Abstract
Human inflammation-related CC chemokine ligand 5 (hCCL5) has significant self-assembly property under physiological conditions. The mechanism and function of hCCL5 oligomerization remain unclear. Different intermolecular interactions, such as E66-K25 or E66-R44/K45, have been reported to mediate hCCL5 oligomerization. This complexity makes structural determination difficult. Based on a K25S mutation to eliminate the E66-K25 interaction, we observed hCCL5 forming a helical-sharped filament in transmission electron microscopy (TEM). The filamentous polymerization is a dominant process when the concentration reaches ~ 100 nM and the filaments further form a higher-order assembly when concentration increases. In this large filament, a combination of X-ray solution scattering and cryo-EM analysis determined the structure; NMR further confirmed the filament packing, in which the interactions of residues R44 and K45 are critically involved. The sequence 43TRKNR47 was found to be essential for CCL5 trafficking inside the cells and for glycosaminoglycan binding outside the cells. The functional aspects of the chemokine filament are discussed.
Similar content being viewed by others
Introduction
Chemotactic cytokines (chemokines) belong to a family of small molecules (8–12 kDa) that have diverse abilities to regulate immune stimulation1. The monomeric chemokine structure comprises an N-terminal loop, a three-antiparallel β-sheet (β1–β3), and a C-terminal α-helix2. There are conserved intramolecular disulfide bonds to stabilize the structure. Under physiological conditions, many chemokines show the ability to assemble into dimers, tetramers, and even higher-order oligomers2,3. The understanding of chemokine structure is mainly based on the monomer and dimer structures, which are mostly determined under acidic conditions, while chemokine oligomer structures are difficult to determine.
Chemokine oligomerization status is related to its functions. Previous studies have shown that chemokines adopt a monomer configuration to bind G protein-coupled receptors (GPCRs). Chemokines modulate homeostatic cell trafficking, immune surveillance, and development by binding GPCRs that are widely expressed on the surfaces of leukocytes and other types of cells4,5. The mechanism has been revealed by receptor-chemokine complex structures6,7. On the other hand, binding with glycosaminoglycans (GAGs) forms a chemokine concentration gradient to guide cell migration8,9,10. A previous report showed that non-oligomerization mutants have no ability to bind GAGs and induce cell migration11,12,13. There are mutually affected and intensified processes between chemokine oligomerization and GAG binding. Specifically, the presence of GAGs modulates chemokine oligomerization.
Human chemokine CCL5 (hCCL5), expressed from activated platelets, epithelial cells, synovial fibroblasts and immune cells14, has a significant self-assembly property. hCCL5 loses the ability to recruit cells if it cannot form oligomers11. In a concentration-dependent experiment, different effects were demonstrated in cells15,16. In vivo studies, a GAG-bound CCL5 complex was observed17 and hCCL5 bound to the surface of human endothelial cells in a regular filamentous pattern18. These monomeric and oligomeric forms of hCCL5 are thought to be in a dynamic equilibrium to carry out their functions.
As mentioned, hCCL5 has the ability to form high-order oligomers at neutral pH19,20. Strategies of low pH or mutagenesis were used to overcome the aggregation problem. The NMR dimer structure has been solved in pH 3 (PDB 1HRJ). The structure shows a typical CC-type dimer formation in which the dimer interface is constituted by a short N-terminal anti-parallel β-sheet. The CCL5 dimer interface consists of residues T8-P9-C10 from two monomeric units to form the short anti-parallel β-sheet21. Furthermore, E26 and E66 have been confirmed as critical residues for self-association. For example, the E66S mutant adopts a stable dimer formation in neutral pH20,22.
In addition to the dimer structure, previous studies have reported two oligomeric structures: one is a tetramer model (PDB 2L9H), and the other is a hexamer structure (PDB 5CMD). The structure of the hCCL5 tetramer was determined at pH 4.5 by using a combination of various methods, including NMR, SAXS, and molecular simulation20. The tetramer structure consists of two CC-type dimers. The interface of the dimer-dimer interaction is composed of the second β strand (β2) and C-terminal helix. The critical intermolecular interactions are through K25 binding E26 or E66 from another CCL5 unit. The structure of the hexamer was determined by X-ray crystallography at pH 7.5 using an N-terminal truncated variant, hCCL5 (4–68)23. In the hexamer, there are three dimers parallelly aligned with each other. The structure can extend to a rod-shaped polymer. The important intermolecular interactions between two dimers were suspected that include salt bridges of E66 with R44/K45 and E26 with R47. An integrative model was proposed to explain how hCCL5 aggregates through E66 binding K25 or R44/K45, and the 12FAY14 motif acts as a secondary effect to derive precipitation24. These studies underline an importance of dimer-dimer interactions directing large oligomers.
In this study, we attempt to elucidate how hCCL5 polymerizes. We designed a mutation, K25S, to eliminate the interaction with E66 (and E26) and keep E66 interacting only with R44/K45. Through different spectral methods, we captured filamentous chemokine images. Complimentary methods were used to reveal the filament structure and its biophysical properties. This information will be important in understanding how the filamentous process regulates CCL5 functions inside and outside the cells.
Results
hCCL5-K25S mutant
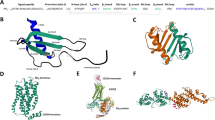
We constructed an hCCL5-K25S mutant to eliminate the intermolecular E66-K25 interaction (Fig. 1A) and preserve the interaction between E66 and R44/K45 (Fig. 1B). The structure of the K25S mutant was evaluated by NMR and compared to the structure of native hCCL5. It has been demonstrated that hCCL5 adopts a dimer structure in an acidic environment and precipitates under neutral conditions. Thus, we tested the hCCL5-K25S dimer under acidic conditions (pH 3.2). The NMR HSQC showed well-dispersive resonances and great similarity to that of hCCL5 (Fig. 1C). The hCCL5-K25S backbone resonances of N, NH, Cα, and Cβ were established. Only the resonance of the mutation site (K25 substituted by S25) showed a significant chemical shift difference (ΔδN+NH ~ 0.11 ppm), while mild differences (ΔδN+NH < 0.02 ppm) were observed in other residues (Fig. S1). Because HSQC is sensitive to protein tertiary structure, we concluded the overall structural similarity between the two proteins. Through the calculation of the chemical shift parameter, ΔδCα – ΔδCβ, which is used to indicate the tendency of α-helix by positive value and β-strand by negative value, hCCL5-K25S and hCCL5 displayed the same secondary structure profiles (Fig. 1D). Residues 20–23 and 55–65 have α-helix tendency, and residues 24–31, 36–43, and 48–54 have β-strand structure. The substitution at position 25 resulted in no significant structural change.
Wound healing assay
We performed a wound healing assay to evaluate hCCL5-K25S activity. In the assay, hCCL5 acted as an agonist, and Met-hCCL5-K25S acted as an antagonist, incorporating an N-terminal methionine that reversed the function of hCCL5 25. After scratching the cells in the well, image evidence showed reduced width of the clear zone after the hCCL5-K25S treatment (Fig. 1E). The cells actively migrated and proliferated when that treated with hCCL5-K25S, while Met-hCCL5-K25S showed an inhibitory effect. In the tests, hCCL5-treated cells were active in repairing the scratch but the effect is not stable. This may be due to the instability of protein aggregation. hCCL5-K25S showed a consistent and enhanced effect in the wound healing assay. The outcome indicates that hCCL5-K25S maintains chemotaxis activity in promoting cell migration.
The hCCL5-K25S mutant. (A) The CCL5 tetramer model determined from SAXS and NMR results. The dimer-dimer interaction is stabilized by the intermolecular salt bridges of K25-E26 or K25-E66. (B) The X-ray hexamer structure determined from the crystal of N-terminal truncated hCCL5 (4–68). The dimer-dimer interaction is the intermolecular salt bridges between E26-R47 and E66-R44/K45. (C)1H-15N HSQC of the wild type hCCL5 dimer in 25 mM sodium acetate, pH 4 (blue) and the hCCL5-K25S dimer in 25 mM sodium acetate, pH 3 (red). The arrow indicates the resonances of K25 of hCCL5 and S25 of hCCL5-K25S. (D) The secondary structure of hCCL5-K25S evaluated by the parameter ΔδCα − ΔδCβ and compared with that of hCCL5. (E) Wound healing assay of hCCL5-K25S. The images show the gap created in the cell layer before (0 h) and after protein treatment (24 h). The time-dependent profiles record the wound healing gap (%) for hCCL5, hCCL5-K25S and Met-hCCL5-K25S.
Soluble massive complex
At pH 3, the hCCL5-K25S dimer had well-dispersive resonances in the HSQC spectrum. When the pH was increased to pH 4, most peaks broadened with decreased intensities (Fig. 2A). When the pH was elevated to 5, the signals completely disappeared, indicating severe aggregation. However, the protein solution was still transparent. The soluble aggregate reversibly dissociated into a dimer when the pH was reduced to pH 3 (Fig. 2A). hCCL5-K25S transitions in different structural phases upon pH changes. A soluble massive complex was formed under neutral conditions, and the NMR signals are too broad to be detected. The NMR sample (~ 0.2 mM) was applied for SEC-MALS and DLS experiments (Fig. S2). In the SEC-MALS experiment, a broad distribution was observed during the separation of the size-exclusion column. At the retention time of the major peak, we did not obtain the corresponding refractive index (dRI) signal. This disproportion of the correlation between the light scattering pattern and dRI signals provides evidence of aggregation and structural heterogeneity. Through the detection of light scattering intensities from different angles, we estimated a molar mass higher than 2000 kDa. DLS analysis yielded a major diameter of ~ 56 nm (Fig. S2).
The analytics of hCCL5-K25S in dimer state and the filament formation. (A) 1H, 15N-HSQC spectra in different pH conditions for defining the reversible association and dissociation of the hCCL5-K25S oligomer. (B) AUC analysis of hCCL5-K25S. The distribution of molecular mass in response to different protein concentrations (50, 25, 10, 5, 2.5 and 1 µM). The fluorescence anisotropy of (C) hCCL5-E66S and (D) hCCL5-K25S in the concentration range of 1 nM to 20 µM (λEX = 280 nm, λEM = 340 nm). Measurement of each concentration was repeated at least 3 times. (E) The fitting curves of hCCL5-K25S and hCCL5-E66S fluorescence anisotropy.
Analytical ultracentrifugation
Analytical ultracentrifugation (AUC) experiments were used to characterize the size of aggregates at different concentrations. Different hCCL5-K25S samples with concentrations from 1 to 50 µM were prepared at pH 6, while concentrations lower than 0.5 µM gave an undistinguished signal from noise. The data showed that there was a 1354 kDa polymer (almost 200 mer) when the concentration was 50 µM (Fig. 2B). The detected polymer sizes were gradually reduced when we used lower protein concentrations. At a concentration of 1 µM, a 108 kDa polymer was predominantly detected in solution. In the AUC-allowed detection range, we observed hCCL5-K25S high-molecular-weight aggregates. The association was gradually enhanced when the protein concentration was increased.
Fluorescence anisotropy experiment
Fluorescence anisotropy is a measurement to report the molecule size by detecting the tumbling of the fluorophore. The increased molecular size reduced the tumbling, resulting in an increased anisotropy. Since hCCL5 contains a single Trp residue, we used Trp as the probe and detected the intrinsic fluorescence anisotropy. We performed a fluorescence anisotropy experiment for hCCL5-E66S, which is known to adopt a predominant dimer structure at a concentration of µM. We investigated the monomer-dimer equilibrium by evaluating samples with concentrations from 20 µM to 1 nM (Fig. 2C). In the analysis, 20 µM hCCL5-E66S adopted a dimer structure with an anisotropy value of 0.05. When the concentration was reduced, the values gradually approached 0, indicating a monomer. We used a sigmoid curve to fit the experimental profile. The critical aggregation concentration (50% hCCL5 dimer dissociated to monomer) is 85 ± 9 nM (Fig. 2E). Noticeably, we observed a slightly higher anisotropy value (~ 0.07) when the concentration was between 0.3 and 1 µM. This may reflect a structural intermediate during the association/dissociation process. The presence of structural intermediate may also lead to larger errors for the data points at which the hCCL5 monomer begins to aggregate.
We tested hCCL5-K25S in which 20 µM protein had a higher anisotropy value of 0.1 (Fig. 2D). The result indicated a larger complex. The value was maintained until the concentration was lower than 1 µM. The values gradually approached 0 as the concentration was further reduced. We fit the experimental profile. The identified critical aggregation concentration was 140 ± 9 nM (Fig. 2E). We did not clearly observe the presence of dimers. Thus, dimeric CCL5 is considered to be a transient population during the oligomerization process.
Transmission electron microscopy
We prepared samples with concentrations from 0.5 to 10 µM and tested them by transmission electron microscopy (TEM). The concentrations of 0.5–2 µM gave clear and adequate images (Fig. 3A). Long and helical-sharped filament structures were observed. The length of a single filament can reach several µm. The average diameter of the individual filaments is ~ 50Å. In some observations, we noticed multiple filaments parallelly aligning with each other. As mentioned, hCCL5 (4–68) has been solved as a hexamer structure (PDB 5CMD) where three CCL5 dimers are stacked in the structure (Fig. 1B). We analyzed the hexamer, which matches the diameter in the filament image. The hexamer structural coordinate can be repeatedly extended to a filament structure.
Filament structure
To clarify the filament structure, a filament model containing 20 dimers built from the X-ray hexamer were compared to the small/wide-angle X-ray scattering (SWAXS) data, collected from the CCL5-K25S filament in solution (Fig. 3B). The calculated SWAXS profile using the structural model adequately overlaps the SWAXS data in a wide q range up to q = 0.8 Å−1 for a spatial resolution of 8 Å, rendering a strong support to the structural model. A small χ2 value of 2.92 between the experimental and predicted profiles was obtained. The model can fit the SWAXS data well in the lower-q region (~ 0.1 Å−1). Cryo-EM was used to further evaluate the filament structure. Cryo-EM images of CCL5 filaments were collected, clustered and overlapped (Fig. 3C). We found two parallel arranged filaments in the 2D classification groups. The phenomena might indicate the interaction between filaments. When filaments are formed and enriched in vitro, the interaction could constitute a high-order assembly. However, only the single filament was well aligned, suggesting that the high-order filaments were not well aligned orderly. This caused difficulty in obtaining high-resolution maps of single or doubly aligned CCL5 filaments. Helical reconstruction was used to generate the EM map of a single filament. In addition, the proximity of many filaments hindered the complete utilization of the images. Finally, a density map with a low resolution of ~ 7.0 Å was obtained. The X-ray hexamer-filament model can dock on the cryo-EM density map with a satisfactory fitting score of 0.85 (Fig. 3C), revealing a helicity comprised of 5.7 layers of dimers in one helical turn (Fig. S3).
The images of hCCL5-K25S filament. (A) Negative-stain electron micrographs of hCCL5-K25S under different conditions. (B) The filament model (20-mer) is extended from the X-ray hexamer structure (PDB 5CMD). The filament model fits the SWAXS experimental results with an χ2 value of 2.92. (C) The classified single filament images are used for the following calculation and the final coulombic potential map is with a resolution of 7.04 Å. The X-ray hexamer filament model fits well in the cryo-EM map by Chimera. The model-map correlation was validated by Phenix with a fitting score of 0.85.
CCL5 tetramer preparation
The resolution of the cryo-EM map is too low to provide detailed structural information for dimer-dimer interface. In the X-ray hexamer structure, it also lacked essential information to explain the interaction because the electron densities of some critical residues in the interface are unclear. Particularly, the positively charged residues R44 and K45 do not have clear side chain densities. The result suggested their flexible orientations. To evaluate the roles of the basic residues, we designed a R44A/K45A mutant. Meanwhile, we considered to use the K25S/E26A/E66S mutant, which showed no aggregation property, to validate dimer-dimer interactions. To constitute the CCL5 filament, we suspect that one protomer contributes basic residues (such as R44/K45) on one side and one protomer contributes acidic residues (such as E26/E66) on the other side. Through electrostatic interactions, the dimer propagates to continuously stack with each other to form the filament. We suspected that R44 and K45 are involved in the polymerization. HSQC spectra showed that hCCL5-R44A/K45A aggregated with a reduced tendency (Fig. S4). After the titration of hCCL5-K25S/E26A/E66S, we were able to detect the hCCL5-R44A/K45A NMR signal (Fig. S4), indicating an oligomer formation with a reduced molecular size. A strategy was therefore customized (Fig. 4A). We mixed the two mutants. Each of the mutant retained only one set of charged residues and lost another set of charged residues. The two mutants adopted the dimer configuration under acidic conditions. The mixing of the two mutants created heterodimers through exchanging their subunits. When the pH was increased, the heterodimer interacted with another heterodimer with the preserved set of the charged residues and formed a tetrameric formation. The aggregation process cannot propagate because of the deficiency of the charge-charge interaction at the mutation side. Here, we prepared a sample of 1:1 molar ratio. The major fraction of the mixture was subjected to SEC-MALS analysis. The results revealed a molecular weight of 32.3–33.9 kDa, indicating a predominant tetramer configuration (Fig. 4B).
The CCL5 tetramer. (A) Scheme of how to prepare the CCL5 tetramer by mixing the R44A/K45A mutant and K25S/E26A/E66S mutant. Residues E26/E66 and R44/K45 are involved in dimer-dimer interactions. Mutating one site and preserving the other creates aggregation-deficient mutants. The mixture of R44A/K45A and K25S/E26A/E66S mutants derives a heterodimer, which has no ability to propagate into an oligomer. Tetramer is expected to be predominantly present in a neutral solution. (B) SEC-MALS experiment of the tetramer complex prepared at a molar ratio of 1:1. (C) 1H-15N HSQCs of the 15N-hCCL5-R44A/K45A dimer (blue) and 15N-hCCL5-R44A/K45A + hCCL5-K25S/E26A/E66S tetramer (red). The spectra are acquired at pH = 3.2 and pH 5. The chemical shift difference (∆δN+NH) derived from the HSQCs is depicted (below). (D) The same evaluation was performed for the 15N-hCCL5-K25S/E26A/E66S dimer (blue) and 15N-hCCL5-K25S/E26A/E66S + hCCL5-R44A/K45A tetramer (red). In the comparison of chemical shift differences, the missing residues (K45 ~ Q48) are shadowed. The perturbed residues are mapped onto the protomers of (E) hCCL5-R44A/K45A and (F) hCCL5-K25S/E26A/E66S in the tetramer model.
Dimer-dimer interaction
We studied the hCCL5 tetramer by NMR. We selectively labeled one mutant with isotopes and left the other mutant unlabeled. Mixing of 2H, 15N, 13C-hCCL5-R44A/K45A and hCCL5-K25S/E26A/E66S allowed us to establish the backbone assignment of hCCL5-R44A/K45A (Fig. 4C, in red). Instead, mixing of the2H, 15N, 13C-hCCL5-K25S/E26A/E66S and hCCL5-R44A/K45A samples allowed us to study hCCL5-K25S/E26A/E66S (Fig. 4D, in red). Compared to the spectra of the respective hCCL5 dimers, the HSQCs contained different resonances (Fig. 4C and D, in blue). The chemical shift difference, therefore, reported the residues perturbed by dimer-dimer interactions. In the case of 2H, 15N, 13C-hCCL5-R44A/K45A, residues S5, T8, Y27, A38, E60, N63, S64, L65, and E66 showed significant differences. In another case of 2H, 15N, 13C-hCCL5-K25S/E26A/E66S, the residues of C11-I15, F41, R44, V49 and C50 were dramatically perturbed, while the resonances of K45 ~ Q48 disappeared. We checked the secondary structural tendency. The parameter ΔδCα – ΔδCβ indicated the presence of an extended C-terminal α-helix in the tetramer (Fig. S5). The C-terminal residues, S64 to S68, adopted a more defined helical structure when a dimer-dimer interaction occurred, implying that the C-terminal helix is stabilized in the tetramer formation. A tetramer unit trimmed from the filament showed that the perturbed residues were well distributed in the dimer-dimer interface (Fig. 4E and F). The NMR result confirmed the filament packing interface in the solution state, in which R44 and K45 dominate the polymerization process.
Stability of CCL5 filament
We tested the structural stability of hCCL5 dimer and hCCL5-K25S filament by circular dichroism (CD) spectra that two dimer formations were respectively prepared by hCCL5-K25S at pH 3.0 and hCCL5-E66S at pH 6.0 (Fig. 5). The CD spectra at different temperatures show that the CCL5 filament adopts a very different secondary structural property than the CCL5 dimer. The CCL5 assembly changed the molar ellipticity at ~ 202 nm. The filament structure significantly increases the chirality. The individual thermal melting temperatures (Tm) were measured at the wavelength of 230 nm of the dimer and 202 nm of the filament. The E66S dimer showed a less cooperative denaturation process (Fig. 5A). There was no sharp transition, indicating a multitude of small energetic barriers between the folded and unfolded states. We therefore characterized the K25S dimer and the experiment reported a sigmoidal curve, showing a Tm of 48 oC (Fig. 5A). For the hCCL5-K25S filament, the high Tm value of 72 oC was observed (Fig. 5B). The comparison reported a complicated unfolding mechanism in CCL5 dimer and filament. The high Tm of CCL5 filament indicated its great stability in solution.
Circular dichroism spectra of hCCL5. The CD spectra of (A) dimer and (B) filament at different temperatures. The corresponding thermal melting temperatures (Tm) are determined at 230 nm and 202 nm, respectively. The protein concentration is 15 µM dissolved in 1 mM citrate phosphate, that are adjusted to pH 3.0 for K25S dimer and pH 6.0 for E66S dimer and K25S filament.
Discussion
Monomeric hCCL5 activates GPCRs, whereas oligomer hCCL5 interacts with GAGs to trigger a GPCR-independent pathway. hCCL5 showed regular binding to the cell surface of human endothelial cells, and oligomerization-deficient mutations of hCCL5, such as E66A and 44AANA47, demonstrated dramatically decreased binding18. Thus, self-association of hCCL5 from monomer to filament is functionally present in the cells. In this study, we studied the hCCL5-K25S mutant and successfully captured images of hCCL5 filament. Under neutral conditions, a low concentration of hCCL5 tends to form a monomer and starts to assemble into a dimer when the concentration is increased. Acting as an intermediate state, the dimers readily self-assemble into filaments.
Based on the experience with hCCL5-K25S, we tested the wild type hCCL5 sample. A high precipitate tendency was observed in hCCL5. We observed the hCCL5 filament occasionally, but not always observed in every examination. However, all the observed filament images of hCCL5 demonstrated the identical structure (Fig. S6). In addition, the determined hCCL5 hexamer structure (PDB 5CMD) is based on a shorter CCL5 construct, hCCL5 (4–68), where the N-terminal three amino acids were truncated. The hCCL5 (4–68) protein is a native variant that is proteolyzed by cathepsin G26. We tested a similar CCL5 construct, Met-hCCL5 (5–68). The TEM images showed that Met-hCCL5 (5–68) also formed filament (Fig. S6). The filament structure forms universally in the native protein and the N-terminal truncated mutant. The removal of K25 causes hCCL5 to form soluble aggregates in a neutral solution. Although the role of K25 is still unclear, this charged residue may be involved in other interaction at higher concentrations24. The closest chemokine relatives, CCL3 and CCL4, do not bring Lys at the same position (Fig. S7). The structural role of K25 might be unique among the CCL5 chemokine, as compared to the filament structures of CCL3 and CCL4.
We combined NMR, SWAXS and cryo-EM to model the CCL5 filament structure. The hCCL5 filament could be considered as the extended conformation of the X-ray hexamer structure. By repeating the hexamer coordinate, this model is consistently supported by the SWAXS result and the cryo-EM map. As shown, the mutations of these R44/K45 residues prevented the CCL5 tetramer further forming a longer filament. Here, we demonstrated the importance of R44/K45 in the dimer-dimer interaction. The interaction propagated to form the filament conformation. We noticed the structural flexibility in the dimer-dimer interface. The result is based on the undetectable resonances of K45 ~ Q48 in the tetramer HSQC spectrum and the previous observation of the absent electron densities of R44 and K45 in the crystal structure23.
On the basis of the above results, we discuss how R44 and K45 involve in the functions inside and outside the cells. Inside the cell, CCL5 is shuttled from the ER to the post-Golgi stage, and accumulated in the motile vesicles27. Once the extracellular stimulation occurs, the CCL5 secretion from the vesicles occurs immediately. The 43TRKN46 motif has been proposed to be indispensable for its secretion. The 43TRKN46-mutated CCL5 changed the ER-to-post-Golgi trafficking pathway. The Ala-substitution mutant had a higher propensity to localize in the ER and had a diffuse and reticulated distribution in the cell supernatant with no significant vesicular organization. Therefore, there was a very substantial inhibition of secretion for the mutated CCL5. The motif carrying R44/K45 residues is proved to be important for constituting the filament formation in this study. We expect that the 43TRKN46-mutated CCL5 has a reduced ability to form filament. It is reasonable to speculate that the filament process could support the process of CCL5 accumulation in the Golgi and further support the transport to the secretory vesicles. The filamentous CCL5 could occur at ~ 100 nM concentration and high-order assembly of the filament occurs at higher concentration. That helps CCL5 to aggregate and reach a very high local concentration in a small volume. In addition, filamentous CCL5 retains better structural stability. These features all provide advantages for “depositing” CCL5 into the secretory vesicles for storage. Additionally, the association between CCL5 molecules is very specific. It provides molecular segregation and helps to separate CCL5 from other chemokines.
Outside of cell, it has been reported that chemokine oligomers have a high affinity to interact with heparin and that heparin also promotes chemokine oligomerization11. It is still unknown how heparin binding changes the filamentous structure on the cell surface. The hCCL5 hexamer structure in complex with octasaccharide was reported in which a heparin fragment interacted with the positively charged cluster 55KKWVR59 23. However, most studies recognized the heparin-binding BBXB motif 44RKNR47, which significantly contributes to GAG binding8. The R44 and K45 residues, responsible for constituting the filament formation, are buried in the convex side of the X-ray hexamer structure. The presence of heparin competes with the interaction and modulates the CCL5 association/dissociation. Considering that hCCL5 starts to oligomerize at a concentration of ~ 100 nM and the affinity between CCL5 and heparin octasaccharide has been determined to be a comparable value of ~ 100 nM28, we suspect that the heparin fragment longer than octasaccharide shows the ability to competitively dissociate the CCL5 oligomer. Previous studies also demonstrated that sulfated tyrosine on the receptor CCR5 N-terminus increased the affinity to CCL5 29,30. The solution structure of monomeric hCCL5 in complex with a sulfated CCR5 N-terminal peptide has been determined31. The sulfated Y10 interacts with hCCL5 L19, H23, K45, and V49 31. Since residue K45 is involved in CCL5 oligomerization, the sulfated CCR5 N-terminal portion may also play a role in modulating CCL5 polymerization.
In this study, we have characterized how hCCL5-K25S forms the filamentous polymer. The dimer-dimer interfacial structure of E66-R44/K45 and the filaments of hCCL5 are elucidated through the combined analysis of TEM, SWAXS, cryo-EM, and NMR. The important filamentous chemokine polymerization features are demonstrated through CCL5, which sheds lights on other CC-type chemokines in their filament formations. We discussed the possible roles of CCL5 filament inside and outside of the cells, which add the understanding of the structure-function correlation of chemokine filaments.
Methods
Protein expression and purification
We constructed hCCL5 variants, including hCCL5-K25S, hCCL5-E66S, hCCL5-R44A/K45A, hCCL5-K25S/E26A/E66S, and N-terminal four-residue truncation, hCCL5 (5–68). Protein expression and purification followed the established procedure32. We refolded hCCL5 proteins, and the folded hCCL5s were further purified by reversed-phase HPLC32. For NMR experiments, the samples were prepared with isotope labeling, and 15NH4Cl (1 g/L) and 13C-labeled D-glucose (2 g/L) were added into M9 minimal medium as the sole nitrogen and carbon sources.
Wound healing assay
The human osteosarcoma U2OS cell line (ATCC) was cultured, and 12-well plates were used to seed the cells. We seeded 2 × 105 cells in each well with fetal bovine serum (FBS) in Dulbecco′s modified Eagle′s medium (DMEM) and incubated the plate in a 37 °C, 5% CO2 incubator for 24 h. After a period of 24 h, a single layer of cells had grown to cover the surface of each well. A gap was created by suction using a P10 tip, and each well was cleaned twice with PBS buffer. CCL5 was added at the desired concentrations, while hCCL5 and Met-hCCL5-K25S were used as the positive and negative controls, respectively33. The protein concentration was 20 ng/ml. All experiments were replicated three times. Digital image processing tools and software ImageJ or Fiji are used to measure wound healing gap. Comparative analyses of cell migration were carried out with the use of the GraphPad Prism v9 program and the unpaired t test for statistical purposes (Mann–Whitney). A significant p value was considered * <0.05, ** <0.01 or *** <0.001.
Transmission electron microscope (TEM) imaging
The protein samples were prepared in PBS buffer for the concentration range of 0.5 ~ 2 µM at pH 5.0 or 6.0 and were applied to a 20 µl drop on freshly glow-discharged grids (Formvar/Carbon film on Copper 300-mesh) for 2 min. Excess fluid was carefully wiped off by filter paper and 10 µl 1% (w/v) uranyl acetate solution was placed on the grid for 1 min to stain the sample. The excess stain was wiped off with filter paper again and the grids were dried for 24 h. Electron micrographs were taken with a Hitachi HT7700 Biotransmission electron microscope operating at 120 kV. We analyzed TEM images by using ImageJ software.
The analytical ultracentrifuge (AUC)
The proteins were prepared at concentrations of 1, 2.5, 5, 10, 25, and 50 µM in PBS buffer, pH 6. The experiments were performed on a Beckman Optima XL-A ultracentrifuge at 25,000 rpm at 20 ℃ and detected by absorption at 280 nm. Every sample was scanned 50 times with a 4-minute delay.
Fluorescence anisotropy
The proteins were prepared at concentrations of 20–0.001 µM in PBS buffer, pH 6. We detected the emission fluorescence at a wavelength of 340 nm by exciting Trp with polarized light at a wavelength of 280 nm at 25 ℃. We measured the intensity of the fluorescence emission passing the parallel (Iǁ) and perpendicular (I┴) polarization filters. We calculated the fluorescence anisotropy (FA) by measuring the intensities of Iǁ and I┴ through the relation FA = (Iǁ − I┴)/(Iǁ + 2I┴).
CCL5 tetramer preparation
We dissolved lyophilized hCCL5 R44A/K45A and hCCL5 K25S/E26A/E66S in 25 mM sodium acetate buffer, pH 3.0, and mixed the two proteins at a 1:1 molar ratio. The protein solution was adjusted to pH 5.0 to allow protein oligomerization. After removing the precipitate by high-speed centrifugation (16000 × g for 20 min), we collected the supernatant and purified it by size-exclusion chromatography (Superdex-75 10/300). The fraction corresponding to the CCL5 tetramer was collected and concentrated for experimental usage.
NMR backbone assignment of hCCL5s
The NMR backbone assignments of the hCCL5 variants were achieved by the standard procedure34. For the hCCL5-K25S dimer, an NMR sample of 0.5 mM protein was prepared in a solution of 25 mM sodium acetate, pH 3.0, and 10% D2O (v/v). All NMR experiments were acquired on a Bruker AVANCE II 600 MHz spectrometer at 298 K, and the chemical shift was referenced to sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) as 0 ppm. The acquisition spectra were processed by NMRPipe35 and analyzed by Sparky36. To identify the protein secondary structure of hCCL5-K25S, the parameter (∆δCα-∆δCβ) was calculated, where ∆δCα and ∆δCβ are the respective differences between the observed Cα or Cβ chemical shifts and the corresponding values observed in a random coil structure. The positive and negative values of (∆δCα-∆δCβ) indicate α-helix and β-strand propensity, respectively.
For the hCCL5-R44A/K45A and hCCL5-K25S/E26A/E66S tetramer, we selectively isotopically labeled one protomer with 15N,13C and 2H and left the other unlabeled. Deuterium labeling was required to enhance the quality of 3D NMR experiments. We compared backbone N and NH chemical shift differences by the equation: (∆δN+NH) = [((∆δNH)2+ (∆δN/5)2)/2]1/2, where ∆δNH is the chemical shift difference of NH and ∆δN is the chemical shift difference of N.
Size exclusion chromatography-coupled multiangle light scattering (SEC-MALS)
A size-exclusion column of Superdex 75 10/300 GL was used for chromatographic separation on the AKTA purifier FPLC system. The column was equilibrated with PBS buffer, pH 6, at a flow rate of 0.5 ml/min. The protein sample was prepared at a concentration of 2 mg/ml in the same buffer. The multiangle light scattering (MALS) detector was a miniDAWN TREOS with QELS (Wyatt), and the differential refractive index detector was an Optilab T-rEX (Wyatt).
Small- and wide-angle X-ray scattering (SWAXS)
SWAXS data were collected at the 13 A BioSWAXS beamline of the Taiwan Photon Source (TPS) of the National Synchrotron Radiation Research Center (NSRRC). The beamline is equipped with two Dectris Eiger X 9 M and 1 M in-vacuum detectors for synchronized SWAXS measurements to cover a wide range of scattering vectors q = 4πλ− 1sinθ defined by the X-ray wavelength λ (0.886 or 0.827 Å with 14 or 15 keV used) and scattering angle 2θ37,38. Due to strong interactions of the sample solution with either polymer- or silica-based size-exclusion columns, all samples were measured in column-bypass mode, with a frame exposure time of 2 s over ca. 300 s sample elution with buffer. The buffer was PBS buffer at pH 6.0 for the CCL5 filament. The CCL5 filament solution showed little sign of radiation damage, and SWAXS data were simultaneously collected, combined, and analyzed. The SWAXS data were processed using the TPS 13 A SWAXS Data Reduction Kit (DRK) Ver. 3.6 and analyzed using the ATSAS package, including PRIME, DAMMIN, SASREF, SREFLEX, and CRYSOL39. The SWAXS data were evaluated with Guinier analysis and DAMMIN to build a bead model for a low-resolution shape. All χ2 values between the models and SWAXS data were calculated by CRYSOL.
Cryogenic electronic microscopy (cryo-EM)
To prepare cryo-EM specimens, 5–50 µM CCL5 filaments were used for grid preparation. The filament was applied to Quantifold holey carbon grids (Cu, 300 mesh, R2.0/2.0 μm) by using a Vitrobot Mark IV system (ThermoFisher) with a blotting time of 3.0–4.0 s at 100% humidity and 4 °C. Grids were rapidly frozen in liquid ethane and stored in a liquid nitrogen reservoir. Cryo-EM data for the CCL5 filaments were acquired on a Titan Krios microscope equipped with a Gatan K3 direct detector. In total, 5146 50-frame micrographs were collected at a physical pixel size of 0.415 Å. The total electron dose and desired defocus ranges were 54.15 e-/Å2 and − 0.8 to − 1.6 μm, respectively. Cryo-EM data were processed by using cryoSPARC 3.239. Movies were aligned using patch-motion correction followed by CTF estimation using patchCTF. We manually picked up ~ 50 CCL5 filaments to provide good templates for automatic filament picking using a filament tracer. A total of 641,756 filament segments (400-pixel box size) were extracted for 2D classifications, and 178,680 segments were used to perform helical reconstruction using the “Helical Refinement” function in cryoSPARC 3.2. The initial model was calculated without applied helical twist, helical rise and symmetry. The helical rise of 45–50 Å and helical twist of 90° based on the crossover in 2D classification was then used to obtain a map at 7.04 Å.
Circular dichroism (CD)
The CD data was collected at 24 A of the Taiwan Light Source (TLS) of the National Synchrotron Radiation Research Center (NSRRC). The instrument model used in the experiment was the Jasco J-815, equipped with a 1 cm pathlength quartz cuvette. The sample was prepared as a 15 µM protein solution in a 1 mM citrate phosphate buffer, with a total volume of 0.3 mL. The wavelength scan range spanned from 190 nm to 260 nm, with measurements taken at 1 nm intervals.
Data availability
All data generated or analyzed during this study is provided within the manuscript and supplementary information files.
References
Rollins, B. J. & Chemokines Blood 90, 909–928, doi:https://doi.org/10.1182/blood.V90.3.909 (1997).
Mayo, K. H. & Chen, M. J. Human platelet factor 4 monomer-dimer-tetramer equilibria investigated by proton NMR spectroscopy. Biochemistry 28, 9469–9478. https://doi.org/10.1021/bi00240a009 (1989).
Jansma, A. L., Kirkpatrick, J. P., Hsu, A. R., Handel, T. M. & Nietlispach, D. NMR analysis of the structure, dynamics, and unique oligomerization properties of the chemokine CCL27. J. Biol. Chem. 285, 14424–14437. https://doi.org/10.1074/jbc.M109.091108 (2010).
Zlotnik, A. & Yoshie, O. Chemokines: a new classification system and their role in immunity. Immunity 12, 121–127. https://doi.org/10.1016/S1074-7613(00)80165-X (2000).
Cardona, S. M., Garcia, J. A. & Cardona, A. E. The fine balance of chemokines during disease: trafficking, inflammation, and homeostasis. Chemokines: Methods Protocols. 1–16. https://doi.org/10.1007/978-1-62703-426-5_1 (2013).
Zheng, Y. et al. Structure of CC chemokine receptor 5 with a potent chemokine antagonist reveals mechanisms of chemokine recognition and molecular mimicry by HIV. Immunity 46, 1005–1017. https://doi.org/10.1016/j.immuni.2017.05.002 (2017).
Proudfoot, A. E. Chemokine receptors: multifaceted therapeutic targets. Nat. Rev. Immunol. 2, 106–115. https://doi.org/10.1038/nri722 (2002).
Proudfoot, A. E., Johnson, Z., Bonvin, P. & Handel, T. M. Glycosaminoglycan interactions with chemokines add complexity to a complex system. Pharmaceuticals 10, 70. https://doi.org/10.3390/ph10030070 (2017).
Middleton, J., Patterson, A. M., Gardner, L., Schmutz, C. & Ashton, B. A. Leukocyte extravasation: chemokine transport and presentation by the endothelium. Blood J. Am. Soc. Hematol. 100, 3853–3860. https://doi.org/10.1182/blood.V100.12.3853 (2002).
Bishop, J. R., Schuksz, M. & Esko, J. D. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446, 1030–1037. https://doi.org/10.1038/nature05817 (2007).
Proudfoot, A. E. et al. Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proceedings of the National Academy of Sciences 100, 1885–1890. https://doi.org/10.1073/pnas.0334864100 (2003).
Lau, E. K. et al. Identification of the glycosaminoglycan binding site of the CC chemokine, MCP-1: implications for structure and function in vivo. J. Biol. Chem. 279, 22294–22305. https://doi.org/10.1074/jbc.M311224200 (2004).
Proudfoot, A. E. et al. The BBXB motif of RANTES is the principal site for heparin binding and controls receptor selectivity. J. Biol. Chem. 276, 10620–10626. https://doi.org/10.1074/jbc.M010867200 (2001).
Aldinucci, D. & Colombatti, A. The inflammatory chemokine CCL5 and cancer progression. Mediat. Inflamm. 2014 https://doi.org/10.1155/2014/292376 (2014).
Roscic-Mrkic, B. et al. RANTES (CCL5) uses the proteoglycan CD44 as an auxiliary receptor to mediate cellular activation signals and HIV-1 enhancement. Blood 102, 1169–1177. https://doi.org/10.1182/blood-2003-02-0488 (2003).
Czaplewski, L. G. et al. Identification of amino acid residues critical for aggregation of human CC chemokines macrophage inflammatory protein (MIP)-1α, MIP-1β, and RANTES: characterization of active disaggregated chemokine variants. J. Biol. Chem. 274, 16077–16084. https://doi.org/10.1074/jbc.274.23.16077 (1999).
Wagner, L. et al. β-chemokines are released from HIV-1-specific cytolytic T-cell granules complexed to proteoglycans. Nature 391, 908–911. https://doi.org/10.1038/36129 (1998).
Øynebråten, I. et al. Oligomerized, filamentous surface presentation of RANTES/CCL5 on vascular endothelial cells. Sci. Rep. 5, 9261. https://doi.org/10.1038/srep09261 (2015).
Skelton, N. J., Aspiras, F., Ogez, J. & Schall, T. J. Proton NMR assignments and solution conformation of RANTES, a chemokine of the CC type. Biochemistry 34, 5329–5342. https://doi.org/10.1021/bi00016a004 (1995).
Wang, X., Watson, C., Sharp, J. S., Handel, T. M. & Prestegard, J. H. Oligomeric structure of the chemokine CCL5/RANTES from NMR, MS, and SAXS data. Structure 19, 1138–1148. https://doi.org/10.1016/j.str.2011.06.001 (2011).
Chung, C., Cooke, R. M., Proudfoot, A. E. & Wells, T. N. The three-dimensional solution structure of RANTES. Biochemistry 34, 9307–9314. https://doi.org/10.1021/bi00029a005 (1995).
Denisov, S. S. et al. Structural characterization of anti-CCL5 activity of the tick salivary protein evasin-4. J. Biol. Chem. 295, 14367–14378. https://doi.org/10.1074/jbc.RA120.013891 (2020).
Liang, W. G. et al. Structural basis for oligomerization and glycosaminoglycan binding of CCL5 and CCL3. Proceedings of the National Academy of Sciences 113, 5000–5005 (2016).
Chen, Y. C. et al. Integrative model to coordinate the oligomerization and aggregation mechanisms of CCL5. J. Mol. Biol. 432, 1143–1157. https://doi.org/10.1016/j.jmb.2019.12.049 (2020).
Wang, S. W. et al. CCL5 and CCR5 interaction promotes cell motility in human osteosarcoma. PloS One. 7, e35101. https://doi.org/10.1371/journal.pone.0035101 (2012).
Lim, J. K., Lu, W., Hartley, O. & DeVico, A. L. N-terminal proteolytic processing by cathepsin G converts RANTES/CCL5 and related analogs into a truncated 4‐68 variant. J. Leukoc. Biol. 80, 1395–1404. https://doi.org/10.1189/jlb.0406290 (2006).
Soria, G. et al. Mechanisms regulating the secretion of the promalignancy chemokine CCL5 by breast tumor cells: CCL5’s 40s loop and intracellular glycosaminoglycans. Neoplasia 14, 1–IN3 (2012).
Segerer, S. et al. The basic residue cluster 55KKWVR59 in CCL5 is required for in vivo biologic function. Mol. Immunol. 46, 2533–2538. https://doi.org/10.1016/j.molimm.2009.05.015 (2009).
Farzan, M. et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96, 667–676. https://doi.org/10.1016/S0092-8674(00)80577-2 (1999).
Ludeman, J. P. & Stone, M. J. The structural role of receptor tyrosine sulfation in chemokine recognition. Br. J. Pharmacol. 171, 1167–1179. https://doi.org/10.1111/bph.12455 (2014).
Abayev, M. et al. The solution structure of monomeric CCL 5 in complex with a doubly sulfated N-terminal segment of CCR 5. FEBS J. 285, 1988–2003. https://doi.org/10.1111/febs.14460 (2018).
Chen, Y. C., Li, K. M., Zarivach, R., Sun, Y. J. & Sue, S. C. Human CCL5 trimer: expression, purification and initial crystallographic studies. Acta Crystallogr. Sect. F: Struct. Biology Commun. 74, 82–85. https://doi.org/10.1107/S2053230X17018544 (2018).
Liang, C. C., Park, A. Y. & Guan, J. L. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2, 329–333. https://doi.org/10.1038/nprot.2007.30 (2007).
Leopold, M., Urbauer, J. L. & Wand, A. J. Resonance assignment strategies for the analysis of NMR spectra of proteins. Mol. Biotechnol. 2, 61–93. https://doi.org/10.1007/BF02789290 (1994).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6, 277–293. https://doi.org/10.1007/BF00197809 (1995).
Lee, W., Tonelli, M. & Markley, J. L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327. https://doi.org/10.1093/bioinformatics/btu830 (2015).
Shih, O. et al. Performance of the new biological small-and wide-angle X-ray scattering beamline 13A at the Taiwan photon source. J. Appl. Crystallogr. 55 https://doi.org/10.1107/S1600576722001923 (2022).
Liu, D. G. et al. Optical design and performance of the biological small-angle X-ray scattering beamline at the Taiwan photon source. J. Synchrotron Radiat. 28, 1954–1965. https://doi.org/10.1107/S1600577521009565 (2021).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. CryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 14, 290–296. https://doi.org/10.1038/nmeth.4169 (2017).
Acknowledgements
The authors are grateful to the National Synchrotron Radiation Research Center, the NMR facility at National Tsing Hua University, the Cryo-EM Facility at Academia Sinica and the Biophysical Instrumentation Laboratory at the Institute of Biological Chemistry, Academia Sinica.
Funding
This work was supported by the Ministry of Science and Technology, Taiwan (grants 110-2113-M-007-025, 111-2113-M-007-001, 111-2327-B-007-002 and 111-2113-M-001-026-MY2), the Academia Core Facility and Innovative Instrument Project (AS-CFII-111-210) and the Taiwan Protein Project (AS-KPQ-109-TPP2).
Author information
Authors and Affiliations
Contributions
Y.T.Y. contributed methodology, formal analysis, conceptualization, investigation, data curation and writing. T.C.G. contributed conceptualization, investigation, data curation and writing. C.Y.W contributed data curation. Y.Q.Y. contributed methodology and data curation. C.T.W. contributed data curation. S.Y.H. contributed data curation. Z.W.W. contributed data curation. M.R.H. contributed data curation. S.T.H. contributed resource. Y.C.C. contributed conceptualization and data curation. C.R.C. contributed data curation and Resource. K.P.W contributed formal analysis, investigation, data curation and writing. U.S.J. contributed resource, supervision and writing. S.C.S. contributed conceptualization, project administration, validation and writing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yuan, YT., Guo, TC., Wu, CY. et al. Filamentous chemokine CCL5 structure and the functional aspects. Sci Rep 15, 13552 (2025). https://doi.org/10.1038/s41598-025-98114-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-98114-9