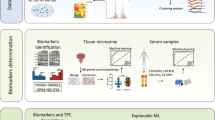
Abstract
Prostate cancer (PCa) is a highly prevalent malignancy among men. Castration-resistant prostate cancer (CRPC) is the main pathological type leading to the death of patients. Due to the lack of effective early diagnostic biomarkers, the treatment of CRPC is often lagging. Ferroptosis is a new discovered pathway of cell death. The diagnostic value of ferroptosis-related genes for CRPC is still uncertain. In this study, we identified 10 differently expressed FRGs between 145 hormone-sensitive PCa (HSPC) and 49 CRPC. A novel ferroptosis-related genes (FRGs) signature was constructed using seven FRGs (ABCC5, ACSL1, AKR1C3, CAMKK2, GDF15, PDSS2, and ZFP36), which could distinguish CRPC from HSPC with area under the ROC curve (AUC) = 0.93. This result was confirmed in an independent validation cohort with AUC = 0.90, F1 score = 0.85. Among the seven FRGs, GDF15 independently predicted CRPC with the good performance (AUC = 0.84). Moreover, GDF15 could effectively distinguish PCa from normal tissues (AUC = 0.79 and 0.83 in two cohorts). Two distinct ferroptosis profiles were discovered, and FRGs signature and GDF15 could also effectively distinguish them. GDF15 expression level shows an inverted U-shape with disease progression (normal, HSPC, CRPC). Although GDF15 expression was decreased in CRPC cells, overexpression of GDF15 still facilitates CRPC cells migration and invasion in vitro. Our findings suggest that this novel FRGs signature and GDF15 could be robust biomarkers for predicting CRPC in clinical practice.
Similar content being viewed by others
Introduction
Prostate cancer (PCa) is the most frequently diagnosed cancer among men1. In 2024, the estimated PCa alone accounts for 29% new diagnosed cancer (299,010/1,029,080) in men in United States1. PCa is also the second cause of death due to malignancy among men (estimated 35,250 deaths annually in the US)1. Through radical prostatectomy and radiotherapy, patients with localized disease have a better prognosis outcome. For patients with advanced PCa, androgen deprivation therapy (ADT) is the commonly used treatment2. Although ADT is initially effective in nearly all patients, the disease relapse eventually occurs due to the emergence of castration-resistant prostate cancer (CRPC), which is the leading cause of mortality in PCa patients3. The diagnosis of CRPC is usually based on the serum prostatic-specific antigen (PSA) level. However, serum PSA does not always correlate with the clinical status of CRPC4. Previous studies have constructed gene signatures to distinguish CRPC from hormone-sensitive PCa (HSPC)5,6. However, there is a problem that cannot be ignored in their analysis process. These studies used the survival data of TCGA cohort to screen candidate genes. However, the prognostic value of genes is not equivalent to their diagnostic value. This resulted in the loss of some genes that were valuable for differentiating CRPC from HSPC, although they were treated as of no prognostic value in the TCGA cohort.
Ferroptosis is a new discovered pathway of cell death and is different from apoptosis, necrosis, and autophagy. Ferroptosis results from accumulation of peroxidation of phospholipids enriched with polyunsaturated fatty acids and reactive oxygen species7. Accumulating studies have suggested that ferroptosis play important roles in many biological processes, including the initiation and progression of cancer8,9. Bordini et al.10 found that iron was toxic for all prostate cancer cells, and strengthened the efficacy of antiandrogen therapy. Flubendazole, a novel P53 inducer, was found that exert anti-proliferation and pro-apoptosis effects in CRPC through hindering the cell cycle and activating the ferroptosis11. Research has confirmed that androgen receptor and its splicing variants play a key role in the development of CRPC. Yang et al.12 demonstrated that erastin, a ferroptosis inducer, downregulated androgen receptor and its splice variants in castration‑resistant prostate cancer and strengthened the growth inhibitory efficacy of docetaxel. The researchers also provided new targeted therapeutic strategies for CRPC based on ferroptosis13,14. These studies indicated that ferroptosis plays a key role in the progression and prognosis of CRPC. Researchers have developed several ferroptosis-related genes (FRGs) signatures to predict prognosis of PCa15,16. The application of FRGs signature in CRPC has not been reported. Therefore, we hope to find a FRGs signature that can effectively identify CRPC.
In this study, we combined 3 Gene Expression Omnibus (GEO) datasets to obtain a CRPC cohort, which included 145 HSPC samples and 49 CRPC samples. We identified 10 differentially expressed FRGs between HSPC and CRPC tumors. We avoided using TCGA survival data to screen them and developed a novel 7-FRGs signature as diagnostic biomarker for CRPC. This signature could distinguish CRPC from HSPC with high specificity and sensitivity and was further validated in an independent dataset. Moreover, growth differentiation factor 15 (GDF15), encoding a secreted protein, could distinguish PCa from normal and CRPC from HSPC with high specificity and sensitivity. The ability of serum and urine GDF15 levels to distinguish PCa from benign prostatic hyperplasia (BPH) has been demonstrated by other studies17,18,19. GDF15 is expected to be an effective non-invasive diagnostic biomarker for both PCa and CRPC.
Materias and methods
Data collection
All human samples involved in this study were obtained from publicly available databases, and that we did not use any tissue samples from our institution. Four GEO datasets (GSE7076820, GSE3226921, GSE537722, GSE3598823) were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). Basic information of the included datasets was shown in Table 1. GSE70768 is derived from a study involving 482 tumor, benign and germline samples from 259 men with primary PCa. The samples were divided into two cohorts: the discovery cohort and the validation cohort. GSE70768 includes only a portion of the discovery cohort samples, comprising 13 CRPC samples, 113 HSPC samples, and 73 matched benign samples, all of which were analyzed using the HumanHT-12 V4.0 platform. We obtained 10 HSPC samples (localized PCa, 7; locally advanced PCa, 3) and 7 CRPC (early stage, 4; late stage, 3) samples from the GSE5377 dataset. Clinical data of the 17 samples can be found in Supplementary Table 1. GSE32269 includes 22 primary PCa (hormone-dependent) and 29 metastatic PCa (CRPC). CRPC samples were obtained through CT-guided bone marrow biopsies and were confirmed by microscopic examination to primarily consist of tumor tissue. HSPC samples were sourced from hormone-naive patients and were extracted from frozen biopsy samples using laser capture microdissection. In GSE35988, gene expression profiling was performed on 12 matched benign prostate tissues, 49 HSPC tissues, and 27 CRPC tissues. The researchers provided clinical data for 1 HSPC samples and 25 CRPC samples (Supplementary Table 2). ‘SVA’ R package was used to adjust batch effects in microarray expression data (GSE70768, GSE32269, GSE5377) to create the integrated GEO dataset. Specifically, the Combat function in the ‘SVA’ package employs empirical Bayes methods to estimate the batch effects for each sample and eliminates the influence of these effects by adjusting the estimated values24. GSE35988 was enrolled as the external validation cohort.
The TCGA prostate adenocarcinoma (adjacent normal tissues, n = 52; PCa tissues, n = 498) dataset was downloaded from the UCSC Xena database platform (http://xena.ucsc.edu/). The 260 experimentally validated human-derived FRGs were obtained at the FerrDb database (supplementary file 1)25. The Human Protein Atlas (HPA) database (https://www.proteinatlas.org/) was used to evaluate the expression level of GDF15 in different prostate cancer cell lines and prostate cancer tissues. HuPSA–MoPSA (https://pcatools.shinyapps.io/HuPSA-MoPSA/) is developed by Drs. Siyuan Cheng and Xiuping Yu26. This platform includes three datasets: Human Prostate Single Cell Atlas (HuPSA), Mouse Prostate Single Cell Atlas (MoPSA), and Prostate Atlas (ProAtlas). ProAtlas compiles bulk RNA sequencing data. Through this app, we assessed the expression patterns of GDF15 at both the tissue and single-cell levels.
Identification of differentially expressed FRGs
Differentially expressed genes (DEGs) between CRPC and HSPC were identified utilizing the ‘LIMMA’ R package27. The cutoff criteria are false discovery rate < 0.05 and |fold change| > 2. To identify the differentially expressed FRGs, the DEGs and FRGs were intersected.
Gene list annotation and analysis
Metascape28 was used to perform gene annotation, pathway and process enrichment analysis, and protein-protein interaction (PPI) enrichment analysis. The Molecular Complex Detection (MCODE) algorithm29 was applied to find molecular complexes in PPI network. MCODE is a graph theoretic clustering algorithm that can identify subnetworks within the PPI network that may represent molecular complexes based on connectivity data.
Construction of the FRGs signature
Least absolute shrinkage and selection operator (LASSO) Cox regression analysis was applied to obtain the characteristic genes of risk signature. LASSO Cox regression analysis compresses the variable coefficients in the regression model by generating a penalty function, which helps to prevent overfitting and address the issue of high collinearity. This compresses certain coefficients to zero to achieve model simplification. The R package ‘glmnet’30 were used to perform LASSO Cox regression analysis. Cross-validation (10-fold) was performed on the model using the ‘cv.glmnet function’ from the ‘glmnet’ package to identify the optimal lambda (λ) value. Lambda.min is the value of λ that minimizes the mean squared error in cross-validation, representing the highest model fitting performance and the best prediction accuracy. In this study, lambda.min was chosen to select model variables. The calculation formula of the model is as follows: risk score = sum (each gene’s expression × corresponding coefficient). The Receiver operating characteristic (ROC) curve and F1 score were used to evaluate the ability of signature to distinguish CRPC and HSPC. The area under the ROC curve (AUC) reflects discriminative ability. There are 3 levels of diagnostic ability (low, 0.5–0.7; medium, 0.7–0.9; high, 0.9-1.0). In TCGA, the Kaplan-Meier method was utilized to examine differences in disease-free survival (DFS) and progression-free survival (PFS) between groups with high and low expression of FRGs.
Identification of ferroptosis molecular subtypes
The R package ‘ConsensusClusterPlus’31 was employed to identify the ferroptosis molecular subtypes. We compared the expression of signature genes and risk scores across molecular subtypes. The ROC curve was used to evaluate the ability of risk score and genes to distinguish ferroptosis molecular subtypes. To explore the biological significance of these molecular subtypes, gene set enrichment analysis (GSEA) was carried out. GSEA software (version 3.0) was obtained from the GSEA website (http://software.broadinstitute.org/gsea/index.jsp). The samples were divided into two groups based on molecular subtypes, and we downloaded the c2.cp.kegg.v7.4.symbols.gmt subset from the Molecular Signatures Database (http://www.gsea-msigdb.org/gsea/downloads.jsp) to assess relevant pathways and molecular mechanisms. P-value of < 0.05 was considered statistically significant.
Comparison of immune infiltrate cells
The analytical tool CIBERSORTx32 can use gene expression data to perform linear support vector regression, thus estimating immune cell infiltration. We calculated 22 immune cell types in patients using CIBERSORTx and analyzed the difference in the proportion of immune cells in different grouping modes. The box plots are used to display the immune cell composition of patients with different immune patterns. The Wilcoxon rank-sum test was employed to assess the differences in immune cell proportions. A statistically significant P-value of < 0.05 was used to determine the significance of the results.
Cell culture
The normal prostate epithelial cell line RWPE-1 cell line was presented by Zhigang Zhao from The First Affiliated Hospital of Guangzhou Medical University. LNCaP, PC-3, and DU145 PCa cell lines were purchased from Wuhan Pricella Biotechnology Co., Ltd. (Wuhan, China). C4-2 PCa cell lines was purchased from BeNa Culture Collection (Henan, China). Cell lines were authenticated by STR profiling, tested negative for mycoplasma. All cells were cultured as described previously33, and grew in 5% CO2 at 37 °C.
RNA extraction, reverse transcription and real-time quantitative PCR (RT-qPCR)
Total RNA from prostate cells was isolated with TRIzol Reagent (Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) following manufacturer’s instructions. The cDNA of mRNA was synthesized with PrimeScript RT reagent Kit with gDNA Eraser (Takara Biomedical Technology (Beijing) Co., Ltd, Beijing, China), and qPCR was conducted using the SYBR green Premix Ex Taq II (Takara Biomedical Technology (Beijing) Co., Ltd, Beijing, China). Q-PCR was performed with Applied Bio-systems 7500 Fast Real-Time RCR System (Applied Biosystems, Foster City, CA, USA). Each measurement was performed in triplicate, and the results were normalized to the internal control of GAPDH. Relative expression of mRNA was determined by 2−△△Ct method. Primer sequences information:
-
GDF15 forward primer: 5′-CCGCCAGCTACAATCCCAT − 3′;
-
GDF15 reverse primer: 5′-AGTGGCAGTCTTTGGCTAACA − 3′;
-
GAPDH forward primer 5′- ACAACTTTGGTATCGTGGAAGG − 3′;
-
GAPDH reverse primer: 5′- GCCATCACGCCACAGTTTC − 3′.
Transfection
The plasmid used to overexpress GDF15 in cells was provided by WZ Biosciences Biotechnology Inc. (Jinan, China). Cells were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
Western blot (WB) analysis
Western blot analysis was performed as described previously34. The total proteins were separated by SDS–PAGE and detected with the primary antibodies against GDF15 (Proteintech, #27455-1-AP). β-actin (Proteintech, #20536-1-AP) and β-tubulin (Proteintech, #10094-1-AP) were used as internal control. The blots were cut prior to hybridization with antibodies during blotting. iBright Analysis Software (Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) was used to visualize and quantify protein bands.
Migration and invasion assays
Cell migration and invasion were evaluated using a Transwell permeable support chamber (Corning Incorporated, Corning, NY, USA) with or without Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). The specific procedure is as follows: Prepare a cell suspension of 2.5 × 10^5/ml using serum-free culture medium. Add 200 µl of the cell suspension to the upper Transwell chamber and 750 µl of culture medium containing 10% FBS to the lower chamber. After 36 h of incubation, fix the cells with 4% paraformaldehyde and stain them with 0.1% crystal violet solution. Gently remove the residual cells on the membrane surface with a cotton swab; the cells adhered to the underside of the membrane are the migrated or invaded cells. Subsequently, photographs of five randomly selected fields of view were taken using an inverted microscope (Olympus DP72) at a magnification of 200×, and the counting was performed using Image J software. The number of cells migrating through the chambers in each group was documented as the average value.
Statistical analysis
All the statistical analyses were carried out utilizing Sangerbox analysis platform35 and GRAPHPAD PRISM version 8.0 (GraphPad Software, San Diego, CA, USA). The independent Student’s t-test (normally distributed variables) and Mann-Whitney U-test (non-normally distributed variables) were conducted to compare two groups of continuous variables. P < 0.05 indicated statistical significance.
Results
Identification and pathway enrichment analysis of DEGs
The integrated GEO dataset, including 73 normal samples, 145 HSPC samples, and 49 CRPC samples, was obtained after removing batch effects (Fig. 1A-D). In total, 196 DEGs were screened out in CRPC samples compared with HSPC samples (supplementary file 2). Out of these, 40 were upregulated, whereas 156 were downregulated (Fig. 1E). Figure 1F presents the expression heatmap of the 50 most significantly changed genes.
Identification of differentially expressed genes between HSPC and CRPC. Sample distribution before de-batching (A) and after de-batching (B). Uniform manifold approximation and projection (UMAP) between datasets before de-batching (C) and after de-batching (D). (E) The volcano plot was constructed based on fold change values and P-adjust, with red and blue dots representing upregulated and downregulated genes, respectively. (F) The heatmap depicting the top 50 genes identified through analysis of differential gene expression.
Figure 2A shows the pathway and process enrichment analysis results of 196 DEGs as a heatmap. The top 5 clusters were ‘response to hormone’, ‘binding and uptake of ligands by scavenger receptors’, ‘vesicle-mediated transport’, ‘response to retinoic acid’, and ‘negative regulation of cell population proliferation’. Based on STRING database, we build a PPI network of 196 DEGs (Fig. 2B). In the PPI network, nodes were color-coded based on the status of DEGs (red, up-regulated; blue, down-regulated). Then, we identified 5 densely connected network components in the PPI network (Fig. 2C). Pathway and process enrichment analysis was applied to each MCODE component independently (Fig. 2D). MCODE1 was enriched in ‘ECM proteoglycans’, ‘binding and uptake of ligands by scavenger receptors’, and ‘miR-509-3p alteration of YAP1/ECM axis’. MCODE2 was enriched in ‘small molecule catabolic process’, ‘amino acid catabolic process’, and ‘carboxylic acid catabolic process’. MCODE 3 was enriched in ‘WNT signaling’, ‘canonical Wnt signaling pathway’, and ‘Wnt signaling pathway’. MCODE 4 was enriched in ‘biological process involved in symbiotic interaction’.
Construction of the ferroptosis-related risk signature
By intersecting DEGs with FRGs, a total of 10 differently expressed FRGs were identified (Fig. 3A). Among these, 2 were upregulated, while 8 were downregulated. The boxplots (Fig. 3B) and heatmaps (Fig. 3C) show their expression levels in samples. We did not further evaluate the prognostic value of these genes but directly used them to construct the risk model. To prevent over fitting of the model, the LASSO Cox regression analysis was performed to screen the 10 FRGs. The lambda.min (λ = 0.02) was obtained using the cv.glmnet function, and finally, 7 genes were identified to construct the risk signature (Fig. 3D, E).
Construction of the FRGs signature. (A) Identification of 10 differently expressed FRGs by intersecting DEGs with FRGs. Boxplots (B) and heatmaps (C) of the expression of 10 differently expressed FRGs between HSPC and CRPC tissues. (D,E) A 7-FRGs signature was constructed by LASSO Cox regression analysis. (F) The expression correlation among 10 differently expressed FRGs. (G) The risk score decreased in HSPC compared with ANT, but increased in CRPC compared with ANT and HSPC. (H) Unsupervised clustering analysis using 7 FRGs that comprise the risk signature. Data are analyzed using the Wilcoxon rank-sum test, N ≥ 3. The P-values are expressed in scientific notation.
Correlations between gene expression levels between 7 FRGs were examined in PCa samples (Fig. 3F). There are 6 groups of genes whose |correlation coefficient| exceeds 0.3: ABCC5 and GDF15 (-0.34), ABCC5 and ZFP36 (-0.31), ACSL1 and GDF15 (0.31), AKR1C3 and CAMKK2 (-0.31), CAMKK2 and GDF15 (0.49), and GDF15 and ZFP36 (0.37). GDF15 appeared the most frequently. The risk score decreased in HSPC compared with ANT, but increased in CRPC compared with ANT and HSPC (Fig. 3G). The differences in risk scores across various samples form the basis for their role as biomarkers. Subsequently, we performed unsupervised clustering analysis (Complete Linkage Clustering) using these 7 genes to reflect their classification potential (Fig. 3H). To evaluate the predictive value of the FRGs signature for CRPC, we also performed ROC curves analysis. The AUC of risk score was 0.93 (95% confidence interval (CI): 0.89–0.97) in the training cohort (merged GSE dataset) (Fig. 4A). Five in 7 FRGs (ABCC5, ACSL1, CAMKK2, GDF15, and ZFP36) could efficiently distinguish CRPC from HSPC samples with an AUC value greater than 0.7 (Fig. 4B–H). In the validation cohort, the risk score also demonstrated promising discriminative power (AUC, 0.90; 95%CI, 0.82–0.99. F1 score, 0.85) (Fig. 4I). Because previous studies17,18,19 have confirmed that GDF15 can be used as a diagnostic marker for PCa, we paid special attention to the value of GDF15 in distinguishing CRPC from HSPC (AUC, 0.84; 95%CI, 0.78–0.90).
Identification of ferroptosis molecular subtypes
Based on 7 signature FRGs, 2 ferroptosis molecular subtypes (cluster 1 and cluster 2) were identified utilizing the R package ‘ConsensusClusterPlus’ (Fig. 5A–C). The boxplots show the expression levels of 7 FRGs in different subtypes (Fig. 5D). The risk score decreased in cluster 2 compared to cluster 1 (Fig. 5E). In addition, cluster 1 included 108 samples (CRPC, 48; HSPC, 60), whereas cluster 2 include 86 samples (CRPC, 1; HSPC, 85) (Fig. 5F). To evaluate the predictive value of the FRGs signature for molecular subtypes, we performed ROC curves analysis. Risk score could efficiently distinguish cluster 1 samples from cluster 2 samples with an AUC value greater than 0.9 (Fig. 5G). Only AKR1C3 and PDSS2 had an AUC lower than 0.7, and GDF15 had an AUC of 0.89 (Fig. 5H–N). To explore the biological significance of distinct subgroups, GSEA was carried out. We found that Base excision repair, Notch signaling pathway, ECM receptor interaction, and taste transduction were activated in cluster 1 compared with cluster 2 (Supplementary Fig. 1A). The metabolism of various substances (such as arginine, proline, tyrosine, fatty acid, and sphingolipid) was inhibited in cluster 1 (Supplementary Fig. 1B).
Identification of ferroptosis molecular subtypes. Two ferroptosis molecular subtypes were obtained using ConsensusClusterPlus (A, cumulative distribution curve; B, the area under cumulative distribution curve; C, cluster heatmap). (D) The expression boxplots of 7 FRGs in molecular subtypes. (E) The risk score decreased in cluster 2 compared to cluster 1. (F) Cluster 1 included 108 samples (CRPC, 48; HSPC, 60), whereas cluster 2 include 86 samples (CRPC, 1; HSPC, 85). (G–N) ROC curves of risk score and 7 FRGs demonstrating their ability to distinguish the ferroptosis molecular subtypes. Data are analyzed using the Wilcoxon rank-sum test, N ≥ 3. The P-values are expressed in scientific notation.
Based on 7 signature FRGs, 2 ferroptosis molecular subtypes were also identified in the validation cohort (Supplementary Fig. 2A-C). Four FRGs (ABCC5, GDF15, PDSS2, and ZFP36) exhibited differential expression across various subtypes (Supplementary Fig. 2D). The risk score also decreased in cluster 2 compared to cluster 1 (Supplementary Fig. 2E). Cluster 1 included 43 samples (CRPC, 26; HSPC, 17), whereas cluster 2 include 33 samples (CRPC, 1; HSPC, 32) (Supplementary Fig. 2F). The risk score could efficiently distinguish cluster 1 from cluster 2 with an AUC value of 0.82 (Supplementary Fig. 2G).
Differences in immune characteristics
Ferroptosis plays an important role in immune cells and immunotherapy36. Therefore, we estimated immune cell infiltration across pathological types and ferroptosis molecular subtypes using CIBERSORTx. Figure 6A displays the proportion of 22 immune cell types in each sample. Figure 6B demonstrates differences in immune cell infiltration among different sample types (adjacent normal tissues, HSPC, and CRPC). Resting NK cells and M2 macrophages were remarkably elevated in CRPC compared to HSPC. Plasma cells, resting memory CD4 T cells, T cells follicular helper, M1 macrophages, and resting dendritic cells were considerably lowered in CRPC compared to HSPC. Then, we analyzed differences in immune infiltration between high and low risk groups (Fig. 6C). CD8 T cells, native CD4 T cells, and M2 macrophages were elevated in high-risk group compared to low-risk group. Plasma cells, T cells follicular helper, and resting dendritic cells were lower in high-risk group compared to high-risk group. The levels of memory B cells, native CD4 T cells, resting NK cells, and M2 macrophages were remarkably elevated in cluster 1 compared to cluster 2. In contrast, the levels of plasma cells and T cells follicular helper were considerably lowered in cluster 1 compared to cluster 2 (Fig. 6D).
Differences in immune characteristics. (A) Immune cell content stacking plot for each sample. 22 immune cell types content histogram in different pathological types (B), risk groups (C), and molecular subtypes (D). Data are analyzed using the Wilcoxon rank-sum test, N ≥ 3. The P-values are expressed in scientific notation or symbol (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
The clinical value of GDF15
Considering that GDF15 showed good discriminatory value for PCa in previous studies and for CRPC in our study, we further analyzed the characteristics of GDF15 in PCa. GDF15 was elevated in HSPC tissues compared with adjacent normal tissues (ANT). However, when the disease progressed to CRPC, the expression of GDF15 decreased (Fig. 7A). Through TCGA pan-cancer analysis, we found that GDF15 was upregulated in various tumors including PCa (Fig. 7B, C, Supplementary Fig. 3A). Subsequently, we assessed the expression pattern of GDF15 using the HuPSA–MoPSA platform. ProAtlas compiles bulk RNA sequencing data from 877 human prostate samples. In ProAtlas, samples were re-classified into distinct subtypes: nonmalignant prostate, PCa with various Gleason Scores (GS), KRT7, progenitor-like, and neuroendocrine prostate cancer (NEPC). KRT7 and progenitor-like PCa subtypes represent dedifferentiated double-negative prostate cancer. Therefore, KRT7, progenitor-like, and NEPC samples demonstrated stronger aggressiveness. We found that GDF15 was elevated in PCa samples with various GS compared to nonmalignant samples (Fig. 7D). Compared to PCa samples with different GS, GDF15 expression was reduced in KRT7, progenitor-like, and NEPC samples. In NEPC samples, GDF15 was even lower than that in nonmalignant samples. Figure 7E shows the 26 cell populations in HuPSA. Cheng et al. provided detailed description of each cell population in Supplementary Note 1 of their article26. PCa cells were primarily located in the region highlighted by the red circles. Feature plot and violin plot show the expression of GDF15 in different cell populations (Fig. 7F–G). Although no statistical analysis was performed, it is evident from the figure that GDF15 is predominantly highly expressed in some subsets of PCa cells.
The clinical value of GDF15. (A,B) The expression level of GDF15 in GEO and TCGA cohort. (C) GDF15 expression levels in the paired PCa tissues compared with that in the ANT in TCGA cohort. (D) GDF15 expression patterns in ProAtlas. (E) UMAP dimensional reduction plot displaying 26-cell populations in HuPSA. This figure was produced by Cheng et al.26 and is licensed under a Creative Commons Attribution 4.0 International License. The red circles in the figure were added later. Feature plot (F) and violin plot (G) show the expression of GDF15 in different cell populations. (H) GDF15 levels in HSPC cells, NEPC cells, and CRPC cells (HPA database). (I,J) GDF15 protein levels in PCa (HPA database). (K) The proportion of patients with moderate and high expression of GDF15 protein (HPA database). (L,M) ROC curves of GDF15 demonstrating its ability to distinguish the PCa and ANT. (N,O) Kaplan-Meier survival analysis of DFS and PFS of PCa patients with different GDF15 levels. Data are analyzed using the Wilcoxon rank-sum test, N ≥ 3. The P-values are expressed in scientific notation or symbol (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). The red * represents the control group as nonmalignant samples, and the green * represents the control group as GS6-9 samples.
GDF15 encodes a secreted ligand of the TGF-beta superfamily of proteins. Querying the HPA database, we found the expression of GDF15 decreased steadily from HSPC cells, neuroendocrine prostate cancer (NEPC) cells, to CRPC cells (Fig. 7H). GDF15 localizes to the Golgi apparatus and extracellular space (Supplementary Fig. 3B-C). According to the results of immunohistochemical staining, GDF15 protein was elevated in PCa (Fig. 7I, J). Compared with other tumors, the proportion of patients with moderate and high expression of GDF15 protein was highest in PCa (Fig. 7K). To evaluate the predictive value of GDF15 for tumor, we performed ROC curves analysis. The AUC of GDF15 was 0.79 (95% CI: 0.72–0.85) in the merged GSE dataset and 0.83 (95% CI: 0.77–0.89) in the TCGA cohort (Fig. 7L, M). Interestingly, although GDF15 is highly expressed in PCa, high GDF15 expression demonstrated that longer DFS and PFS in TCGA cohort (Fig. 7N, O). The prognostic value for the other 6 FRGs can be found in Supplementary Fig. 4.
GDF15 facilitates CRPC cells migration and invasion in vitro
We further examined GDF15 expression by cellular experiments. Compared to the normal prostate epithelial cells (RWPE-1), the mRNA expression levels of GDF15 in LNCap (HSPC cells) significantly increased, while in CRPC cell lines (PC-3, DU145, and C4-2), its expression levels exhibited a notable decrease compared to LNCap (Fig. 8A). Similar results were also confirmed by WB (Fig. 8B). Although GDF15 expression was decreased in CRPC cells compared with that in RWPE-1 and HSPC cells, we further explored whether GDF15 still has the effect of promoting migration and invasion in CRPC cells by Transwell assays. We increased GDF15 expression in PC-3 and DU145 by plasmid transfection. Successful overexpression of GDF15 was confirmed by qPCR and WB (Fig. 8C, D). As shown in Fig. 8E, GDF15 can markedly facilitated migrating and invading capacity of CRPC cells.
GDF15 facilitates CRPC cells migration and invasion in vitro. Compared to the RWPE-1 cell line, the mRNA (A) and protein (B) expression levels of GDF15 in LNCap (HSPC cells) significantly increased, while in CRPC cell lines (PC-3, DU145, and C4-2), its expression levels exhibited a notable decrease compared to LNCap. Successful overexpression of GDF15 in PC-3 and DU145 was confirmed by qPCR (C) and WB (D). (E) Transwell assays were performed to assess the migration and invasion ability in GDF15 overexpressing CRPC cells. Data are shown as the mean ± SD, N ≥ 3. **p < 0.01, ***p < 0.001, ****p < 0.0001. Quantitative comparisons between samples were employed on the same blot. original blots are presented in Supplementary Fig. 5.
Discussion
PCa is a highly prevalent malignancy and the second cause of cancer-related deaths due to malignancy among men in US1. In China, the special situation is that most patients are advanced prostate cancer at the time of initial diagnosis, and hormonal therapy (ADT) is the main therapy37. Not surprisingly, after several years of ADT, the HSPC progressed to CRPC. The second-generation anti-androgens (enzalutamide, abiraterone), taxane-based chemotherapy (docetaxel), immunotherapy, and targeted therapies (olaparib, pazopanib) are the main treatment options for CRPC38,39,40. Unfortunately, PSA cannot fully meet the clinical needs, and we often fail to detect the progression of patients to CRPC in time. As a result, our treatment of CRPC is lagging. Hence, identifying novel diagnostic markers for CRPC is necessary.
Ferroptosis is a form of cell death which is morphologically, biochemically, and genetically distinct from apoptosis, necrosis, and autophagy. Although ferroptosis was formally reported only in 20127, its role in cancer has been extensively studied. Ferroptosis is a novel method for the destruction of cancer cells. Wang et al.41 found that Enzalutamide-resistant (EnzaR) CRPC cells was characterized by ferroptosis resistance. ASCL1-mediated ferroptosis resistance enhanced the progress of CRPC to NEPC. Inhibiting ASCL1-mediated ferroptosis resistance presents a new therapeutic target for treating CRPC and NEPC. Phosphoglycerate dehydrogenase (PHGDH), the rate-limiting enzyme in the serine synthesis pathway, is frequently overexpressed in various types of cancers. Deng et al.42 discovered that high levels of PHGDH correlate with enzalutamide resistance in CRPC cells. PHGDH promoted cell growth by inhibiting ferroptosis in EnzaR CRPC cells. In vivo experiments demonstrated that silencing PHGDH significantly reduced the tumor volume and weight of C4-2-EnzaR xenografts, effects that were further amplified by enzalutamide treatment. Zhuo et al.43 also reported that Polyphyllin I induces ferroptosis in CRPC cells via the ERK/DNMT1/ACSL4 axis and can inhibit the growth of xenografts in nude mice by inducing ferroptosis. This suggests that ferroptosis may serve as a potential therapeutic target for CRPC and could become a novel strategy for combination therapy.
Moreover, several studies have constructed FRGs signature, which can be used to predict the prognosis of cancer (uveal melanoma44, lung adenocarcinoma45, hepatocellular carcinoma46, and so on). Lv et al.47 identified a nine FRGs signature for predicting biochemical recurrence of PCa. Wei et al.48 constructed an eight FRGs signature to predict prognosis and immune microenvironment in PCa. But no studies have explored the ferroptosis-related genes signature in CRPC. Our results show the potential of using FRGs signature as biomarkers to predict CRPC development.
In this study, we obtained a CRPC cohort by merging 3 GEO datasets. We identified 40 up-regulated and 156 down-regulated DEGs between CRPC and HSPC. ‘Response to hormones’ was the most significantly enriched biological process. This is consistent with the pathological features of hormone resistance in CRPC. We identified 5 densely connected network components in the PPI network. They were mainly enriched in catabolic process and Wnt signaling. The Wnt pathway has been shown to interact with AR to promote CRPC transition49. By intersecting DEGs with FRGs, a total of 10 differently expressed FRGs were identified. Based on these FRGs, a 7-FRGs signature were constructed. This signature could efficiently distinguish CRPC from HSPC samples with an AUC value greater than 0.9 in both training cohort and validation cohort. Then, we identified the ferroptosis molecular subtypes using the signature. The signature could also distinguish cluster 1 (high risk score) patients with high specificity and sensitivity. This certainly increases its potential for clinical application.
Ferroptosis can regulate immune tolerance of cancer36,50,51. Therefore, we estimated immune cell infiltration in different group samples using CIBERSORTx. Macrophages and T cells are among the immune cells of greatest interest. M2 macrophages and Tregs play an important role in inhibiting anti-tumor immune responses. Like previous studies52,53, M2 macrophages and Tregs were significantly increased in PCa samples, especially in CRPC samples. M2 macrophages also significantly increased in high-risk group and cluster 1 samples compared to low-risk group and cluster 2 samples. Fang et al.54 found M2 macrophages produce Taurine to repress ferroptosis in PCa (LNCaP and DU145). In Hepatocellular carcinoma, induction of ferroptosis in macrophages can promote the transformation of M2 phenotype to M1 phenotype, which is an effective scheme to improve the efficacy of immunotherapy55. Xu et al.56 found that loss of glutathione peroxidase 4 (Gpx4) induced ferroptosis in Tregs. Although this did not lead to the death of Tregs, it caused them to lose their immunosuppressive capacity. Therefore, inducing ferroptosis in macrophages and Tregs is a potential therapeutic strategy to improve CRPC treatment. Moreover, the research by Mao et al. discovered that CD8 + T cells could induce ferroptosis in CRPC cells through STAT1/SLC7A11/GPX4 signaling axis57. Ferroptosis, as a form of immunogenic cell death, plays a critical role in triggering anti-tumor immune responses in the tumor microenvironment. Jin et al.58 successfully induced ferroptosis in CRPC cells using irradiated microparticles (RC@RMPs) and restructured the tumor immune microenvironment of PCa by DC activation and M1 macrophage polarization, thereby improving the immune desert state of PCa. From these evidences, it is clear that there is a complex interplay between CRPC cells, immune cells, and ferroptosis: (1) immune cells such as CD8 + T cells can inhibit tumor progression by inducing ferroptosis in tumor cells; (2) ferroptosis in tumor cells can improve the immune desert state of prostate cancer; (3) inducing ferroptosis in immune cells such as M2 macrophages and Treg cells can effectively alleviate local immune suppression.
As GDF15 is a secreted protein, both its mRNA and protein can be detected in blood and urine. Therefore, whether GDF15 can be used as a non-invasive biomarker for PCa has been concerned by researchers. Brown et al.59 found that GDF15 serum levels was elevated in PCa and could predicted poor cancer-specific survival of PCa patients. GDF15 serum levels could effectively distinguish between benign hyperplasia and PCa17,18. Zhao et al.60 reported that GDF15 serum levels could be used as a candidate biomarker for docetaxel treatment response in mCRPC patients. Urinary GDF15 mRNA levels were also found to be elevated in PCa19. The investigators constructed a 6-gene signature including GDF15 and found that this urine mRNA signature effectively differentiated PCa from BPH with PSA between 3 ng/ml and 10 ng/ml. Recently, Tekin et al.61 detected urinary GDF15 levels in 8 PCa and 10 normal subjects by ELISA. They found that urinary GDF15 level in patients with PCa was higher, but the difference was not statistically significant (P = 0.07). Because the sample size of this study was too small, it is possible that additional findings could be obtained with a larger cohort. In this study, we confirmed the potential of tissue GDF15 mRNA in distinguishing CRPC from HSPC through bioinformatics analysis. In addition, GDF15 can differentiate PCa patients from non-PCa patients with high specificity and sensitivity. In future studies, we plan to examine GDF15 mRNA and protein levels in urine and blood of BPH, HSPC and CRPC patients to further evaluate its potential as a non-invasive biomarker for PCa.
Exploring the biological background of biomarkers not only helps reveal the molecular basis and pathological processes of diseases but also enhances their clinical application value and promotes the development of personalized treatment. Growth differentiation factor 15 (GDF15), also known as macrophage inhibitory cytokine 1 (MIC-1), is a member of the transforming growth factor β (TGF-β) superfamily62. As a stress response cytokine, GDF15 is overexpressed in various malignant tumors, including prostate cancer63, ovarian cancer64, malignant gliomas65, gastric cancer66. Through bioinformatics analysis, we discovered that as the disease progresses (from normal to HSPC to CRPC), the mRNA expression level of GDF15 in tumor tissues first increases and then decreases. This characteristic was confirmed in cellular experiments. Compared to the RWPE-1 cell line, the mRNA and protein expression levels of GDF15 in LNCap (HSPC cells) significantly increased, while in CRPC cell lines (PC-3, DU145, and C4-2), its expression levels exhibited a notable decrease compared to LNCap. Karan et al.67 reported that, compared to the androgen-sensitive LNCaP-C33 cells, the GDF15 gene is highly expressed in the androgen-independent LNCaP-C81 cells and their metastatic variant LNCaP-Ln3. Furthermore, GDF15 is found at very low to undetectable levels in androgen-independent PC3 and DU145 cells. These results partially overlap with our findings. The mechanism underlying this characteristic variation in GDF15 expression levels remains unclear. During the progression of prostate cancer, fluctuations in endogenous androgen levels and dysregulation of various transcription factors, including the androgen receptor, are commonly observed. Multiple experimental evidences indicate that androgens mediate the regulation of GDF15 expression in PCa cells67,68,69. This may partially explain the observed phenomenon. The characteristic variation of GDF15 expression levels in relation to disease status is one of the reasons it can serve as a biomarker.
The role of GDF15 in cancer biology is contradictory in different malignancies70. Its role in prostate cancer is also subject to controversy. Senapati et al.71 found that GDF15 was overexpression in metastatic variants of PCa cells (PC-3 M, PC-3MLN4, and LNPCa-LN3) compared to parental PCa cells (PC3 and LNPCa). GDF15 promoted motility and invasiveness of PC-3 and PC-3 M cells by activating FAK and RhoA. Similarly, Mimeault et al.63 discovered that the overexpression of GDF15 could enhance the invasion and metastatic capabilities of prostate cancer cells, promote epithelial-mesenchymal transition (EMT), and increase the resistance of tumor cells to docetaxel. Our study also confirmed that although the expression of GDF15 is decreased in CRPC cells compared to HSPC cells, the overexpression of GDF15 still facilitates CRPC cells migration and invasion in vitro. On the other hand, Cheng et al.72 reported that wild-type p53 attenuated the p53-null PCa cancer cells motility by inducing GDF15 expression. Husaini et al.73 found overexpression of GDF15 could inhibit the progression of spontaneous PCa in C57BL/6 background TRAMP mice and suppress tumor development in the orthotopic TRAMP tumor engraftment model. PCa is a complex endocrine-related tumor with a long disease course and multiple stages, involving various transcription factors in its progression. This suggests that we should investigate the pro-cancerous or anti-cancerous roles of GDF15 in a context-dependent manner.
In various non-tumor diseases, GDF15 is regarded as an inhibitor of ferroptosis. For instance, GDF15 inhibited ferroptosis in cardiomyocytes through the SOCS1/GPX4 signaling pathway, thereby alleviating sepsis-induced myocardial dysfunction74. There is limited research on the relationship between GDF15 and ferroptosis in malignant tumors. A total of three studies have demonstrated through cellular experiments that GDF15 can inhibit ferroptosis in nasopharyngeal carcinoma75, colorectal cancer76, and gastric cancer77. However, GDF15 expression was decreased in clear cell renal cell carcinoma and inhibited ferroptosis78. There are currently no research reports on the regulatory role of GDF15 in ferroptosis related to PCa and CRPC in PubMed. This is also one of the aspects we aim to evaluate in the future.
GDF15 has significant potential as a diagnostic biomarker, with the characteristic of ferroptosis inhibitor, which may have important implications for the management of PCa and CRPC. On one hand, as a diagnostic marker, GDF15, when used alone or in conjunction with PSA, may enhance the early detection rates of patients with PCa and CRPC, thereby optimizing personalized treatment plans. On the other hand, as a ferroptosis inhibitor, GDF15 provides a novel therapeutic avenue for patients. By promoting the ferroptosis pathway, GDF15 inhibitors may significantly impact the tumor microenvironment, immune cell behavior, and resistance to treatment. Future therapeutic strategies could consider combining GDF15 inhibitors with existing therapies, such as PARP inhibitors or immune checkpoint inhibitors, thereby improving treatment efficacy.
It should be recognized that this study has limitations. First, the integrated dataset comes from different microarray platforms, which may affect the consistency of the data and the reliability of the analysis. Second, in GSE32269, the 22 hormone-dependent PCa samples refer to tumor samples from hormone-naive patients, while the 29 metastatic PCa samples refer to bone metastasis specimens from CRPC patients. Patients who have not received hormone treatment may also exhibit “De Novo Resistance”, and the signaling pathways in CRPC bone metastases may differ significantly from those in primary tumors. These uncertainties could potentially confound the study results. Third, although the bioinformatics analysis demonstrates that the 7-FRGs signature and GDF15 have good diagnostic performance, there is a lack of larger and more diverse datasets, clinical samples, or prospective cohorts for external validation. Additionally, it is important to note that this study presents the findings of GDF15 in tissue samples. The potential of GDF15 as a serum or urine marker requires further validation through clinical trials. Fourth, PSA is currently the most widely used biomarker for PCa and CRPC. Due to the lack of PSA data from the CRPC cohort, we are unable to compare the diagnostic accuracy of the FRG signature and GDF15 with that of PSA. Fifth, the reason for the decline in GDF15 expression in CRPC remains unclear, and further research is needed to explore the contradiction between its expression and its biological function in CRPC.
Conclusion
In conclusion, we identified differently expressed FRGs in CRPC and generated a novel diagnostic FRGs signature for CRPC. Moreover, GDF15 could distinguish CRPC from HSPC with high specificity and sensitivity. GDF15 is expected to be an effective non-invasive diagnostic biomarker for CRPC.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 74, 12–49 (2024).
Denis, L. J. & Griffiths, K. Endocrine treatment in prostate cancer. Semin Surg. Oncol. 18, 52–74 (2000).
Sharifi, N. et al. A retrospective study of the time to clinical endpoints for advanced prostate cancer. BJU Int. 96, 985–989 (2005).
Mizokami, A. et al. Understanding prostate-specific antigen dynamics in monitoring metastatic castration-resistant prostate cancer: implications for clinical practice. Asian J. Androl. 19, 143–148 (2017).
Zhang, B., Zhang, Z., Hu, H. & Dong, J. T. Novel gene signatures predictive of patient recurrence-free survival and castration resistance in prostate cancer. Cancers. 13. (2021).
Li, S. et al. Establishment of a novel combined nomogram for predicting the risk of progression related to castration resistance in patients with prostate cancer. Front. Genet. 13, 823716 (2022).
Dixon, S. J. et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012).
Lei, G., Zhuang, L. & Gan, B. Targeting ferroptosis as a vulnerability in cancer. Nat. Rev. Cancer. 22, 381–396 (2022).
Nakamura, T. & Conrad, M. Exploiting ferroptosis vulnerabilities in cancer. Nat. Cell. Biol. (2024).
Bordini, J. et al. Iron induces cell death and strengthens the efficacy of antiandrogen therapy in prostate cancer models. Clin. Cancer Res. 26, 6387–6398 (2020).
Zhou, X. et al. Flubendazole, FDA-approved anthelmintic, elicits valid antitumor effects by targeting P53 and promoting ferroptosis in castration-resistant prostate cancer. Pharmacol. Res. 164, 105305 (2021).
Yang, Y. et al. Ferroptosis inducer Erastin downregulates androgen receptor and its splice variants in castration–resistant prostate cancer. Oncol. Rep. 45. (2021).
Wu, L. et al. The establishment of polypeptide PSMA-targeted chimeric antigen receptor-engineered natural killer cells for castration-resistant prostate cancer and the induction of ferroptosis-related cell death. Cancer Commun. (Lond). 42, 768–783 (2022).
Li, Y. et al. Genetically engineered membrane-Coated nanoparticles for enhanced prostate-Specific membrane antigen targeting and ferroptosis treatment of Castration-Resistant prostate cancer. Adv. Sci. (Weinh) e2401095. (2024).
Liu, C. et al. The ferroptosis-related long non-coding RNAs signature predicts biochemical recurrence and immune cell infiltration in prostate cancer. BMC Cancer. 22, 788 (2022).
Wang, Y. et al. A novel ferroptosis-related gene prognostic index for prognosis and response to immunotherapy in patients with prostate cancer. Front. Endocrinol. (Lausanne). 13, 975623 (2022).
Li, J., Veltri, R. W., Yuan, Z., Christudass, C. S. & Mandecki, W. Macrophage inhibitory cytokine 1 biomarker serum immunoassay in combination with PSA is a more specific diagnostic tool for detection of prostate cancer. PLoS One. 10, e0122249 (2015).
Bansal, N. et al. Relevance of MIC-1 in the era of PSA as a serum based predictor of prostate cancer: A critical evaluation. Sci. Rep. 7, 16824 (2017).
Kang, H. W. et al. A novel urinary mRNA signature using the droplet digital polymerase chain reaction platform improves discrimination between prostate cancer and benign prostatic hyperplasia within the prostate-specific antigen gray zone. Investig Clin. Urol. 61, 411–418 (2020).
Ross-Adams, H. et al. Integration of copy number and transcriptomics provides risk stratification in prostate cancer: A discovery and validation cohort study. EBioMedicine 2, 1133–1144 (2015).
Cai, C. et al. ERG induces androgen receptor-mediated regulation of SOX9 in prostate cancer. J. Clin. Invest. 123, 1109–1122 (2013).
Stoss, O. et al. Transcriptional profiling of transurethral resection samples provides insight into molecular mechanisms of hormone refractory prostate cancer. Prostate Cancer Prostatic Dis. 11, 166–172 (2008).
Grasso, C. S. et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487, 239–243 (2012).
Johnson, W. E., Li, C. & Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007).
Zhou, N. et al. FerrDb V2: update of the manually curated database of ferroptosis regulators and ferroptosis-disease associations. Nucleic Acids Res. 51, D571–d582 (2023).
Cheng, S. et al. Unveiling novel double-negative prostate cancer subtypes through single-cell RNA sequencing analysis. NPJ Precis Oncol. 8, 171 (2024).
Ritchie, M. E. et al. Smyth GK: Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Bader, G. D. & Hogue, C. W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 4, 2 (2003).
Engebretsen, S. & Bohlin, J. Statistical predictions with Glmnet. Clin. Epigenetics. 11, 123 (2019).
Wilkerson, M. D. & Hayes, D. N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573 (2010).
Rusk, N. Expanded CIBERSORTx. Nat. Methods. 16, 577 (2019).
Zhu, Z. et al. Identifying the key genes and microRNAs in prostate cancer bone metastasis by bioinformatics analysis. FEBS Open. Bio. 10, 674–688 (2020).
Zhu, Z. et al. MiRNA-671-5p promotes prostate cancer development and metastasis by targeting NFIA/CRYAB axis. Cell. Death Dis. 11, 949 (2020).
Shen, W. et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. Imeta 1, e36 (2022).
Wiernicki, B. et al. Cancer cells dying from ferroptosis impede dendritic cell-mediated anti-tumor immunity. Nat. Commun. 13, 3676 (2022).
Ma, C. G. et al. [Epidemiology of prostate cancer from three centers and analysis of the first-line hormonal therapy for the advanced disease]. Zhonghua Wai Ke Za Zhi. 46, 921–925 (2008).
Antonarakis, E. S. et al. Pembrolizumab for treatment-refractory metastatic castration-resistant prostate cancer: multicohort, open-label phase II KEYNOTE-199 study. J. Clin. Oncol. 38, 395–405 (2020).
Jiang, Y. et al. Prospect of prostate cancer treatment: armed CAR-T or combination therapy. Cancers (Basel) 14. (2022).
Vogelzang, N. J. et al. Efficacy and safety of autologous dendritic Cell-Based immunotherapy, docetaxel, and prednisone vs placebo in patients with metastatic Castration-Resistant prostate cancer: the VIABLE phase 3 randomized clinical trial. JAMA Oncol. (2022).
Nie, J., Zhang, P., Liang, C., Yu, Y. & Wang, X. ASCL1-mediated ferroptosis resistance enhances the progress of castration-resistant prostate cancer to neurosecretory prostate cancer. Free Radic Biol. Med. 205, 318–331 (2023).
Wang, J. et al. Inhibition of phosphoglycerate dehydrogenase induces ferroptosis and overcomes enzalutamide resistance in castration-resistant prostate cancer cells. Drug Resist. Updat. 70, 100985 (2023).
Zou, P., Chen, Z., He, Q. & Zhuo, Y. Polyphyllin I induces ferroptosis in castration-resistant prostate cancer cells through the ERK/DNMT1/ACSL4 axis. Prostate 84, 64–73 (2024).
Luo, H. & Ma, C. A novel Ferroptosis-Associated gene signature to predict prognosis in patients with uveal melanoma. Diagnostics (Basel) 11. (2021).
Zhang, A., Yang, J., Ma, C., Li, F. & Luo, H. Development and validation of a robust ferroptosis-related prognostic signature in lung adenocarcinoma. Front. Cell. Dev. Biol. 9, 616271 (2021).
Liu, Y. et al. Development and validation of a combined ferroptosis and immune prognostic classifier for hepatocellular carcinoma. Front. Cell. Dev. Biol. 8, 596679 (2020).
Lv, Z. et al. Identifying a ferroptosis-related gene signature for predicting biochemical recurrence of prostate cancer. Front. Cell. Dev. Biol. 9, 666025 (2021).
Wang, H. et al. Ferroptosis-related gene signature predicts prognosis and immune microenvironment in prostate cancer. Transl Androl. Urol. 13, 2092–2109 (2024).
Khurana, N. & Sikka, S. C. Interplay between SOX9, Wnt/β-Catenin and androgen receptor signaling in Castration-Resistant prostate cancer. Int. J. Mol. Sci. 20. (2019).
Dang, Q. et al. Ferroptosis: a double-edged sword mediating immune tolerance of cancer. Cell. Death Dis. 13, 925 (2022).
Bao, Y. et al. BIBR1532 combined with radiotherapy induces ferroptosis in NSCLC cells and activates cGAS-STING pathway to promote anti-tumor immunity. J. Transl Med. 22, 519 (2024).
Erlandsson, A. et al. M2 macrophages and regulatory T cells in lethal prostate cancer. Prostate 79, 363–369 (2019).
Andersen, L. B. et al. Immune cell analyses of the tumor microenvironment in prostate cancer highlight infiltrating regulatory T cells and macrophages as adverse prognostic factors. J. Pathol. 255, 155–165 (2021).
Xiao, H. et al. Taurine inhibits ferroptosis mediated by the crosstalk between tumor cells and tumor-Associated macrophages in prostate cancer. Adv. Sci. (Weinh). 11, e2303894 (2024).
Hao, X. et al. Inhibition of APOC1 promotes the transformation of M2 into M1 macrophages via the ferroptosis pathway and enhances anti-PD1 immunotherapy in hepatocellular carcinoma based on single-cell RNA sequencing. Redox Biol. 56, 102463 (2022).
Xu, C. et al. The glutathione peroxidase Gpx4 prevents lipid peroxidation and ferroptosis to sustain Treg cell activation and suppression of antitumor immunity. Cell. Rep. 35, 109235 (2021).
Zhou, X. et al. Abrogation of HnRNP L enhances anti-PD-1 therapy efficacy via diminishing PD-L1 and promoting CD8(+) T cell-mediated ferroptosis in castration-resistant prostate cancer. Acta Pharm. Sin B. 12, 692–707 (2022).
Deng, Z. et al. Irradiated microparticles suppress prostate cancer by tumor microenvironment reprogramming and ferroptosis. J. Nanobiotechnol. 22, 225 (2024).
Brown, D. A. et al. Macrophage inhibitory cytokine 1: a new prognostic marker in prostate cancer. Clin. Cancer Res. 15, 6658–6664 (2009).
Zhao, L. et al. Identification of candidate biomarkers of therapeutic response to docetaxel by proteomic profiling. Cancer Res. 69, 7696–7703 (2009).
Tekin, B. et al. Mass spectrometry-based assessment of prostate cancer-associated crystalloids reveals enrichment for growth and differentiation factor 15. Hum. Pathol. 135, 35–44 (2023).
Luan, H. H. et al. GDF15 is an Inflammation-Induced central mediator of tissue tolerance. Cell 178, 1231–1244e1211 (2019).
Mimeault, M., Johansson, S. L. & Batra, S. K. Marked improvement of cytotoxic effects induced by docetaxel on highly metastatic and androgen-independent prostate cancer cells by downregulating macrophage inhibitory cytokine-1. Br. J. Cancer. 108, 1079–1091 (2013).
Staff, A. C. et al. Growth differentiation factor-15 as a prognostic biomarker in ovarian cancer. Gynecol. Oncol. 118, 237–243 (2010).
Roth, P. et al. GDF-15 contributes to proliferation and immune escape of malignant gliomas. Clin. Cancer Res. 16, 3851–3859 (2010).
Joo, M., Kim, D., Lee, M. W., Lee, H. J. & Kim, J. M. GDF15 promotes cell growth, migration, and invasion in gastric cancer by inducing STAT3 activation. Int. J. Mol. Sci. 24. (2023).
Karan, D. et al. Dysregulated expression of MIC-1/PDF in human prostate tumor cells. Biochem. Biophys. Res. Commun. 305, 598–604 (2003).
Paralkar, V. M. et al. Cloning and characterization of a novel member of the transforming growth factor-beta/bone morphogenetic protein family. J. Biol. Chem. 273, 13760–13767 (1998).
Chen, S. J. et al. Prostate-derived factor as a paracrine and autocrine factor for the proliferation of androgen receptor-positive human prostate cancer cells. Prostate 67, 557–571 (2007).
Siddiqui, J. A. et al. Pathophysiological role of growth differentiation factor 15 (GDF15) in obesity, cancer, and cachexia. Cytokine Growth Factor. Rev. 64, 71–83 (2022).
Senapati, S. et al. Overexpression of macrophage inhibitory cytokine-1 induces metastasis of human prostate cancer cells through the FAK-RhoA signaling pathway. Oncogene 29, 1293–1302 (2010).
Cheng, J. C., Chang, H. M. & Leung, P. C. Wild-type p53 attenuates cancer cell motility by inducing growth differentiation factor-15 expression. Endocrinology 152, 2987–2995 (2011).
Husaini, Y. et al. Growth differentiation factor-15 slows the growth of murine prostate cancer by stimulating tumor immunity. PLoS One. 15, e0233846 (2020).
Li, X. et al. GDF15 attenuates sepsis-induced myocardial dysfunction by inhibiting cardiomyocytes ferroptosis via the SOCS1/GPX4 signaling pathway. Eur. J. Pharmacol. 982, 176894 (2024).
Wu, Z. & Qu, Q. Mechanism of Luteolin induces ferroptosis in nasopharyngeal carcinoma cells. J. Toxicol. Sci. 49, 399–408 (2024).
Huang Fu, Z. M. et al. Suppressing GDF15 enhances the chemotherapeutic effect of 5 FU on MSI-H CRC by regulating the ferroptosis pathway SLC7A11/GSH/GPX4. J. Cancer Res. Clin. Oncol. 151, 6 (2024).
Chen, L., Qiao, L., Bian, Y. & Sun, X. GDF15 knockdown promotes erastin-induced ferroptosis by decreasing SLC7A11 expression. Biochem. Biophys. Res. Commun. 526, 293–299 (2020).
Yang, D. et al. Downregulation of GDF15 suppresses ferroptosis and predicts unfavorable prognosis in clear cell renal cell carcinoma. Cell. Div. 18, 21 (2023).
Funding
This research was funded by China Postdoctoral Science Foundation (2023M731309), Guangdong Basic and Applied Basic Research Foundation (2020A1515110946), PhD Research Foundation of Affiliated Hospital of Jining Medical University (2022-BS-001), Research Fund for Academician LinHe New Medicine (JYHL2022FMS09) and Jining Key Research and Development Foundation (2023YXNS023).
Author information
Authors and Affiliations
Contributions
The conception and design of the study: XL, FL and ZZ; the acquisition of data: YS, JL; analysis and interpretation of data: ZQ, TZ and ZZ; performing the experiments: NL, JH; drafting the article: YS, XL, and JL; revising the article: FL, ZZ; providing funding: ZZ. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shao, Y., Li, X., Liu, J. et al. Identification of a ferroptosis related genes signature and GDF15 contributing to a new perspective for the diagnosis of CRPC. Sci Rep 15, 21624 (2025). https://doi.org/10.1038/s41598-025-97045-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-97045-9