Abstract
To explore the role of RBM10 in colorectal cancer (CRC) and the regulatory mechanism of CRC invasion through alternative splicing (AS) of long non-coding RNA (lncRNA) SNHG17 by RBM10. Samples were collected from sixty cases of CRC and their corresponding adjacent normal tissues. Immunohistochemistry and Western blot were performed to analyze the expression of RBM10. A Transwell invasion assay was conducted to evaluate the effect of RBM10 on the invasion of HCT116 cells, and a Western blot was performed to detect the expression of EMT-related proteins. Moreover, CLIP-seq and RIP experiments were performed to explore the interaction between RBM10 and SNHG17. The expression of RBM10 was significantly decreased in CRC tissues and cells compared to the normal adjacent tissues. Overexpression of RBM10 inhibited CRC invasion, while knockdown of RBM10 had the opposite effect. RBM10 was found to interact with SNHG17 and regulate its splice isoform balance. Specifically, the splice variant SNHG17_2 regulated by RBM10 was upregulated in CRC and was positively correlated with CRC invasion. RBM10 inhibits CRC invasion by regulating the alternative splicing of SNHG17, providing new research directions and potential targets for CRC treatment.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) has emerged as the third most common malignant tumor globally, with approximately 1.9 million new cases and nearly 960,000 deaths annually1. In China, the incidence of CRC is increasing, with a trend towards younger patients2. Despite advances in treatment, the survival rate for advanced-stage patients remains below 50% due to the high invasiveness and metastasis rate of CRC3. Therefore, exploring the molecular mechanisms of invasion and metastasis in CRC holds great significance. In recent years, RNA binding proteins (RBPs) have gained attention in tumor research. Among them, RBM10 was found to play an essential role in CRC, as it participates in the regulation of multiple signaling pathways and is closely related to metastatic CRC4,5,6,7. Alternative splicing (AS) is a post-transcriptional gene expression regulation mechanism that plays a crucial role in various biological behaviors of tumors8,9. Our preliminary research revealed that RBM10 has binding sites with long non-coding RNA SNHG17, suggesting that RBM10 may affect the invasion and metastasis of CRC by regulating the AS of SNHG17. Based on these preliminary findings, this study aimed to explore the in-depth mechanism underlying the regulatory effects of RBM10 on CRC invasion and metastasis through SNHG17 AS. This research provides a better understanding of the mechanisms of tumor occurrence and new ideas for future therapeutic strategies.
Methods
Clinical samples
From October 2021 to October 2022, tumor samples were collected from surgical resections of 60 CRC patients from our hospital and collaborating units, along with their corresponding adjacent normal tissues. Among these samples, 37 were colon cancer cases and 23 were rectal cancer cases. Clear inclusion and exclusion criteria were established to ensure the quality and representativeness of the samples.
Inclusion criteria:
CRC patients who visited our hospital or collaborating units within the specified date range.
Availability of surgically resected tumor samples and corresponding adjacent normal tissues (distance from tumor > 3 cm).
No chemotherapy or radiotherapy before surgery.
Met the CRC diagnostic criteria defined by the World Health Organization.
Patients or their families have provided written informed consent.
Exclusion criteria:
Patients who have previously received or are currently receiving chemotherapy or radiotherapy.
Patients with other concurrent malignant tumors or systemic diseases.
Samples with insufficient quantity or poor quality that are unsuitable for subsequent experiments.
Patients who did not provide written informed consent.
This study followed the principles outlined in the Helsinki Declaration and was approved by the Ethics Committee of Daqing People’s Hospital (Approval No.:KY2024-018). All tissue samples were immediately frozen in liquid nitrogen post-surgery and stored at -80 °C for subsequent research. To ensure sample accuracy, all tissue samples were independently evaluated and confirmed by two experienced pathologists.
Cell culture
Human colon cancer cell line HCT116 cells (ATCC, CCL-247) were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Human normal intestinal epithelial cell line HIEC-6 cells (ATCC, XY-XB2311) were cultured in DMEM medium containing 20% FBS and 1% penicillin-streptomycin. All cells were cultured at 37 °C and 5% CO2. When the cell density reached 80–90%, passaging was performed in new culture flasks using 0.25% trypsin-EDTA solution for digestion followed by suspension in RPMI-1640 medium containing 10% FBS at a 1:3 ratio.
Cell transfection
Stable overexpression and knockdown of RBM10 lentiviruses, as well as negative control vectors, were constructed by Shanghai GeneChem Co.Ltd. (Shanghai, China). HCT116 cells were cultured in six-well plates. When the cell density reached 70-80%, they were passaged and then washed with phosphate-buffered saline (PBS). Following the manufacturer’s instructions, the lentiviruses and negative control virus were transfected into the cells. After 24 h of lentiviral transfection, cells were selected in a puromycin-containing medium to establish stable overexpression and knockdown cell lines of the RBM10 gene. Subsequently, SNHG17 interference fragments were constructed by Suzhou GenePharma Co., Ltd. (Suzhou, China). HCT116 cells were cultured in six-well plates; when cell density reached 70-80%, they were passaged and then washed with PBS. Plasmids were transfected into cell culture wells using a Lipofectamine 2000 reagent kit (Invitrogen) according to the manufacturer’s instructions. Cells were cultured for 24 and 48 h and then used for subsequent experiments.
Western blot
The cells were fully lysed in an immunoprecipitation assay buffer containing protease and phosphatase inhibitors (Beyotime, Jiangsu, China), and protein concentrations were determined using the BCA protein assay kit (Beyotime, Jiangsu, China). Protein samples (30 µg) were separated by 8-15% SDS-PAGE gel electrophoresis and transferred to polyvinylidene difluoride membranes (PIVH00010, Merck Millipore, Burlington, MA, USA). After blocking for 2 h, membranes were incubated overnight at 4 °C with specific primary antibodies (diluted in PBS containing Tween-20), followed by 1.5 h of incubation at room temperature with the corresponding secondary antibodies. The ECL detection kit (Merck Millipore, Burlington, MA, USA) was used for protein blotting, and Image J software (http://rsb.info.nih.gov/ij/, Bethesda, MD, USA) was employed for quantification. The primary antibodies included anti-RBM10 (14423-1-AP, 1:500), GAPDH (60004-1-lg, 1:3000) from Proteintech Group (Wuhan, China), and anti-N-cadherin (ab76011, 1:1000), anti-Vimentin (ab92547, 1:1000), and anti-E-cadherin (ab40772, 1:1000) from Abcam (Cambridge, MA, USA). Quantification of protein bands was conducted using ImageJ (v1.53t; National Institutes of Health) by measuring the integrated density normalized to β-actin loading control.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and cDNA was obtained by reverse transcription using a kit (RR047A, Takara, Japan). RT-qPCR was performed using the SYBR Premix Ex TaqTM II (Perfect Real Time) kit (DRR081, Takara, Japan) on an ABI QuantStudio5 real-time PCR instrument (ABI, Foster City, CA, USA). Primers were synthesized by Shanghai Biotechnology Co., Ltd. (Shanghai, China), as shown in Table 1. Ct values for each well were recorded using β-actinas as an internal reference. The relative expression was calculated using the 2−ΔΔCt method.
Immunohistochemistry
CRC tumor samples and adjacent paraffin-embedded specimens were sectioned into 4 μm slices using a slicer. The sections were stained using the Bond-Max automated immunohistochemistry staining system (Leica, Wetzlar, Germany). Thereafter, Image J software (v1.53t; National Institutes of Health) was used for quantitative analysis of randomly captured 40x field images.
Transwell invasion assay
Matrigel (356234, Corning, Corning, NY, USA) was diluted with serum-free medium (1:8) and added to the upper chamber of transwell inserts. The inserts were placed in a 24-well plate and incubated at 37 °C with 5% CO2 for 2 h. Cell density was adjusted to 105 cells/100 µL, with 100 µL added to the upper chamber and 500 µL of medium containing 15% serum added to the lower chamber, followed by incubation for 24 h. The inserts were removed, and the Matrigel and cells in the upper chamber were wiped off with a cotton swab. The cells were fixed with 4% formaldehyde for 10 min, and stained with 0.1% crystal violet for 10 min. Invasion cells were counted using Image J software(v1.53t; National Institutes of Health) under a high-power microscope.
CLIP-Seq method
CLIP-Seq can reveal RNA-RBP interactions at the whole-genome level. CLIP-Seq technology involves coupling processed cell or tissue samples under 254 nm UV irradiation, capturing RNA through specific antibodies against RBPs, and forming protein-RNA complexes. Within the complex, RNA fragments protected by protein molecules are retained, while the remaining fragments are degraded to protect the target fragments. The ends of RNA fragments are labeled with radioactive phosphates, allowing separation of protein and RNA fragment complexes by denaturing SDS-PAGE gel, followed by autoradiography. Proteinase K degrades the protein components of protein-RNA complexes, preserving RNA components. RNA fragments are extracted and subjected to high-throughput sequencing. The CLIP-seq experiment was jointly completed by our research group and Appreciate The Beauty of Life, lnc (Wuhan, China).
RIP method
During the study, logarithmic growth phase HCT116 cells were collected and incubated for 1 h at 4 °C for dispersion. The cells were further incubated with magnetic beads and antibody mixtures for 2 h to purify the final product. Following the instructions of Guangzhou Gene Seed Biotechnology Co.Ltd, RNA binding protein immunoprecipitation (RIP) experiments were conducted using the RIP kit (product number: P0101). The antibodies used in this experiment include anti-RBM10 antibodies (14,423-1-AP, Proteintech, Wuhan, China, dilution ratio: 1:500) and Goat Anti-Rabbit IgG (ab205718, Abcam, Cambridge, MA, USA, dilution ratio: 1:50).
Dual-luciferase reporter assay
The dual-luciferase reporter assay was performed to investigate the interaction between miR-375 and CBX3 3’UTR in HCT116 and SW480 cells. The binding sequence between miR-375 and CBX3 was predicted using the Starbase database (http://starbase.sysu.edu.cn/index.php). Wild-type and mutant CBX3 mRNA 3’UTR sequences (lacking miR-375 binding sites) were cloned and inserted into the pSI-Check2 vector downstream of the firefly luciferase (fLUC) reporter gene. These constructs were then co-transfected with miR-375 mimics into shNC or shRBM10 stable cell lines. Following 48 h of transfection, cells were lysed and analyzed using the Dual-Luciferase Reporter Assay System (E1980, Promega, Madison, WI, USA) according to the manufacturer’s protocol. Luminescence measurements were conducted on a Promega GloMax Multi + Detection System (Madison, WI, USA), with Renilla luciferase signals normalized to firefly luciferase activity for quantitative analysis.
Statistical methods
In this study, all data were collected from at least 3 independent experiments and presented in the form of mean ± standard deviation. All statistical analyses were performed using Microsoft Excel (Version 2310 Build 16.0.16924.20054) and GraphPad Prism v6.01 software (GraphPad Software, Inc, USA). Differences between two groups were evaluated by unpaired Student’s t-test. Comparisons among multiple groups were analyzed with a one-way analysis of variance (ANOVA). In this study, a P value less than 0.05 was considered statistically significant.
Results
Significantly decreased expression level of RBM10 in CRC tissues
In this study, tissue samples were collected from 60 CRC patients, including 37 cases of colon cancer and 23 cases of rectal cancer. Moreover, corresponding adjacent normal tissue samples were collected. The H&E and immunohistochemical staining revealed the morphology of colorectal cancer tissues and the gradient expression of RBM10 (Fig. 1A). The expression of RBM10 was significantly decreased in colorectal cancer tissues by Western blot (Fig. 1B-C). In cellular experiments, the CRC cell lines (HCT116 and SW480) and control cell lines (HIEC-6 and NCM460) were selected for comparison. The results indicated that the expression level of RBM10 in CRC cell lines were significantly lower than in control cell lines (Fig. 1D-E).
RBM10 expression was down-regulated in CRC. (A) The representative images of H&E and IHC staining of RBM10 expression in four CRC tissues with negative, low, moderate and high expression. Scale bar = 200 μm. (B-C) The expression levels of RBM10 in CRC tissues and corresponding adjacent normal tissues were detected by Western blot. (D-E) The expression levels of RBM10 in colorectal cancer cell lines (HCT116 and SW480) and corresponding normal intestinal mucosal cells (HIEC6 and NCM460) were detected by Western blot. All values are expressed as mean ± SD from > 3 independent experiments. *P < 0.05, **P < 0.01, *** P < 0.001, **** P < 0.0001.
Overexpression of RBM10 inhibits migration and EMT of HCT116 cells
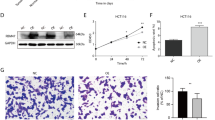
To further investigate the functional role of RBM10 in CRC migration, a series of in vitro functional experiments were conducted. Using lentiviral transduction technology, RBM10 was successfully overexpressed and knocked down in HCT116 cells, and the transfection efficiency was validated by Western blot (Fig. 2A). During the process of epithelial-mesenchymal transition (EMT), the overexpression of RBM10 significantly inhibited the expression of mesenchymal markers N-cadherin and Vimentin in colorectal cancer cell lines (HIC116 and SW480), and promoted the expression of the epithelial marker E-cadherin (Fig. 2B-E). While, the opposite results were obtained when RBM10 was knocked down (Fig. 2B-E). The Transwell invasion assay revealed that overexpression of RBM10 significantly inhibited the migration ability of CRC cells (HIC116 and SW480) compared to the control group (Fig. 2F-G). In contrast, the knockdown of RBM10 significantly increased the migration ability of cells (Fig. 2F-G). Overall, these results suggest that RBM10 plays a crucial regulatory role in the migration of CRC cells, providing strong evidence for RBM10 as a potential therapeutic target for CRC.
RBM10 inhibited the migration and EMT of CRC cells. (A) The overexpression and knockdown efficiency of RBM10 in CRC cells were detected by Western blot. (B-E) Protein levels of epithelial marker (E-cadherin) and mesenchymal marker (N-cadherin and vimentin) were measured by Western blot in HCT116 and SW480 cells after overexpression and knockdown of RBM10. (F) The invasion ability of colorectal cancer cell lines (HCT116 and SW480) after overexpression and knockdown of RBM10 were detected by Transwell assay. (G) Quantification of the Transwell assay showing the number of migrated HCT116 and SW480 cells. Scale bar = 100 μm. All values are expressed as mean ± SD from > 3 independent experiments. *P < 0.05, **P < 0.01, *** P < 0.001, **** P < 0.0001.
RBM10 regulates the alternative splicing of SNHG17
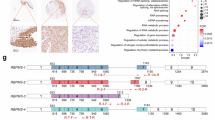
CLIP-seq analysis revealed the binding relationship between RBM10 and 353 lncRNAs, among which the highest part in 10 peaks, RBM10 has 3 significant binding sites with SNHG17 (as shown in Table 2; Fig. 3G). RIP experiments further confirmed the binding between RBM10 and SNHG17 and SNHG17-2 in CRC cell lines (HICT116 and SW480) (Fig. 3A-B and E-F). Interestingly, the overexpression of RBM10 was negatively correlated with the expression level of SNHG17, as evidenced by the decreased expression level of SNHG17 after RBM10 overexpression, in contrast, after RBM10 knockdown, the expression level of SNHG17 demonstrated a significant increase (Fig. 3C-D).
Due to the lack of specific primers for SNHG17-1, its expression was inferred based on the expression changes of SNHG17 and SNHG17-2 and the proportion of SNHG17-2 in SNHG17. RBM10 overexpression was found to inhibit the expression of SNHG17-2, leading to a significant decrease in the proportion of SNHG17-2 in SNHG17. However, RBM10 knockdown promoted the expression of SNHG17-2, resulting in a significant increase in the proportion of SNHG17-2 in SNHG17 (Fig. 3H-K). The results indicated that RBM10 overexpression induced an increased expression of SNHG17-1; conversely, after RBM10 knockdown, the expression of SNHG17-1 significantly decreased (Fig. 3L-M). Collectively, these data revealed that RBM10 balances the two splicing variants of SNHG17 via alternative splicing mechanisms.
The alternative splicing of SNHG17 was regulated by RBM10. (A-B) The binding between RBM10 and SNHG17 was detected by RIP-qPCR in HCT116 and SW480 cells. (C-D) The expression of SNHG17 was detected by qRT-PCR in CRC cells (HCT116 and SW480) after overexpression and knockdown of RBM10. (E-F) The binding between RBM10 and SNHG17-2 was detected by RIP-qPCR in HCT116 and SW480 cells. (G) CLIP-seq analysis confirmed that the main binding site of RBM10 was located at 38,411,123–38,433,908 and concentrated on SNHG17-2. (H-I) The expression of SNHG17-2 was detected by qRT-PCR in CRC cells (HCT116 and SW480) after overexpression and knockdown of RBM10. (J-K) The proportion of SNHG17-2 in SNHG17 after overexpression and knockdown of RBM10. (L-M) The effects of RBM10 overexpression and knockdown on the expression level of SNHG17-1. All values are presented as the mean ± SD. *P < 0.05, **P < 0.01, *** P < 0.001, **** P < 0.0001.
SNHG17–2 is remarkably upregulated in colorectal cancer cells and tissues, exhibiting a negative correlation with RBM10
Real-time quantitative PCR detection revealed that SNHG17-2 is significantly upregulated in the CRC cell line HCT116, while its expression is significantly decreased in normal colonic epithelial cells HIEC-6 (Fig. 4A). Furthermore, compared to adjacent normal tissues, SNHG17-2 was significantly upregulated in CRC tissues (Fig. 4B). Pearson correlation analysis was conducted and revealed a significant negative correlation between RBM10 and SNHG17-2 in CRC tissues (Fig. 4C). These research findings suggest a negative regulatory relationship between RBM10 and SNHG17-2 during the pathogenesis of CRC.
SNHG17-2 was remarkably upregulated in colorectal cancer cells and tissues, exhibiting a negative correlation with RBM10. (A) RT-qPCR was employed to quantify SNHG17-2 expression in HCT116 and HIEC-6 CRC cell lines. (B) SNHG17-2 expression in CRC tissues was measured by RT-qPCR and compared to adjacent normal tissues. (C) The relationship between RBM10 and SNHG17-2 expression levels in CRC tissues was examined using Pearson correlation analysis. The scatter plot, representing data from CRC tissues, demonstrates a negative correlation between the two genes. All values are presented as the mean ± SD. *P < 0.05, **P < 0.01, *** P < 0.001, **** P < 0.0001.
RBM10 inhibits the migration of CRC cells lines by suppressing SNHG17-2
To clarify whether SNHG17-2 exerts an influence on migration and EMT of colorectal cancer cell lines (HCT116 and SW480), we used siRNA to suppress the expression of SNHG17-2 (Fig. 5A). SNHG17-2 silencing was shown to effectively inhibit both EMT (Fig. 5B-E) and cell migration ability (Fig. 5F-G). To further validate whether RBM10 affects EMT and migration of CRC cells via SNHG17-2, a stable transfected cell line with sh-RBM10 was established. It was then found that while RBM10 was inhibited, knockdown of SNHG17-2 could reverse the enhanced EMT and migration induced by RBM10 downregulation.
RBM10 Regulated tumor cell migration by modulating SNHG17-2 Expression in colorectal cancer. (A) RT-qPCR was used to determine SNHG17-2 expression levels in HCT116 and SW480 cells following transfection with si-SNHG17-2a and si-SNHG17-2b, with si-NC as a control. (B-E) Western blot analysis and quantification of N-cadherin, E-cadherin, and vimentin protein levels in HCT116 and SW480 cells after si-SNHG17-2a transfection, normalized to GAPDH. (F) Images from Transwell migration assay of HCT116 and SW480 cells transfected with si-SNHG17-2a or si-NC, Scale bar = 100 μm. (G) Quantification of the Transwell assay showing the number of migrated HCT116 and SW480 cells. (H-I) RT-qPCR was used to assess SNHG17-2 expression in HCT116 and SW480 cells co-transfected with sh-RBM10 and si-SNHG17-2a, with si-NC as a control. (J-M) Western blot analysis and quantification of N-cadherin, E-cadherin, and vimentin protein levels in HCT116 and SW480 cells co-transfected with sh-RBM10 and si-SNHG17-2a or si-NC, normalized to GAPDH. (N) Images from a Transwell migration assay of HCT116 and SW480 cells co-transfected with sh-RBM10 and si-SNHG17-2a or si-NC, Scale bar = 100 μm. (O) Quantification of the Transwell assay showing the number of migrated HCT116 and SW480 cells. All values are presented as the mean ± SD. *P < 0.05, **P < 0.01, *** P < 0.001, **** P < 0.0001.
RBM10 negatively regulates CBX3 expression in colorectal cancer cells
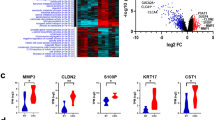
Based on previous findings that lncRNA SNHG17 promotes colorectal cancer progression through the miR-375/CBX3 axis19, we further investigated whether RBM10 regulates CBX3 expression via the lncSNHG17/miR-375 pathway. First, PCR analysis demonstrated that RBM10 negatively modulates CBX3 mRNA levels in colorectal cancer cell lines (HCT116 and SW480) (Fig. 6A and D). Consistent with this observation, western blotting revealed similar regulatory effects of RBM10 on CBX3 protein expression (Fig. 6B-C and E-F). To explore whether RBM10 influences the ceRNA function of lncSNHG17 and subsequently affects CBX3 mRNA stability, we predicted miR-375 binding sites within the CBX3 mRNA 3’UTR using bioinformatics tools and constructed corresponding mutant vectors (Fig. 6G). Dual-luciferase reporter assays demonstrated that miR-375 significantly suppressed CBX3 mRNA stability, and that mutation of the predicted binding sites abolished this suppression. Furthermore, knockdown of RBM10 expression abolished miR-375-mediated suppression of CBX3 mRNA stability (Fig. 6H).
RBM10 regulated the expression of CBX3 in colorectal cancer cells. (A) The expression of CBX3 was detected by qRT-PCR in HCT116 cells after overexpression and knockdown of RBM10. (B-C) The expression of CBX3 was detected by western blot in HCT116 cells after overexpression and knockdown of RBM10. (D) The expression of CBX3 was detected by qRT-PCR in SW480 cells after overexpression and knockdown of RBM10. (E-F) The expression of CBX3 was detected by western blot in SW480 cells after overexpression and knockdown of RBM10. (G) Schematic of miR-375 binding sites in CBX3 3’UTR. CBX3-WT contains the binding sites, while CBX3-MUT lacks them. (H) Dual-luciferase assay in HCT116 cells and SW480 cells. Relative luciferase activity in shNC and shRBM10 cells transfected with miR-375, CBX3-WT, or CBX3-MUT. All values are expressed as mean ± SD from > 3 independent experiments. *P < 0.05, **P < 0.01, *** P < 0.001, **** P < 0.0001.
Expression levels of RBM10 and SNHG17_2 and analysis of clinical pathological data
This study explored the relationship between the expression of RBM10 and SNHG17_2 and the clinical pathological features of CRC patients in detail (see Table 3). Firstly, among patients over 60 years old, RBM10 was upregulated in 19 cases and downregulated in 11 cases (p = 0.0707), while SNHG17_2 was upregulated in 15 cases and downregulated in 20 cases. Regarding gender distribution, no significant difference was observed between high and low expressions of RBM10 and SNHG17_2 in both males and females. Regarding tumor location, for patients with right colon cancer, 12 cases demonstrated high expression of RBM10 and 8 cases low expression, while 10 cases showed high expression of SNHG17_2, and another 10 cases low expression. Among rectal cancer patients, RBM10 showed high expression in 15 cases and low expression in 8 cases, whereas 8 cases demonstrated high expression of SNHG17_2 and 15 cases low expression. In terms of tumor size, patients with tumors smaller than 5 cm were associated with a significantly higher rate of RBM10 overexpression compared to low expression (25 cases vs. 5 cases, p = 0.00000093); in contrast, low SNHG17_2 expression was significantly more common than high expression (20 cases vs. 10 cases, p = 0.0201). For patients with tumors larger than or equal to 5 cm, both high expression of RBM10 and SNHG17_2 were less common than low expression. In terms of tumor invasion depth, in stage T1, 20 cases demonstrated a high expression of RBM10, which was significantly more than the 10 cases with low expression (p = 0.0242); however, high expression of SNHG17_2 was observed in 6 cases, with 5 cases of low expression. Among patients in stage T3, 18 cases presented with high expression of SNHG17_2, which was significantly higher than the 8 cases with low expression. In terms of lymph node metastasis, both high expression of RBM10 and SNHG17_2 were found in 20 cases without lymph node metastasis, while only 10 cases were detected in patients with lymph node metastasis (p-value of 0.0201 for both), showing a significant correlation with lymph node metastasis. Lastly, in terms of the AJCC stage, 20 cases showed high expression of RBM10 in stage I patients, which was significantly more than the 10 cases of low expression (p = 0.0242). Both high and low expression of SNHG17_2 included 5 cases, with a P value of 0.00894. Overall, these results revealed the potentially important roles of RBM10 and SNHG17_2 in the progression of CRC, especially in terms of tumor size, invasion depth, and lymph node metastasis.
Discussion
RBM10 is a splicing factor closely related to the occurrence and development of various malignant tumors. However, its specific mechanism in CRC remains unclear. This study aimed to explore the function of RBM10 in CRC and its potential mechanism and to investigate whether RBM10 can inhibit the invasion of CRC by regulating the alternative splicing of SNHG17.
Currently, multiple studies have revealed frequent mutations of RBM10 in CRC9,10,11. Bioinformatics studies further indicated that low RBM10 expression was associated with reduced survival rates in CRC patients12. More importantly, single-gene cell mutations of RBM10 in late-stage CRC patients were closely related to a brief progression-free survival period13. Consistently, a decrease in RBM10 expression was observed in CRC tissues, while overexpression of RBM10 significantly inhibited the invasion of CRC at the cellular level. During the invasion and metastasis of tumor cells, cells often lose epithelial characteristics and exhibit mesenchymal features, a process known as epithelial-mesenchymal transition (EMT). EMT is also accompanied by an increase in N-cadherin and Vimentin and a decrease in E-cadherin14,15,16. Our findings demonstrate that RBM10 can inhibit the expression of N-cadherin and Vimentin while promoting the expression of E-cadherin, further confirming the anticancer effect of RBM10 in CRC. Clinical data analysis of CRC patients revealed that the expression of RBM10 was significantly correlated with tumor size, depth of tumor invasion, presence of lymph node metastasis, and AJCC stage. Patients with low expression of RBM10 often exhibit poorer pathological characteristics, consistent with our experimental findings, further confirming the important role of RBM10 in the development, invasion, and metastasis of CRC.
As an RNA binding protein, RBM10 plays a crucial role in alternative splicing, identifying and regulating multiple RNA motifs. Our experimental data demonstrate that RBM10 can bind to 4040 types of RNA, including 353 types of lncRNA. Specifically, RIP experiments further confirmed the binding of RBM10 to SNHG17.
Notably, SNHG17 is considered an oncogene in CRC, and its abnormal expression is significantly associated with poor patient survival16,17,18. Liu et al.19 reported that SNHG17 promotes the proliferation, migration, and invasion of colon cancer cells and enhances their malignant characteristics through the miR-375/CBX3 axis. In addition, other studies have also shown that SNHG17 is universally upregulated in various tissues and is significantly associated with tumor size, stage, and lymph node metastasis20,21,22. Our study further reveals that RBM10 can regulate the alternative splicing of SNHG17, thereby influencing the behavior of CRC cells. Specifically, overexpression of RBM10 reduces the expression of SNHG17_2, while silencing RBM10 increases the expression of SNHG17_2.
Conclusions
The main contributions of this study are twofold. First, the findings confirm the role of RBM10 in CRC as a tumor suppressor, which is mediated by regulating the alternative splicing of SNHG17 to inhibit CRC invasion; second, the interaction between RBM10 and SNHG17 is clarified, providing a deeper understanding of the molecular mechanisms of CRC. Nevertheless, the limitations of the present study should be acknowledged. No in vivo experiments were carried out, and are required to further validate our findings; moreover, the detailed mechanism of action between RBM10 and SNHG17 remains incompletely understood. In the future, we plan to further study the role of RBM10 and SNHG17 in CRC in animal models to explore the interaction mechanism between the two.
Data availability
Data supporting this study are available in the Supplementary Information to the paper.
Change history
26 August 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41598-025-14511-0
Abbreviations
- CRC:
-
Colorectal cancer
- AS:
-
Alternative splicing
- lncRNA:
-
Long non-coding RNA
- PBS:
-
Phosphate-buffered saline
- RT-qPCR:
-
Reverse transcription-quantitative polymerase chain reaction
- RIP:
-
RNA binding protein immunoprecipitation
- ANOVA:
-
A one-way analysis of variance
- EMT:
-
Epithelial-mesenchymal transition
References
Siegel, R. L., Wagle, N. S., Cercek, A., Smith, R. A. & Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J Clin. 73 (3), 233–254. https://doi.org/10.3322/caac.21772 (2023). PMID: 36856579.
Sedlak, J. C., Yilmaz, Ö. H. & Roper, J. Metabolism and colorectal Cancer. Annu. Rev. Pathol. 18, 467–492. https://doi.org/10.1146/annurev-pathmechdis-031521-041113 (2023). PMID: 36323004; PMCID: PMC9877174.
Cervantes, A. et al. Electronic address: clinicalguidelines@esmo.org. Metastatic colorectal cancer: ESMO clinical practice guideline for diagnosis, treatment and follow-up. Ann. Oncol. 34 (1), 10–32 (2023). PMID: 36307056.
Rigaud, V. O. C. et al. RNA-Binding protein LIN28a regulates new myocyte formation in the heart through long noncoding RNA-H19. Circulation 147 (4), 324–337. https://doi.org/10.1161/CIRCULATIONAHA.122.059346 (2023). PMID: 36314132; PMCID: PMC9870945.
Jung, J. H., Lee, H., Zeng, S. X. & Lu, H. RBM10, a new regulator of p53. Cells 9 (9), 2107. https://doi.org/10.3390/cells9092107 (2020). PMID: 32947864; PMCID: PMC7563659.
Cao, Y. et al. RNA-binding motif protein 10 represses tumor progression through the Wnt/β- Catenin pathway in lung adenocarcinoma. Int. J. Biol. Sci. 18 (1), 124–139. https://doi.org/10.7150/ijbs.63598 (2022). PMID: 34975322; PMCID: PMC8692145.
Xu, T. Germline profiling and molecular characterization of early onset metastatic colorectal Cancer. Front. Oncol. 19, 10:568911. https://doi.org/10.3389/fonc.2020.568911 (2020). PMID: 33194656; PMCID: PMC7604404.
Wan, L. et al. .SRSF6-regulated alternative splicing that promotes tumour progression offers a therapy target for colorectal cancer. Gut 68 (1), 118–129. https://doi.org/10.1136/gutjnl-2017-314983 (2019). PMID: 29114070.
Wu, L. et al. Multiple omics analysis of the role of RBM10 gene instability in immune regulation and drug sensitivity in patients with lung adenocarcinoma (LUAD).Biomedicines.2023;11(7),1861.https://doi.org/10.3390/biomedicines11071861
Liu, Y. et al. Noncoding RNAs regulate alternative splicing in Cancer. J. Exp. Clin. Cancer Res. 6 (1), 11. https://doi.org/10.1186/s13046-020-01798-2 (2021). PMID: 33407694; PMCID: PMC7789004.
Wang, H. et al. The comparison of risk factors for colorectal neoplasms at different anatomical sites. Int. J. Colorectal Dis., 38(1), 26. https://doi.org/10.1007/s00384-022-04296-3
Jung, J. H., Lee, H., Zeng, S. X. & Lu, H. RBM10, a new regulator of p53. Cells 9 (9), 2107 (2020).
Guo, L. et al. Molecular Profiling Provides Clinical Insights into Targeted and Immunotherapies as well as Colorectal Cancer Prognosis. Gastroenterology.
Tang, Q. et al. TM4SF1 promotes EMT and cancer stemness via the Wnt/β-catenin/SOX2 pathway in colorectal cancer. J. Exp. Clin. Cancer Res. 39 (1), 232. https://doi.org/10.1186/s13046-020-01690-z (2020). PMID: 33153498; PMCID: PMC7643364.
Tan, Z. et al. Current Progress of EMT: A New Direction of Targeted Therapy for Colorectal Cancer with Invasion and Metastasis. Biomolecules, 12(12), 1723. https://doi.org/10.3390/biom12121723
Bian, Z. et al. SNHG17 promotes colorectal tumorigenesis and metastasis via regulating Trim23-PES1 axis and miR-339-5p-FOSL2-SNHG17 positive feedback loop. J. Exp. Clin. Cancer Res. 40 (1), 360. https://doi.org/10.1186/s13046-021-02162-8 (2021).
Editorial Office. Erratum to long non-coding RNA SNHG17 promotes gastric cancer progression by inhibiting P15 and P16. Translational cancer Res. 12 (6), 1647–1648. https://doi.org/10.21037/tcr-2023-01 (2023).
Ma, L. et al. Long noncoding RNA SNHG17: a novel molecule in human cancers. Cancer Cell Int., 22(1), 104. https://doi.org/10.1186/s12935-022-02529-7
Liu, J. et al. lncRNA-SNHG17 promotes colon adenocarcinoma progression and serves as a sponge for miR-375 to regulate CBX3 expression. Am. J. Translational Res., 12,9 5283–5295. https://doi.org/10.1186/s12935-020-01621-0
Pan, X. et al. STAT3-Induced LncRNA SNHG17 exerts oncogenic effects on ovarian Cancer through regulating CDK6. Molecular therapy. Nucleic Acids, 22, 38–49. https://doi.org/10.1016/j.omtn.2020.08.006
Li, X., Yuan, Y., Pal, M. & Jiang, X. Identification and validation of lncRNA-SNHG17 in lung adenocarcinoma: A novel prognostic and diagnostic Indicator. Front. Oncol. 12, 929655. https://doi.org/10.3389/fonc.2022.929655 (2022).
Ma, Z. et al. Long non-coding RNA SNHG17 is an unfavourable prognostic factor and promotes cell proliferation by epigenetically silencing P57 in colorectal cancer. Mol Biosyst. 13(11), 2350–2361. (2017). https://doi.org/10.1039/c7mb00280g. Erratum in: Mol Omics. 2020; 16(2):174–175.
Funding
Heilongjiang Provincial Health Commission Research Fund (20220404011047); Scientific Research Fund of Harbin Medical University Affiliated Fifth Hospital (2023-002).
Author information
Authors and Affiliations
Contributions
LX An and P Chen completed the basic experimental part of the cells; M He and HF Li complete data analysis and data organization; YS Zhou and HW Zhao proofreading and editing the article; CH Geng completed the draft writing.All authors read and approved the final manuscript and agree to publish.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This experiment is for cellular basic research ethics approval not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error in Figure 5F in which the image of the si-SNHG17-2a group of SW480 cells is duplicated with that of the sh-RBM10+si-SNHG17-2a group of SW480 cells in Figure 5N.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
An, L., Chen, P., He, M. et al. RBM10 suppresses colorectal cancer invasion by regulating LncRNA SNHG17 alternative splicing. Sci Rep 15, 12761 (2025). https://doi.org/10.1038/s41598-025-97869-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-97869-5