induced to some degree in our organoids. Importantly, during human renal development the UB displays some plasticity as it can express some urothelial markers such as uroplakins55. While we observed the expression of some UB and ureter markers in our organoids, given the lack of some lineage markers, as well as visible dichotomous branching, we defined them as tubular epithelial organoids.
Unlike some UB-like organoid differentiation methods31,32,33,57 our organoids are cultured without the addition of Matrigel or an ECM and form tubules internally that branch repeatedly. This important development greatly facilitates manipulation of the organoids and even allows high throughput culture for future drug screening efforts.
We have shown that tubular organoids generated from PKD1−/− hiPSCs spontaneously develop cystic tubules resembling early tubular dysregulation in ADPKD. PKD1−/− showed more proliferation in the tubules, consistent with previous studies highlighting the role of proliferation in cystogenesis58. Our finding that culturing PKD1−/− organoids in suspension without ECM promotes spontaneous cystogenesis is in line with the discovery by Cruz and colleagues38. Having a spontaneous model of cystogenesis in ADPKD is crucial in that the omission of forskolin will limit the bias towards cAMP inhibitors, which would neglect other significant pathways that may promote cystogenesis. Nevertheless, it is interesting to note that our PKD1−/− organoids can respond even to cAMP stimulation.
Previous studies analysed organoid morphology by light and immunofluorescence microscopy. Here, to analyse the organoids at a deeper level, we performed ultrastructural analysis with SEM, an invaluable tool to characterize the organoids’ complex 3D architecture, at extremely high spatial resolution. SEM observations confirmed the finding of severely enlarged tubular lumen in PKD1−/− organoids, but also revealed that the largest cystic tubules lack primary cilia and microvilli. Notably, non-cystic tubules do present primary cilia, comparable to the tubules of control organoids. In our organoids, we can speculate that the loss of cilia occurs when the tubules become cystic and the luminal diameter expands significantly. Recently, other groups have reported that in nephron organoids cysts from PKD1−/− organoids display defects in polarisation as seen by the expression of primary cilia outwards, rather than towards the lumen, while normal tubules expressed cilia at the apical side59,60. Our finding of loss of cilia in PKD1−/− cysts therefore seem to confirm that in organoid models of ADPKD cyst-lining cells are incorrectly polarised, although the causal relationship between cystogenesis and dysregulated apicobasal polarity is uncertain. These findings, however, warrant further investigation into how the loss of PC1 affects the function primary cilia.
PC1 is thought to play an important role in mechanosensation in the primary cilium and may inhibit a process called cilia-dependent cyst activation (CDCA), as concomitant loss of PC1 and the primary cilium prevents cystogenesis61. This was uncovered by Ma and colleagues62 who showed that in conditional mouse models, the loss of either PC1 or PC2 concomitantly with the ciliary proteins Kif3a or IFT20 slows cyst growth. The timing between the loss of polycystins and the loss of cilia determines disease severity. The implication of these findings would be that in the absence of polycystins, cilia promote rapid cystogenesis and the inhibition of such CDCA pathway could be therapeutic. In a mouse model of PKD1 haploinsufficiency, Harris and colleagues used SEM analysis and determined that cilia are elongated in both cystic and non-dilated CDs54. In our model of cystogenesis it would appear that the loss of cilia occurs exclusively in cystic tubules, while non-dilated tubules present normal cilia. This evidence, together with the transition zone we have observed, may suggest that in our model the loss of cilia is a dynamic process that takes place only when the tubule diameter becomes abnormally large. Differences in the genetic background of our cells, which completely lack any functional PC1, and the inherent developmental defects in the formation of tubules, may explain the different observations compared to previous studies. Additionally, interspecies differences cannot be ruled out, as the majority of studies on cilia length were carried out on mouse models. Ultimately, the most conclusive evidence regarding cilia size would come from SEM analysis of patient biopsies.
In the present study we have addressed numerous challenges associated with hiPSC-derived ADPKD models. Firstly, our methodology generates tubular organoids with high efficiency, without using Matrigel or other ECM components and does not require cell sorting at any stage, facilitating organoid generation and manipulation. Additionally, culture conditions could be readily upscaled, enabling future drug screening experiments. Finally, PKD1−/− organoids spontaneously develop dilated tubules, representing an early ADPKD process. This model could therefore serve as an accessible platform to study ADPKD, as it not technically challenging, does not require long term culture and minimises the use of expensive materials.
To further explore our model for the study of ADPKD, cystogenesis in organoids with heterozygous mutations in PKD1 (PKD1+/−) will need to be studied to determine whether there is a gene dosage effect. So far, Nishinakamura and colleagues are the only group to have used CD organoids to study ADPKD in patient-derived cells, however cAMP stimulation was required over long periods of time and the cyst induction rate was very low37. As a step forward, we will apply our protocol to patient-derived hiPSCs to determine whether the tubular epithelial organoids can model patient-specific disease processes and serve as a suitable drug screening platform.
In conclusion, in this study we describe an efficient method for generating tubular epithelial organoids from hiPSCs. We showed that PKD1−/− organoids model early tubular dilation in ADPKD and established a scalable culture condition. Our protocol has addressed the challenges of current ADPKD in vitro models and may prove to be a valuable research tool.
Methods
Supplementary Tables S1–S3 contain all the cell lines and reagents utilised.
Human iPSC culture
The Gibco Episomal hiPSC line (A18945, Thermo Fisher Scientific) is a zero-footprint, viral-integration-free hiPSC line generated using cord blood-derived CD34+ progenitors.
Control hiPSCs (IRFMNi001-B) were derived from commercially available Normal Human Dermal Fibroblasts Neonatal (NHD-Neo, CC-2509, Lonza) while IRFMNi003-A25 were generated from peripheral blood mononuclear cells (PBMCs) of a healthy subject who provided written informed consent for this study. Both lines were generated in our laboratory using the Sendai virus Technology and registered in the Human pluripotent stem cell registry https://hpscreg.eu. The PKD1 knockout hiPSC line IRFMNi003-A-4 was obtained from the IRFMNi003-A clone by targeting the PKD1 gene with the CRISPR-Cas9 system. It is a compound heterozygous clone carrying a 1 bp deletion (c.10745_10745delC, P3582Rfs*3) on one allele and a 2 bp deletion (c.10744_10745delCC, P3582Gfs*44) on the other44. The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Bergamo on September 2019. hiPSCs were maintained in mTeSR1 (Stem Cell Technologies) on Cultrex UltiMatrix RGF BME (R&D Systems) coated plates and cultured in 5% CO2 at 37°C. Cells were passaged twice a week or when they reached 80–90% confluence using Accutase.
Ureteric bud-like spheroid induction
hiPSCs were seeded as single cells on U-bottom 96-well ultra-low attachment plates at a density of 104 cells per well in mTeSR1 medium supplemented with 10 µM Y-27632 and centrifuged at 200 g for 5 min. hiPSCs were differentiated in basal medium consisting of DMEM/F12 GlutaMAX supplemented with 17.5 mg/ml stem cell grade BSA, 17.5 µg/ml human insulin, 275 µg/ml human holo-transferrin, 450 µM 1-thioglycerol, 0.1 mM non-essential amino acids and 1% (vol/vol) penicillin/streptomycin. After 2 days medium was changed to basal medium supplemented with 30 ng/ml BMP4 and 50 ng/ml FGF2. On day 2, spheroids were treated with 10 ng/ml Activin A, 100 ng/ml BMP2 and 1 µM Retinoic acid. On day 4, spheroids were cultured in the presence of 5 µM CHIR99021, 100 ng/ml FGF9, 1 µM Retinoic acid and 10 nM LDN193189. On day 6 spheroids were dissociated in 0.25% Trypsin, seeded on U-bottom 96 well low attachment plates at a density of 104 cells per well in medium containing 5 µM CHIR99021, 100 ng/ml FGF9, 1 µM Retinoic acid, 10 nM LDN193189, 2 ng/ml GDNF and 10 µM Y-27,632 and centrifuged at 200×g for 5 min. After 2 days spheroids were cultured in medium containing 1 µM CHIR99021, 5 ng/ml FGF9, 100 ng/ml FGF1, 1 µM Retinoic acid, 10 nM LDN193189 and 2 ng/ml GDNF for 48 h.
Generation of tubular epithelial organoids
Spheroids differentiated for 10 days were cultured on U-bottom 96 well ultra-low attachment plates until day 24 in branching medium containing 5 µM CHIR99021, 100 ng/ml FGF1, 30 ng/ml FGF7, 1 µM Retinoic acid, 10 nM LDN193189 and 100 ng/ml GDNF. Medium was changed every 2 days with freshly supplemented branching medium. Alternatively, spheroids were transferred to 24 well low attachment plates and cultured with branching medium on a Fisherbrand Multi Function 3D Rotator (Fisher Scientific) at 30 rpm for 10 s, orbital rotation for 10 s and vibration for 5 s. For experiments involving the addition of Forskolin, organoids were treated with branching medium containing 25 µM Forskolin from day 16 until day 18.
RNA extraction and quantitative real time PCR
Spheroids were harvested, homogenised and RNA was extracted using the PureLink RNA Mini Kit (Thermo Fisher). RNA was reverse transcribed with SuperScript VILO cDNA synthesis kit (Thermo Fisher). Real Time PCR was carried out with SYBR Green Master Mix or Taqman Gene Expression Master Mix using a ViiA 7 Real-Time PCR System (Applied Biosystems). Relative mRNA levels were calculated using the ΔCT method and standardised to HPRT1 expression. Primer sequences and Taqman probe list are available in Table S2.
Wholemount immunostaining
For wholemount immunofluorescence samples were fixed in 4% PFA in PBS for 30 min at room temperature and were then washed in PBS. All incubations and washing were performed under mild agitation with an orbital shaker.
Spheroids were incubated with penetration buffer (PBS1X, 0.2% Triton, 20% DMSO, 3 M glycine) for 30 min at room temperature and blocked with blocking solution (PBS1X, 0.2% Tween, 10% DMSO, 1% BSA) for 30 min at room temperature. Primary antibodies were diluted in antibody buffer (PBS1X, 0.2% Tween, 5% DMSO, 1% BSA) and incubated with the samples overnight at 4°C. After 3 washes in wash buffer (PBS1X, 0.2% Tween, 1% BSA) secondary antibodies and DAPI (1:500) were diluted in antibody buffer and incubated with the samples for 1 h at RT. Samples were then washed 3 times in wash buffer and cleared with an 88% glycerol aqueous solution for 1 h at RT. Samples were then mounted on a glass coverslip in 88% glycerol aqueous solution and imaged with a Leica SP8 confocal microscope.
Organoids were stained according to Klingberg et al.63. This procedure was only utilised for organoids, as spheroids are fragile and sensitive to the harsh buffers and the dehydration procedure. Briefly, samples were incubated with PBS-TxDB (PBS1X, 2% Triton X-100, 20% DMSO, 5% BSA) for 4 h for permeabilisation and blocking, then incubated with primary antibodies in PBS-TxDB for 1 day. Samples were then washed 3 times in PBS-TxDB for 30 min and incubated with secondary antibodies in PBS-TxDB with DAPI (1:500) for 1 day. Samples were washed 3 times in PBS-TxDB for 30 min then post-fixed in 4% PFA for 30 min. After fixing, samples were dehydrated in a series of 30%, 50%, 70% and twice in 100% ethanol diluted in PBS adjusted to pH 9.0–9.5 with triethylamine. Dehydration was performed at 4°C and for 1 h each step. After dehydration, samples were transferred to ethyl cinnamate for at least 1 h, then imaged on 96 well µ-Plates uncoated (Ibidi) with a Leica SP8 confocal microscope. All immunostaining was performed on at least 3 biological samples from 3 independent experiments. Negative controls omitting 1° antibody as well as validation of AQP2 staining are shown in Fig. S4.
Immunohistochemical and histological analysis
Formalin-fixed samples were embedded in paraffin and 3 μm thin sections were cut. Antigen retrieval was performed with DIVA decloaking buffer (Biocare Medical) in a decloaking chamber (DCNxGen 220 V; Biocare Medical) at 110°C for 15 min and then endogenous peroxidases were quenched. Non-specific sites were blocked with Background Punisher (Biocare Medical), then samples were incubated with primary antibodies for 2 h. MACH4-HRP Polymer kit (Biocare Medical) was used according to manufacturer’s instructions, slides were then counterstained with Meyer’s hematoxylin, dehydrated in an alcohol gradient and imaged. Periodic acid Schiff (PAS) staining was performed on formalin-fixed 3 μm thin sections. Samples were then counterstained with Meyer’s hematoxylin, mounted and imaged. All immunohistochemical analysis was performed on at least 3 organoids from 3 independent experiments.
RNA sequencing analysis
Day 10 spheroids and day 24 organoids were harvested, homogenised and RNA was extracted using the PureLink RNA Mini Kit. RNA was sent to GenomeScan BV (Leiden, Netherlands) for RNA sequence library preparation and sequencing. Briefly, RNA quality was determined with a Fragment Analyzer (Agilent) and the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina was used to process the samples according to manufacturer’s instructions. Clustering and DNA sequencing using the NovaSeq 6000 (Illumina) was performed according to manufacturer’s protocols. A concentration of 0.8 nM of DNA was used. Reads were then mapped to the GRCh38 human reference genome and differential gene expression analysis was performed between the day 10 and day 24 groups. Each group consisted of RNA samples from 3 independent experiments.
Tubular lumen quantification
Tubular lumen area of the organoids was measured in brightfield images using ImageJ software, similar to Kuraoka et al.,37. The areas of multiple lumens and/or cysts were then pooled to give the total lumen area as a proportion of the organoid area.
Scanning electron microscopy
For scanning electron microscopy (SEM) observation, samples were fixed in 2.5% glutaraldehyde in 0.1 mol/l sodium cacodylate buffer for 1 h, washed in cacodylate buffer, and then post-fixed in 1% osmium tetroxide for an additional hour. Fixed samples were dehydrated in an ethanol gradient and dried in pure hexamethyldisilazane (HMDS, Fluka Chemie AG, Buchs, Switzerland). Finally, samples were mounted on stubs, coated with gold in a sputter coater (Agar Scientific Ltd., Stansted, England) and then examined on a Cross-Beam 1540EsB electron microscope (Carl Zeiss Microscopy, Oberkochen, Germany). After the surface observation, samples were then physically fractured to analyse their internal structure. Acceleration voltage was set to 0.8-2.0 kV and enlargement up to 35 kx. The working distance was set to 5.0–9.0 mm.
Statistical analysis
Statistical analysis was performed using an unpaired t-test with Welch’s correction in GraphPad Prism. Values of p < 0.05 were considered statistically significant.
Data availability
Raw transcriptomic data generated during the current study are available in NCBI’s Gene Expression Omnibus (GEO: GSE286070). The data generated in the present article is available upon request to the corresponding author.
References
Willey, C. J. et al. Prevalence of autosomal dominant polycystic kidney disease in the European union. Nephrol. Dial Transpl. 32, 1356–1363 (2017).
Spithoven, E. M. et al. Renal replacement therapy for autosomal dominant polycystic kidney disease (ADPKD) in Europe: prevalence and survival—an analysis of data from the ERA-EDTA registry. Nephrol. Dial. Transplant. 29, iv15–iv25 (2014).
Torres, V. E., Harris, P. C. & Pirson, Y. Autosomal dominant polycystic kidney disease. Lancet 369, 1287–1301 (2007).
Hateboer, N. et al. Comparison of phenotypes of polycystic kidney disease types 1 and 2. European PKD1-PKD2 study group. Lancet 353, 103–107 (1999).
Saigusa, T. & Bell, P. D. Molecular pathways and therapies in autosomal-dominant polycystic kidney disease. Physiol. (Bethesda). 30, 195–207 (2015).
Nauli, S. M. et al. Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 33, 129–137 (2003).
Xu, C. et al. Human ADPKD primary cyst epithelial cells with a novel, single codon deletion in the PKD1 gene exhibit defective ciliary polycystin localization and loss of flow-induced Ca2 + signaling. Am. J. Physiol. Ren. Physiol. 292, F930–945 (2007).
Torres, V. E. & Harris, P. C. Strategies targeting cAMP signaling in the treatment of polycystic kidney disease. J. Am. Soc. Nephrol. 25, 18–32 (2014).
Torres, V. E. et al. Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl. J. Med. 367, 2407–2418 (2012).
Torres, V. E. et al. Tolvaptan in Later-Stage autosomal dominant polycystic kidney disease. N Engl. J. Med. 377, 1930–1942 (2017).
Caroli, A. et al. Effect of longacting somatostatin analogue on kidney and cyst growth in autosomal dominant polycystic kidney disease (ALADIN): a randomised, placebo-controlled, multicentre trial. Lancet 382, 1485–1495 (2013).
Perico, N. et al. Octreotide-LAR in later-stage autosomal dominant polycystic kidney disease (ALADIN 2): A randomized, double-blind, placebo-controlled, multicenter trial. PLoS Med. 16, e1002777 (2019).
Chebib, F. T. et al. A practical guide for treatment of rapidly progressive ADPKD with Tolvaptan. J. Am. Soc. Nephrol. 29, 2458–2470 (2018).
Pisani, A. et al. Long-term effects of octreotide on liver volume in patients with polycystic kidney and liver disease. Clin. Gastroenterol. Hepatol. 14, 1022–1030e4 (2016).
Spinelli, L. et al. Left ventricular dysfunction in ADPKD and effects of octreotide-LAR: A cross-sectional and longitudinal substudy of the ALADIN trial. Int. J. Cardiol. 275, 145–151 (2019).
Trillini, M. et al. Effects of Octreotide-Long-Acting release Added-on Tolvaptan in patients with autosomal dominant polycystic kidney disease: pilot, randomized, Placebo-Controlled, Cross-Over trial. Clin. J. Am. Soc. Nephrol. 18, 223–233 (2023).
Chebib, F. T., Sussman, C. R., Wang, X., Harris, P. C. & Torres, V. E. Vasopressin and disruption of calcium signalling in polycystic kidney disease. Nat. Rev. Nephrol. 11, 451–464 (2015).
Koslowski, S., Latapy, C., Auvray, P., Blondel, M. & Meijer, L. An overview of in vivo and in vitro models for autosomal dominant polycystic kidney disease: A journey from 3D-Cysts to Mini-Pigs. Int. J. Mol. Sci. 21, 4537 (2020).
Happé, H. & Peters, D. J. M. Translational research in ADPKD: lessons from animal models. Nat. Rev. Nephrol. 10, 587–601 (2014).
Sharma, M., Reif, G. A. & Wallace, D. P. In vitro cyst formation of ADPKD cells. Methods Cell. Biol. 153, 93–111 (2019).
Torres, V. E. & Harris, P. C. Autosomal dominant polycystic kidney disease: the last 3 years. Kidney Int. 76, 149–168 (2009).
Benedetti, V. et al. Engineered kidney tubules for modeling Patient-Specific diseases and drug discovery. EBioMedicine 33, 253–268 (2018).
Takahashi, K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007).
Karagiannis, P. et al. Induced pluripotent stem cells and their use in human models of disease and development. Physiol. Rev. 99, 79–114 (2019).
Ciampi, O. et al. Generation of functional podocytes from human induced pluripotent stem cells. Stem Cell. Res. 17, 130–139 (2016).
Yoshimura, Y. et al. Manipulation of Nephron-Patterning signals enables selective induction of podocytes from human pluripotent stem cells. J. Am. Soc. Nephrol. 30, 304–321 (2019).
Taguchi, A. et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell. Stem Cell. 14, 53–67 (2014).
Takasato, M. et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015).
Morizane, R. et al. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193–1200 (2015).
Mae, S. I. et al. Expansion of human iPSC-Derived ureteric bud organoids with repeated branching potential. Cell. Rep. 32, 107963 (2020).
Taguchi, A. & Nishinakamura, R. Higher-Order kidney organogenesis from pluripotent stem cells. Cell. Stem Cell. 21, 730–746e6 (2017).
Zeng, Z. et al. Generation of patterned kidney organoids that recapitulate the adult kidney collecting duct system from expandable ureteric bud progenitors. Nat. Commun. 12, 3641 (2021).
Shi, M. et al. Human ureteric bud organoids recapitulate branching morphogenesis and differentiate into functional collecting duct cell types. Nat. Biotechnol. 41, 252–261 (2023).
Mae, S. I. et al. Human iPSC-derived renal collecting duct organoid model cystogenesis in ADPKD. Cell. Rep. https://doi.org/10.1016/j.celrep.2023.113431 (2023).
Freedman, B. S. et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 6, 8715 (2015).
Shimizu, T. et al. A novel ADPKD model using kidney organoids derived from disease-specific human iPSCs. Biochem. Biophys. Res. Commun. 529, 1186–1194 (2020).
Kuraoka, S. et al. PKD1-Dependent renal cystogenesis in human induced pluripotent stem Cell-Derived ureteric bud/collecting duct organoids. J. Am. Soc. Nephrol. 31, 2355–2371 (2020).
Cruz, N. M. et al. Organoid cystogenesis reveals a critical role of microenvironment in human polycystic kidney disease. Nat. Mater. 16, 1112–1119 (2017).
Xia, Y. et al. Directed differentiation of human pluripotent cells to ureteric bud kidney progenitor-like cells. Nat. Cell. Biol. 15, 1507–1515 (2013).
Costantini, F. & Shakya, R. GDNF/Ret signaling and the development of the kidney. Bioessays 28, 117–127 (2006).
Gilbert, T. Vitamin A and kidney development. Nephrol. Dial Transpl. 17 Suppl 9, 78–80 (2002).
Batourina, E. et al. Vitamin A controls epithelial/mesenchymal interactions through Ret expression. Nat. Genet. 27, 74–78 (2001).
Kawakami, T., Kumasaka, M., Kato, M., Mizoguchi, M. & Soma, Y. BMP-4 down-regulates the expression of Ret in murine melanocyte precursors. J. Dermatol. Sci. 63, 66–69 (2011).
Romano, E., Trionfini, P., Ciampi, O., Benigni, A. & Tomasoni, S. Generation of PKD1 mono-allelic and bi-allelic knockout iPS cell lines using CRISPR-Cas9 system. Stem Cell. Res. 47, 101881 (2020).
Qiao, J. et al. Multiple fibroblast growth factors support growth of the ureteric bud but have different effects on branching morphogenesis. Mech. Dev. 109, 123–135 (2001).
Bridgewater, D. et al. Canonical WNT/beta-catenin signaling is required for ureteric branching. Dev. Biol. 317, 83–94 (2008).
Wang, P. et al. Dissecting the global dynamic molecular profiles of human fetal kidney development by Single-Cell RNA sequencing. Cell. Rep. 24, 3554–3567e3 (2018).
Grassmeyer, J. et al. Elf5 is a principal cell lineage specific transcription factor in the kidney that contributes to Aqp2 and Avpr2 gene expression. Dev. Biol. 424, 77–89 (2017).
Chen, L. et al. Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq. Proc. Natl. Acad. Sci. 114, E9989–E9998 (2017).
Terryn, S., Ho, A., Beauwens, R. & Devuyst, O. Fluid transport and cystogenesis in autosomal dominant polycystic kidney disease. Biochimi. Biophys. Acta 1812, 1314–1321 (2011).
Larkins, C. E., Aviles, G. D. G., East, M. P., Kahn, R. A. & Caspary, T. Arl13b regulates ciliogenesis and the dynamic localization of Shh signaling proteins. MBoC 22, 4694–4703 (2011).
Devuyst, O. et al. Expression of aquaporins-1 and – 2 during nephrogenesis and in autosomal dominant polycystic kidney disease. Am. J. Physiol. 271, F169–183 (1996).
Starremans, P. G. et al. A mouse model for polycystic kidney disease through a somatic in-frame deletion in the 5’ end of Pkd1. Kidney Int. 73, 1394–1405 (2008).
Hopp, K. et al. Functional polycystin-1 dosage governs autosomal dominant polycystic kidney disease severity. J. Clin. Invest. 122, 4257–4273 (2012).
Lopes, F. M. & Woolf, A. S. Kidney development. in Heptinstall’s Pathology of the Kidney 169–214 (Wolters Kluwer, 2023).
Jafari, N. V. & Rohn, J. L. The urothelium: a multi-faceted barrier against a harsh environment. Mucosal Immunol. 15, 1127–1142 (2022).
Howden, S. E. et al. Plasticity of distal nephron epithelia from human kidney organoids enables the induction of ureteric tip and stalk. Cell. Stem Cell. 28, 671–684e6 (2021).
Parker, E. et al. Hyperproliferation of PKD1 cystic cells is induced by insulin-like growth factor-1 activation of the Ras/Raf signalling system. Kidney Int. 72, 157–165 (2007).
Tran, T. et al. A scalable organoid model of human autosomal dominant polycystic kidney disease for disease mechanism and drug discovery. Cell. Stem Cell. 29, 1083–1101e7 (2022).
Li, S. R. et al. Glucose absorption drives cystogenesis in a human organoid-on-chip model of polycystic kidney disease. Nat. Commun. 13, 7918 (2022).
Ma, M. Cilia and polycystic kidney disease. Semin. Cell. Dev. Biol. 110, 139–148 (2021).
Ma, M., Tian, X., Igarashi, P., Pazour, G. J. & Somlo, S. Loss of cilia suppresses cyst growth in genetic models of autosomal dominant polycystic kidney disease. Nat. Genet. 45, 1004–1012 (2013).
Klingberg, A. et al. Fully automated evaluation of total glomerular number and capillary tuft size in nephritic kidneys using lightsheet microscopy. J. Am. Soc. Nephrol. 28, 452–459 (2017).
Acknowledgements
The authors would like to thank Prof. Adrian S. Woolf for his helpful and inspiring discussions regarding the manuscript. We also acknowledge Dr. Norberto Perico for contribution to the writing and preparation of the manuscript and Paola Staffiere for technical support in transcriptomic data analysis.
Funding
This research was funded by the European Union Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 860977. This study was partially funded by Fondazione Aiuti per la Ricerca sulle Malattie Rare (ARMR), Bergamo, Italy. A.S. was supported by the Horizon 2020 Marie Skłodowska-Curie programme as an early-stage researcher.
Author information
Authors and Affiliations
Contributions
Conceptualization, A.S., C.X., A.B. and S.T.; Generation of organoids, A.S., L.Longhi; confocal analysis, A.S. and P.R.; hiPSC culture and real time expression analysis, L.Longaretti and A.S.; SEM analysis, S.C.; RNA sequencing data analysis, M.B.; investigation, A.S.; data curation, A.S.; writing—original draft preparation, A.S.; writing—review and editing, A.S., S.T, A.B., G.R., P.T.; supervision, S.T., A.B.; All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Scarlat, A., Trionfini, P., Rizzo, P. et al. PKD1 mutation perturbs morphogenesis in tubular epithelial organoids derived from human pluripotent stem cells. Sci Rep 15, 10375 (2025). https://doi.org/10.1038/s41598-025-94855-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-94855-9