Abstract
The high incidence of gastric cancer in China is strongly associated with widespread Helicobacter pylori infection. While the bacterium’s role in gastric cancer initiation and progression is well-established, the precise molecular mechanisms remain incompletely characterized. Current clinical challenges include limited early detection methods and poor therapeutic efficacy in advanced stages. Immune checkpoint inhibitors have shown clinical benefits in subsets of patients; however, many exhibit primary resistance or acquire secondary resistance, though the mechanisms underlying this resistance remain poorly understood. Emerging evidence suggests that H.pylori infection may remodel the tumor microenvironment, thereby influencing gastric cancer pathogenesis, progression, and therapeutic response. This study investigates the CagA virulence factor-mediated signal-transduction pathway during H.pylori infection, elucidating its role in NLRP3 inflammasome activation and subsequent pathological modulation of gastric epithelial cells. We further analyze correlations between NLRP3 expression and clinicopathological features, evaluating its prognostic value in predicting clinical outcomes. Additionally, we examine how this signaling axis regulates immune cell infiltration and modulates molecular pathology within the tumor immune microenvironment, laying a foundation for novel diagnostic and immunotherapeutic strategies.
Similar content being viewed by others
Introduction
Helicobacter pylori (H. pylori), a Gram-negative, microaerophilic spiral bacterium, is globally ubiquitous, infecting approximately 50% of the world’s population. While most infected individuals remain asymptomatic or develop mild gastritis, a subset progresses to atrophic gastritis or gastric cancer. Chronic H. pylori infection and advanced age are established as primary risk factors for gastric carcinogenesis. Advances in medical diagnostics and therapeutics have improved early-stage gastric cancer management, achieving a 5-year survival rate of 95% for early-diagnosed cases. However, most patients present with advanced-stage disease at diagnosis, contributing to poor prognoses and a low overall 5-year survival rate. This disparity underscores the critical need for sensitive, specific, and clinically feasible screening tools to enable early detection1. Current treatment for advanced gastric cancer involves multimodal approaches, including targeted therapy, immunotherapy, and neoadjuvant chemotherapy. Although immunotherapy has revolutionized cancer treatment by overcoming tumor microenvironment-mediated immunosuppression to reactivate innate antitumor responses2, its efficacy remains limited by primary or acquired resistance. Immune checkpoint inhibitor-based therapies highlight the importance of identifying predictive biomarkers for personalized treatment strategies3,4.
Approximately 75% of gastric cancer cases are linked to chronic H. pylori-induced inflammatory lesions, primarily driven by the cytotoxin-associated gene A (CagA) virulence factor encoded by the cagA pathogenicity island5. CagA, a key oncogenic protein, facilitates gastric carcinogenesis by translocating into host cells via the type IV secretion system (T4SS), disrupting intracellular signaling pathways, and promoting aberrant cell proliferation, polarity loss, and malignant transformation6. Clinical and preclinical studies confirm that CagA-positive H. pylori strains significantly elevate gastric cancer risk compared to CagA-negative strains7. Upon T4SS-mediated entry into gastric mucosal cells, CagA activates pro-inflammatory signaling cascades and induces molecular dysregulation, fostering a carcinogenic microenvironment. The molecular mechanisms underlying CagA-driven carcinogenesis involve multifaceted interactions with host signaling networks8.
The NLRP3 inflammasome, a cytosolic multiprotein complex, orchestrates immune responses to pathogenic threats by detecting extracellular danger signals, initiating inflammatory cascades, and modulating cytokine release. Chronic NLRP3 activation contributes to cancer progression by sustaining inflammation, a known tumor-promoting factor. NLRP3 influences cancer pathogenesis through immune cell modulation, pyroptosis regulation, and cytokine dynamics9. In gastric cancer, NLRP3 expression correlates with clinical outcomes, yet its impact on immune cell infiltration within the tumor microenvironment remains unexplored.
This study systematically investigates CagA-mediated NLRP3 upregulation in H. pylori-infected gastric tissues using public databases (TIMER, TCGA) and Kaplan–Meier survival analysis. Through in vitro gene transduction models in gastric mucosal and cancer cell lines, we delineate CagA’s role in NLRP3-dependent pyroptosis and downstream signaling pathways. These findings provide mechanistic insights into how H.pylori infection drives gastric carcinogenesis via NLRP3-mediated inflammatory dysregulation, offering potential biomarkers for early diagnosis and therapeutic targeting.
Materials and methods
Bioinformatics analysis of databases
High-throughput sequencing data, RNA-seq expression profiles, and clinical records of gastric cancer patients were retrieved from The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov/) and Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/gds/). These datasets included clinical staging, pathological grading, survival status, age, gender, tumor stage, histological grade, initial treatment response, progression-free survival (PFS), and overall survival (OS). Cases with incomplete metadata were excluded. From TCGA, we analyzed 412 gastric cancer tissue samples (157 H. pylori-positive, 20 H. pylori-negative) and 36 normal gastric tissues. Data normalization was performed using log2-transformed counts per million (CPM). The GEO dataset GSE36968 served as an independent validation cohort. NLRP3 mRNA expression across tumor and normal tissues was assessed via TIMER2.0 (https://cistrome.shinyapps.io/timer/), a platform enabling immune infiltration analysis across 32 cancer types. Differential gene expression was evaluated using TIMER2.0’s Diff-Exp module. UALCAN (http://ualcan.path.uab.edu/index.html) was employed to compare inflammasome family gene expression (including NLRP3) between gastric cancer and normal tissues within the TCGA cohort. Additionally, H. pylori-associated gastric cancer datasets in UALCAN were analyzed to identify infection-specific transcriptional changes.
Cell culture and H. pylori infection
The normal gastric mucosal cell line GES-1 and human gastric adenocarcinoma cell line AGS were obtained from Haixing Biotechnology (Shanghai, China). GES-1 cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. AGS cells were cultured in Ham’s F-12 medium containing 10% FBS under standard conditions (37 °C, 5% CO2, humidified atmosphere). The CagA-positive H. pylori strain ATCC 700,392 was provided by the Central Laboratory of Kunshan First People’s Hospital. Bacteria were cultured on Columbia agar plates enriched with 10% defibrinated sheep blood and incubated in microaerophilic conditions using anaerobic jars with gas-generating packs (Mitsubishi MGC, Japan) at 37 °C for 48–72 h. Colonies exhibiting characteristic morphology (transparent, pinpoint, flat, and moist) were harvested for infection. For co-culture experiments, H.pylori and gastric cancer cells were incubated at a multiplicity of infection (MOI) of 1:100 in medium containing 2% FBS at 37 °C under 5% CO2. After 48 h, cells were harvested for downstream analyses. Uninfected cells under identical conditions served as negative controls.
Expression of plasmid construction and transfection
The pcDNA3.1-CagA plasmid was commercially synthesized by Shanghai Haixing Biotechnology (Shanghai, China), with pcDNA3.1-MOI serving as the negative control plasmid (sequence schematic provided below). Transfected AGS cells were selected via puromycin screening (6-well plate format, antibiotic concentration optimized based on preliminary dose–response assays). Total protein was extracted from exponentially growing stable transfectants and analyzed by Western Blotting to quantify CagA expression levels. Band intensity was normalized to β-actin for efficiency calculations.
Cell proliferation and scratch assay
AGS cells in logarithmic growth phase were resuspended and seeded into 96-well plates at a density of 2 × 103 ~ 2 × 104 cells/well (200 μL/well), with five technical replicates per sample. Plates were incubated at 37 °C under 5% CO2 for 24, 48, and 72 h. Blank controls (culture medium only) were included, with all experimental conditions standardized. Following a 4–8 h post-seeding adhesion period, 20 μL of CCK-8 reagent was added to each well, ensuring uniform distribution at the well base. Alternatively, fresh medium containing 10% CCK-8 was substituted. Plates were returned to the incubator for 1–4 h until formazan formation reached optimal levels. Absorbance was measured at 450–490 nm (reference wavelength: 600 nm) using a microplate reader. Transfected cells were seeded into 6-well plates (5 × 105 cells/well) and cultured for 24 h to achieve 90% confluency. A sterile 100 μL pipette tip was used to create uniform linear scratches along the central axis of each well. Cells were washed thrice with phosphate-buffered saline (PBS) and maintained in serum-free medium (1 mL/well). Plates were incubated under standard conditions (37 °C, 5% CO2). Wound closure was monitored at 24 –48 h post-scratch using an inverted microscope, and migration rates were quantified using ImageJ software.
Cell migration and invasion assay
Migration and invasion assays were performed using 24-well Transwell® inserts (8-µm pore size; Corning, NY, USA). Cells (1.0 × 105 cells/well) suspended in serum-free medium were seeded into the upper chamber, while the lower chamber contained 600 μL RPMI-1640 supplemented with 25% fetal bovine serum (FBS). Following 24- or 48-h incubation at 37 °C under 5% CO2, non-migratory/non-invasive cells on the upper membrane surface were removed with a sterile cotton swab. Migrated/invaded cells were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and quantified using a TC20 automated cell counter (Bio-Rad, USA).
Measurement of cell apoptosis
Adherent cells were detached using EDTA-free trypsin and centrifuged (300 × g, 5 min). After discarding the supernatant, cells were washed twice with ice-cold phosphate-buffered saline (PBS; 300 × g, 5 min) and resuspended in fluorochrome-binding buffer at a density of 1 ~ 5 × 106 cells/mL. Following the manufacturer’s protocol, 100 μL of cell suspension was stained with Annexin V-FITC and incubated in the dark (room temperature or 2 –8 °C, 5 –15 min). Propidium iodide (PI) was then added, and samples were incubated under identical conditions (1– 5 min). Cells were diluted with 400 μL PBS, filtered through a 200-mesh sieve, and analyzed immediately by flow cytometry (BD Biosciences, USA).
Dual-luciferase target validation
A dual-luciferase reporter assay was performed to validate the interaction between miRNA-1290 and the 3’-untranslated region (3'-UTR) of NKD1. Wild-type (WT) and mutant (MUT) DNA fragments spanning the predicted miRNA-1290 binding site in the NKD1 3'-UTR were synthesized and cloned into the pGL3 dual-luciferase reporter vector. Constructs were co-transfected into cells with either miRNA-1290 mimics or negative control (NC) oligonucleotides. Following 48-h incubation, firefly and Renilla luciferase activities were quantified using the Dual-Glo® Luciferase Assay System (Promega, USA), with Renilla values normalized to firefly signals for transfection efficiency.
qRT-PCR analysis
Total RNA was reverse transcribed into complementary DNA (cDNA) using the following protocol, 1 μg of RNA template, 1 μL of mRNA RT primer, 2 μL of 5 × Reverse Transcription Buffer, and 2 μL of reverse transcriptase RTase Mix were combined in a total reaction volume of 10 μL with nuclease-free water. The reaction was performed at 42 °C for 60 min, followed by enzyme inactivation at 70 °C for 10 min. For quantitative PCR (qPCR), NLRP3 mRNA levels were analyzed using a reaction mixture containing 2 μL of cDNA template, 10 μL of 2 × SYBR Green Mix, 1 μL each of NLRP3 forward and reverse primers (10 μM), and nuclease-free water to adjust the final volume to 20 μL. Amplification conditions included an initial denaturation at 95 °C for 5 min, followed by 40 cycles of 95 °C for 2 s (denaturation), 60 °C for 20 s (annealing), and 70 °C for 10 s (elongation). GAPDH served as the endogenous reference gene, and relative mRNA expression levels were calculated using the 2−ΔΔCT method 10.
Gastric cancer-induced macrophage polarization in co-culture
To investigate macrophage polarization induced by gastric cancer cells, two experimental approaches were employed: (1) treatment with conditioned medium from gastric cancer cell lines and (2) indirect co-culture using Transwell chambers. The following experimental groups were established. AGS + NLRP3(+) + Mac: Macrophages treated with AGS cells overexpressing NLRP3. AGS + Mac: Macrophages treated with wild-type AGS cells. AGS + shNLRP3 + Mac: Macrophages treated with AGS cells with NLRP3 silenced. This grouping strategy was replicated for a secondary gastric cancer cell line. For positive controls, macrophages were polarized to an M2 phenotype via 24-h treatment with recombinant IL-4 (20 ng/mL) and IL-13 (20 ng/mL).
Immunochemistry analysis
Gastric cancer tissues were fixed in 10% neutral buffered formalin, paraffin-embedded, and sectioned onto glass slides. Immunohistochemical staining was performed using antibodies targeting immune cell surface markers. Sections were baked at 60 °C for 2 h, dewaxed in xylene, and rehydrated through a graded ethanol series. Antigen retrieval was conducted by heating slides in citrate buffer (pH 6.0) at 95 °C for 20 min. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide (H₂O₂) for 15 min, followed by blocking with 1% bovine serum albumin (BSA) to reduce nonspecific binding. Primary antibodies against immune cell markers were applied and incubated at 4 °C overnight. After washing with phosphate-buffered saline (PBS), horseradish peroxidase (HRP)-conjugated secondary antibodies were added and incubated at room temperature for 20 min. Staining was visualized using 3,3′-diaminobenzidine (DAB) substrate, and nuclei were counterstained with hematoxylin. Slides were dehydrated in ethanol, cleared in xylene, and mounted for microscopic analysis. Immune cells positive for specific surface markers were quantified in five randomly selected high-power fields (HPFs, 400 × magnification) within the tumor center. Staining intensity was scored as: 0 (none), 1 (weak/light yellow), 2 (moderate/yellow–brown), and 3 (strong/dark brown). The proportion of positive cells was graded as: 0 (≤ 5%), 1 (6 –25%), 2 (26 –50%), and 3 (≥ 51%). A combined score (intensity × proportion) was calculated, with high expression defined as a total score ≥ 4.
Western blotting analyses
Total protein was extracted from samples using Enhanced RIPA Lysis Buffer (BIOSS, Cat. No. C5029), and protein concentrations were quantified using a bicinchoninic acid (BCA) assay (Beyotime Institute of Biotechnology, Cat. No. P00105). Protein samples (20–40 μg per lane) were separated on 8% SDS–polyacrylamide gels (Beyotime Institute of Biotechnology, Cat. No. P0012AC) via SDS-PAGE and electrophoretically transferred to polyvinylidene fluoride (PVDF) membranes (MilliporeSigma, Immobilon®-P). Membranes were blocked with 5% non-fat milk in Tris-buffered saline with Tween-20 (TBST) for 1 h at room temperature and subsequently incubated overnight at 4 °C with the following primary antibodies: rabbit anti-NLRP3 (1:500 dilution; Abcam, Cat. No. ab263899) and rabbit anti-β-actin (1:1,000 dilution; Abcam, Cat. No. ab8226). β-Actin served as a loading control. After three 10-min washes with TBST, membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibodies (1:1,000 dilution; Origene, www.origene.com) for 1 h at room temperature. Protein bands were visualized using a BeyoECL Plus chemiluminescence kit (Beyotime Institute of Biotechnology, Cat. No. P00185) and imaged with a ChemiDoc™ MP Imaging System (Bio-Rad Laboratories, Inc.). Band intensity was quantified using Image Lab™ software (Bio-Rad).
Statistical analysis
Bioinformatics analyses were conducted using R 4.1.2. Immune cell infiltration proportions in gastric cancer tissues were compared between high- and low-NLRP3 expression groups using the Mann–Whitney U test. Correlations between NLRP3 expression and differentially expressed genes across immune cell infiltration levels were assessed using Pearson correlation coefficients. Experimental data were analyzed with SPSS 22.0 and visualized using GraphPad Prism 9.0. Normality of continuous variables was evaluated via the Kolmogorov–Smirnov test; normally distributed data are reported as mean ± SD. Intergroup differences were analyzed by one-way ANOVA, with statistical significance defined as P < 0.05.
Results
NLRP inflammasome expression profiling and H. pylori infection promotes NLRP3 overexpression in gastric cancer
The NLRP inflammasome family, comprising members such as NLRP1, NLRP2, NLRP3, NLRP4, NLRP5, NLRP6, NLRP7, NLRP8, NLRP9, NLRP10, NLRP11, NLRP12, NLRP13 and NLRP14, is implicated in inflammatory pathogenesis across multiple diseases. Using RNA-seq data from TCGA (407 gastric cancer and 32 adjacent normal tissues), we analyzed NLRP inflammasome expression in gastric cancer. As shown in Fig. 1A, paired sample comparison analysis showed that only NLRP3 exhibited significantly elevated mRNA levels in tumor tissues. However, unpaired t—test analysis showed that the expression levels of NLRP1, NLRP3, NLRP5, NLRP8, NLRP9, NLRP12 and NLRP13 in gastric cancer tissues were higher than those in normal tissues (Fig. 1B). To assess relevance in pan-cancer, NLRP3 expression was evaluated across 33 malignancies using TIMER2.0. Differential expression patterns emerged (Fig. 1C): NLRP3 mRNA was downregulated in bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), and rectal adenocarcinoma (READ). Conversely, renal clear cell carcinoma (KIRC) and gastric cancer (STAD) displayed significant NLRP3 upregulation relative to matched normal tissues. RNA sequencing data from 157 H.pylori-positive and 20 H.pylori-negative gastric cancer cases were retrieved from The Cancer Genome Atlas (TCGA) database. Differential gene expression analysis between the two groups revealed significant disparities, with a heatmap illustrating the top 50 differentially expressed genes (Fig. 1D). Notably, H.pylori infection induced dysregulation of inflammatory signaling pathways, including the NLRP3 inflammasome pathway, and was associated with elevated NLRP3 expression in gastric cancer tissues(Fig. 1E). Immunohistochemical (IHC) analysis of 128 gastric cancer tissues (74 H. pylori-positive, 54 H. pylori-negative) revealed significantly higher NLRP3 protein expression in H. pylori-positive versus negative groups (Fig. 1F). RT-PCR further demonstrated elevated NLRP3 mRNA levels in tumor tissues compared to adjacent normal tissues (Fig. 1G). These findings collectively implicate H. pylori infection as a significant driver of NLRP3 overexpression in gastric carcinogenesis.
H. pylori infection promotes NLRP3 overexpression in gastric cancer. (A) Comparison and analysis results of the expression of each inflammasome between gastric cancer and normal tissues in the TCGA database by paired t-test. (B) Comparison and analysis results of the expression of each inflammasome between gastric cancer and normal tissues in the TCGA database by unpaired t-test. (C) The expression differences of NLRP3 between pan-cancer tissues and normal tissues shown by TIMER 2.0 data. (D) Heatmap of gene expression differences in H. pylori-infected positive and negative gastric cancer tissues from the TCGA database (top 50 genes). (E) Volcano plot of differentially expressed genes in gastric cancer tissues. (F) IHC detection results of NLRP3 expression in H. pylori-infected positive and negative gastric cancer tissues. (G) NLRP3 mRNA expression levels in H. pylori-infected positive and negative gastric cancer tissues. *P < 0.05, ** P < 0.01, *** P < 0.001; GC, gastric cancer; H. pylori( +), H.pylori-infected positive; H.pylori(-), H.pylori-infected negative.
The H. pylori virulence factor CagA promotes pyroptosis through the axis of miRNA -1290/NKD1/NLRP3 in gastric cancer
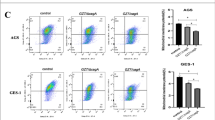
H.pylori strains carrying the CagA virulence factor (H. pylori/CagA +) or lacking CagA (H.pylori/CagA-) were co-cultured with the gastric mucosal cell line GES-1 and gastric cancer cell line AGS (multiplicity of infection [MOI] = 1:100; Fig. 2A). After 48 h, Western blot confirmed CagA expression exclusively in cells co-cultured with H.pylori/CagA + (Fig. 2B). To further investigate CagA’s role, an overexpression plasmid (pcDNA3.1/CagA +) was transfected into GES-1 and AGS cells (Fig. 3C, D). Successful transfection induced morphological changes, including irregular polarity and cell swelling (Fig. 2E). Stable CagA expression was verified by Western blot (Fig. 2F). Flow cytometry revealed significantly elevated apoptosis in over-expressed CagA cells (27.59%) and H. pylori/CagA⁺ co-cultured cells (22.67%) compared to control group (17.97%, Fig. 2G). Pyroptosis-associated markers were markedly increased in these groups: reactive oxygen species (ROS) levels and cytokine release (Fig. 2H–I). These findings demonstrate that CagA, whether delivered via H. pylori infection or transfection, drives pyroptosis in gastric cancer cells by activating inflammatory pathways and cytokine release. Non-coding microRNAs (miRNAs) play a pivotal role in gene regulation and are critically involved in cancer pathogenesis. H. pylori infection alters the expression of specific miRNAs in gastric mucosal tissues, modulating associated signaling pathways. Differential expression analysis identified 306 miRNAs (208 upregulated, 98 downregulated) in gastric cancer tissues compared to normal controls (Fig. 2J). Notably, hsa-miR-1290, a miRNA linked to NLRP3 inflammasome activity, exhibited significant upregulation in gastric cancer tissues and correlated with the H.pylori virulence factor CagA (Fig. 2K). TCGA-based bioinformatics analysis confirmed elevated miR-1290 expression in tumor tissues (Fig. 2L), underscoring its potential role in H.pylori-driven carcinogenesis. Following bacterial infection, CagA translocates into host cells and alters critical signaling pathways. RT-PCR analysis revealed significantly elevated miRNA-1290 levels in gastric tumor compared to normal tissues (Fig. 2M). Similarly, NLRP3 mRNA expression in CagA transfected cells (pcDNA3.1/CagA +) markedly exceeded levels in empty plasmid transfected (pcDNA3.1/CagA-) and untreated control groups with no significant difference between the latter two (Fig. 2N). Immunoblotting confirmed increased NLRP3 protein expression in pcDNA3.1/CagA + transfected cells (Fig. 2O). These findings demonstrate that CagA directly upregulates both miRNA-1290 and NLRP3 in gastric cancer cells (Fig. 2P).
Analysis of differential miRNAs related to NLRP3 expression caused by H. pylori infection in gastric cancer and the expression level of miRNA 1290. (A) Co-culture of H. pylori and AGS. (B) Co-culture with CagA-positive H. pylori leads to the expression of CagA in AGS and GES-1. (C, D) Fluorescent protein expression in stable cell lines transfected with plasmids and cell morphology under bright-field illumination (E). (F) Western blot verification of CagA overexpression in plasmid-transfected cells. (G, H) Flow cytometry detection images and bar charts of apoptosis data. Bar charts of pyroptosis’s ROS (I), cytokines IL-1β (J), and IL-18 (K) detection data. (L) Volcano plot of differentially expressed miRNAs in gastric cancer. (M) Differential expression analysis of miRNA-1290 in gastric cancer and normal tissues. NLRP3 mRNA expression (N), NLRP3 protein levels (O) and miRNA-1290 levels (P) in AGS cells after CagA overexpression. (Q) Schematic diagram of the binding sites between miRNA-1290 and the 3’ UTR of NKD1. (R, S) Western blot verification and protein grayscale bar charts of NKD1 protein expression in AGS lines transfected with miRNA-1290 or inhibitors. (T) Bar chart of relative luciferase activity detection in each group of cells in the luciferase assay.
NLRP3 promotes malignant degree and drives M2 macrophage polarization via IL-6/IL-10 activating JAK2/STAT3 in cancer cells. (A) Overexpressed NLRP3 promotes the proliferation of AGS. (B) Transwell assay shows that NLRP3 overexpression promotes the invasive ability of AGS. (C) Scratch assay shows that NLRP3 overexpression promotes the migration ability of AGS. (D) Overexpression of NLRP3 in AGS induces M2 macrophage polarization. (E) The effect of IL-6/IL-10 on macrophage polarization. (F) The effect of adding IL-6/IL -10 neutralizing antibodies to the culture supernatant on macrophage polarization. (G–I) IL-6/IL-10 affects the gene expression activation of the macrophage JAK2/STAT3 signaling pathway, Western blot detection images and their protein expression level gray value analysis histograms. (J–O) The flow cytometry typing detection results of THP-1 monocytes induced to differentiate into macrophages and the cell proportion histograms (P).
Bioinformatic analysis (TargetScan) identified NKD1 as a putative miRNA-1290 target, with 60.9% sequence complementarity (14/23 bases) in the NKD1 3’UTR (Fig. 3Q). Functional validation showed that miRNA-1290 mimics suppressed NKD1 expression in wild-type AGS cells (AGS/NKD1-WT) but not in cells harboring a mutated 3’UTR (AGS/NKD1-MUT; Fig. 2R, S). Conversely, miRNA-1290 inhibitors increased endogenous NKD1 levels. Dual-luciferase assays confirmed direct targeting, as miRNA-1290 mimics reduced luciferase activity in NKD1-WT reporters but not in mutants (Fig. 2T). These results establish NKD1 as a key miRNA-1290 target. NKD1, a cytoplasmic inhibitor of the Wnt/β-catenin pathway (commonly suppressed in gastric cancer), regulates NLRP3 inflammasome assembly, linking miRNA-1290 to pyroptotic signaling.
High expression of NLRP3 promotes malignant degree and drives M2 macrophage polarization via IL-6/IL-10 activating JAK2/STAT3 in cancer cells
NLRP3 overexpression in AGS cells (AGS + NLRP3( +)) increased proliferation (CCK-8 assay, Fig. 3A), invasion (Transwell assay, Fig. 3B) and migration (scratch assay, Fig. 3C), compared to NLRP3-silenced (AGS + shNLRP3) and untreated cells (AGS). NLRP3 overexpressing AGS cells exhibited elevated pyroptosis-associated cytokines (IL-18, IL-1β) and increased IL-6, IL-10, and IFN-γ in supernatants (Table 1). Co-culture with macrophages in Transwell systems revealed NLRP3-driven polarization shifts: M1 macrophages (CD86 +) decreased (13.3% vs. 43.3%), while M2 macrophages (CD206 +) increased (28.9% vs. 5.8%; Fig. 3D). This polarization shift was recapitulated by treating monocultured macrophages with supernatants from NLRP3( +) cells, which elevated M2-type populations (CD206 + : 4.4% → 18.1%) and suppressed M1-type populations (CD86 + : 38.6% → 14.2%) (Fig. 3E) . Neutralizing IL-6 and IL-10 in these supernatants abolished these effects, with no significant changes in M1 (40.25% vs. 38.11%) or M2 (4.57% vs. 6.31%) proportions (Fig. 3F), confirming IL-6/IL-10 as key mediators of NLRP3-driven macrophage polarization. Western blot analysis demonstrated elevated JAK2/STAT3 protein expression and phosphorylation in macrophages exposed to IL-6/IL-10-enriched supernatants from NLRP3-overexpressing cells (Fig. 3G–I). This activation was suppressed by IL-6/IL-10 neutralizing antibodies, which concurrently blocked M2 polarization. These findings indicate that IL-6 and IL-10 drive macrophage polarization via JAK2/STAT3 signaling, with NLRP3 overexpression indirectly promoting this pathway through cytokine secretion.
Undifferentiated THP-1 cells (THP-1 group) were maintained in RPMI-1640 medium supplemented with 10% FBS. At logarithmic growth phase, cells were treated with 50 ng/mL PMA to induce differentiation into M0-type macrophages (untreated control group; Fig. 3J). Flow cytometry revealed a significant increase in M1-polarized macrophages (CD86 + : 26.15%) post-M1 differentiation compared to undifferentiated THP-1 cells, while M2 markers (CD206) remained negligible (Fig. 3K). M2 differentiation elevated CD206 + populations to 11.16% (Fig. 3L). Co-culture of NLRP3-overexpressing gastric cancer cells (AGS + NLRP3( +)) with untreated macrophages increased M2 polarization (CD206 + : 2.18% vs. 0.093% in AGS + macrophages and 0.71% in AGS + shNLRP3 controls; Fig. 3M–O). Conversely, M1 populations (CD86 +) decreased in the NLRP3( +) group (34.31%) relative to AGS untreated (34.99%) and AGS + shNLRP3 groups (49.15%). These findings demonstrate that NLRP3 overexpression in gastric cancer cells drives macrophage polarization toward an M2 phenotype, while NLRP3 inhibition shifts polarization toward M1(Fig. 3P).
NLRP3 modulates CD4+ T Cell and M2 macrophage infiltration in the gastric cancer microenvironment
CIBERSORT analysis revealed that NLRP3-high tumors showed pronounced enrichment of Tregs, M0 macrophages, naïve B cells, helper T cells, M1 macrophages, activated mast cells, and memory-activated CD4 + T cells relative to normal gastric tissues, alongside suppressed infiltration of plasma cells, resting mast cells, and monocytes (Fig. 4A). Stratification by NLRP3 expression levels further revealed its dual regulatory role: NLRP3-high tumors exhibited strong positive correlations with activated mast cells, neutrophils, NK cells, and M2 macrophages, but negative associations with Tregs and CD8 + T cells (Fig. 4B). Immune cell distribution analysis identified CD8 + T cells and CD4 + resting memory T cells as dominant populations in gastric cancer, followed by M2 macrophages, Tregs, M0 macrophages, plasma cells, and naïve B cells (Fig. 4C). Compared to normal tissues, gastric tumors exhibited higher proportions of Tregs, follicular helper T cells (TFH), and M1 macrophages. Correlation studies demonstrated significant negative associations between CD8 + T cells and CD4 + resting memory T cells, while positive correlations linked CD8 + T cells to activated CD4 + T cells and M1 macrophages (Fig. 4D). NLRP3 expression exhibited weak correlation with M1 macrophage markers but strongly associated with M2 macrophage and monocyte markers in gastric cancer tissues (Fig. 4E–H), indicating its potential role in polarizing tumor-associated macrophages (TAMs) toward an M2 phenotype. Immunohistochemical analysis of 128 gastric cancer samples were stratified into high-expression and low-expression groups based on median NLRP3 immunohistochemical scores. Comparative analysis revealed significantly elevated CD3 + , CD4 + and CD206 + cell counts in NLRP3-high tumors compared to NLRP3-low counterparts, whereas CD19 + and CD21 + cell counts were reduced (Fig. 4I–K). These findings demonstrate NLRP3-mediated remodeling of the tumor immune microenvironment, favoring immunosuppressive M2 macrophages and CD4 + T cell infiltration.
NLRP3 high expression remodulates immune cell infiltration in the Gastric Cancer Microenvironment. (A) Boxplot of immune cell types and their proportions in gastric cancer tissue compared to normal tissue. (B) Heatmap of immune cell infiltration types. (C) Boxplot of the distribution proportions of different types of immune cells in gastric cancer tissue. (D) Heatmap of the correlation of immune cell infiltration in gastric cancer tissue. (E–H) Scatter plots showing the correlation between NLRP3 expression levels and macrophage polarization, monocytes. (I) Immunohistochemical images of representative markers of high immune infiltration in the high NLRP3 expression group. (J) Immunohistochemical image of the M2 macrophage marker CD206. (K) Bar chart of immunohistochemical staining scores of representative markers. (The depth of color blocks represents the degree of correlation coefficients; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
NLRP3 high-expression promotes cancer progression and M2 macrophage infiltration predicting poor prognosis in gastric cancer
Gastric cancer patients were stratified into high- and low-NLRP3 expression groups based on median NLRP3 mRNA levels in tumor tissues. No significant differences in NLRP3 mRNA expression were observed across patient age, sex, histological subtype, or tumor location (P > 0.05). However, NLRP3 mRNA levels were significantly elevated in tumors ≥ 5 cm (vs. < 5 cm), those with lymph node metastasis (vs. non-metastatic), TNM stage III-IV (vs.I-II), and moderately/poorly differentiated tumors (vs. well-differentiated).These findings suggest that NLRP3 overexpression is associated with aggressive tumor behavior, including advanced TNM stage and higher histological grade (Table 2). Collectively, elevated NLRP3 inflammasome protein expression may drive malignant progression in gastric cancer.
Survival curve analysis using the survminer package revealed that immune cell infiltration patterns mirrored the prognostic impact of NLRP3. High infiltration levels of CD3+ , CD4 + , and CD206 + cells correlated with significantly reduced overall survival compared to low-infiltrating cohorts (Fig. 5A–C). Mechanistically, NLRP3 overexpression drives immune suppression by remodeling the tumor microenvironment through increased infiltration of regulatory T lymphocytes (Tregs) and M2-polarized macrophages. This immunosuppressive shift likely contributes to tumor immune evasion and diminished response to immunotherapy. Survival analysis was performed using data from 875 gastric cancer patients in the Kaplan–Meier Plotter database. Patients with high NLRP3 mRNA expression exhibited significantly reduced overall survival (OS) compared to the low-expression group (HR = 1.35, P = 0.00053; Fig. 5D). Similarly, progression-free survival (PFS) and post-progression survival (PPS) were markedly shorter in the high-expression cohort (PFS: HR = 1.23, P = 0.048; PPS: HR = 1.58, P = 0.048; Fig. 5E, F). These findings indicate that elevated NLRP3 mRNA levels correlate with adverse survival outcomes in gastric cancer. To validate these results, we analyzed RNA-seq data from 370 gastric cancer patients in the TCGA cohort. Consistent with the initial findings, the high-NLRP3 expression group demonstrated a significantly lower 10-year survival rate compared to the low-expression group (HR = 1.62, P = 0.005; Fig. 5G). Collectively, both datasets confirm NLRP3 overexpression as a robust prognostic indicator of poor survival in gastric cancer patients.
NLRP3 High-expression and immune infiltration predicts poor prognosis in gastric cancer. (A) Survival curve of patients with CD3 + biomarker expression. (B) Survival curve of patients with CD4 + biomarker expression. (C) Survival curve of patients with CD206 + biomarker expression. Kaplan–Meier Plotter analysis of NLRP3 mRNA expression and its impact on survival prognosis in gastric cancer patients, including (D) overall survival, (E) progression-free survival, and (F) post-progression survival. (G) Survival curve analysis of 10-year survival rates in gastric cancer patients with high and low NLRP3 mRNA expression based on TCGA data.
Discussion
Gastric carcinogenesis is a multifactorial process influenced by genetic, environmental, and microbial factors. Approximately 50% of the global population is infected with H. pylori, though most remain asymptomatic11. The clinical outcomes of infection depend critically on bacterial virulence determinants12. Notably, 90 –95% of East Asian H. pylori strains encode the oncogenic CagA protein, compared to only 50 –60% in Western populations13. CagA-positive strains are strongly associated with acute gastritis, peptic ulcers, and gastric adenocarcinoma14. Mechanistically, CagA translocates into host cells, disrupting signaling pathways to induce dysregulated gene expression, cellular damage, and hyperproliferation15.
Our study confirmed CagA’s pivotal role in modifying cellular morphology and its involvement in H. pylori's pathogenic mechanisms. Through plasmid-mediated CagA transfection in gastric cancer cells, we observed significant upregulation of miRNA-1290 expression compared to control groups. This overexpression of miRNA-1290 was found to suppress multiple tumor suppressor genes while reactivating or amplifying the expression of previously inhibited oncoproteins. Flow cytometry analyses revealed substantial release of inflammatory mediators, including pyroptosis-specific cytokines IL-1β and IL-18. Characteristic pyroptotic features such as increased membrane swelling were consistently observed. Western blot analysis identified modifications in the NLRP3-mediated canonical signaling pathway, with corresponding changes detected at the mRNA level.
The observed overexpression of NLRP3 in gastric carcinoma tissues shows strong correlation with persistent H. pylori infection. The NLRP3 inflammasome, a central mediator of inflammatory and immune responses, has emerged as a key player in tumor-associated inflammation16. Its activation drives inflammatory cascades linked to numerous pathologies, including cancer. H. pylori modulates NLRP3 activity to subvert host immunity. Backert.S demonstrated that H. pylori upregulates NLRP3 via microRNA-223-3p, stimulating IL-10 secretion in co-cultured gastric cancer cells17. Similarly, Kim.S identified synergistic interactions between CagA and innate immune receptors (TLR2, NOD2, NLRP3) that amplify IL-1β production during infection18. In gastric cancer, immunohistochemical and Western blot analyses confirmed elevated NLRP3 levels in tumor tissues, further exacerbated by H.pylori infection. Functionally, NLRP3 activation drives tumor progression by modulating inflammatory ligand-receptor interactions and cytokine networks. Our experiments revealed that CagA H. pylori infection or CagA transfection induces morphological shifts (e.g., spindle-like transformation) in AGS cells, enhancing proliferation, invasion, and migration. These effects correlate with miRNA dysregulation: H. pylori upregulates oncogenic miRNA-1290 in gastric tissues19, which suppresses tumor suppressors like NKD1, a negative regulator of the Wnt/β-catenin pathway20. Since Wnt/β-catenin signaling directly activates NLRP321, this creates a feedforward loop promoting malignancy. NLRP3 overexpression amplified tumor aggressiveness in vitro, while NLRP3 knockdown reversed these phenotypes, implicating NLRP3 as a critical mediator of CagA-driven invasion. Clinically, high NLRP3 expression correlated with advanced tumor size, lymph node metastasis, TNM stage, and poor differentiation, while survival analysis confirmed its prognostic value for poor outcomes.
Flow cytometry detected elevated levels of pyroptosis-associated inflammatory factors (e.g., IL-1β, IL-18) in culture supernatants of NLRP3-high gastric cancer cells. Western blotting revealed NLRP3-mediated activation of pyroptotic pathways, triggering release of IL-6, IL-10, TNF-α, and other cytokines that establish a pro-tumorigenic microenvironment. These cytokines recruit immune cells, amplifying inflammatory cascades that drive immune infiltration, immunosuppression, and metastasis. Notably, IL-6 and IL-10 synergistically activate the JAK/STAT pathway in macrophages, polarizing them toward pro-tumor M2 phenotypes. NLRP3 inhibition reduced IL-6/IL-10 secretion and shifted macrophage polarization toward anti-tumor M1 subtypes, underscoring NLRP3’s role in shaping tumor-associated macrophage (TAM) behavior. TAMs predominantly adopt M2 phenotypes in tumors, fostering immunosuppressive microenvironments (TMEs) that facilitate cancer progression22. M2 TAMs express PD-L1, secrete TGF-β, and alter metabolic pathways to suppress cytotoxic T-cell activity while promoting angiogenesis and immune evasion23,24. NLRP3 further exacerbates immunosuppression by upregulating PD-L1 in lymphoma25 and driving Treg cell expansion via macrophage polarization in colitis26, and hepatocellular carcinoma 27. In gastric cancer, high NLRP3 expression correlates with elevated Treg markers (CD3D, CD4) and M2 TAM infiltration—both predictors of poor survival.
Within the gastric cancer microenvironment, chronic inflammation-derived cytokines critically fuel neoplastic proliferation. H.pylori infection initiates pathogenic interactions through its virulence factor CagA, which engages host cell membrane receptors to activate the NLRP3 inflammasome. This activation triggers pyroptotic signaling cascades, resulting in substantial release of IL-18, IL-1β, IL-6, IL-10, and TNF-α cytokines that collectively establish a pro-tumorigenic niche conducive to cancer cell proliferation and metastasis28. These pyroptosis-associated cytokines further recruit immune cells, amplifying inflammatory responses through cascade effects that exacerbate tumor progression and malignant transformation. Experimental evidence confirms that tumor cells promote M2 macrophage polarization, which collaborates with tumor-infiltrating lymphocytes and dendritic cells to establish an immunosuppressive microenvironment favoring cancer progression and poor clinical outcomes22. To investigate NLRP3’s role in this process, we co-cultured NLRP3-overexpressing gastric cancer cells (AGS) with monocyte-derived macrophages. NLRP3 overexpression markedly enhanced AGS proliferative and migratory capacities while elevating IL-6/IL-10 secretion. Conversely, NLRP3 knockdown via shRNA attenuated cytokine production and shifted macrophage polarization from M2 to M1 phenotypes. Mechanistically, NLRP3-driven IL-6/IL-10 secretion activates the JAK/STAT pathway in tumor-associated macrophages (TAMs), evidenced by increased p-JAK2 and p-STAT3 expression, thereby driving M2 polarization.
Immune profiling revealed that elevated NLRP3 expression correlates with intensified immune infiltration, particularly involving T lymphocytes (Tregs and Th subsets) and M2 macrophage conversion. While CD3+ T cell infiltration generally associates with improved survival, functional heterogeneity exists among CD4 + subsets (Th1, Th2, Th17, Tregs), with conflicting reports on their tumor-modulatory roles29. Notably, dendritic cells serve as critical antigen-presenting cells initiating T-cell-mediated antitumor immunity, though their infiltration patterns influence therapeutic responses30. Survival analyses demonstrate that high expression of immunosuppressive markers (CD3D, CD4, MCR1) indicative of Tregs and M2 macrophage infiltration correlates with reduced survival in gastric cancer patients. Integrated analysis of Kaplan–Meier microarray data and TCGA RNA-Seq datasets revealed significantly poorer survival in NLRP3-high gastric cancer patients. NLRP3 overexpression strongly associates with lymph node metastasis, poor differentiation, advanced TNM staging, and diminished 10-year survival rates. Multivariate Cox regression identifies NLRP3 overexpression, age, and initial treatment response as independent prognostic risk factors. Across multiple databases, NLRP3 demonstrates consistent correlations with aggressive tumor behavior and adverse clinical outcomes, underscoring its role as a critical mediator of gastric cancer progression.
Targeting NLRP3 may recalibrate immune cell dynamics in the TME. By modulating macrophage polarization (e.g., reducing M2/M1 ratios) or disrupting NLRP3-mediated cytokine networks, therapies could counteract immunosuppression and enhance anti-tumor immunity. Emerging strategies aim to regulate TAM recruitment31 or exploit metabolic shifts (e.g., ROS-mediated NLRP3 activation32) to reprogram macrophage function33. Such approaches, combined with immune checkpoint inhibitors, could synergize to restore T-cell surveillance and improve clinical outcomes.
In conclusion, NLRP3 orchestrates crosstalk between H. pylori virulence, inflammatory signaling, and immune evasion in gastric cancer. Its overexpression predicts aggressive disease and poor survival, positioning NLRP3 as both a prognostic biomarker and a therapeutic target. Precision modulation of NLRP3 activity—or its downstream immune effects—may reshape the TME to favor anti-tumor responses, offering novel avenues for immunotherapy in gastric cancer.
Data availability
The datasets generated and analyzed during the current study are available in the UCSC database: UCSC Xena (https://xenabrowser.net/hub/).
References
Heuvers, M. E., Wisnivesky, J., Stricker, B. H. & Aerts, J. G. Generalizability of results from the National Lung Screening Trial. Eur. J. Epidemiol. 27, 669–672. https://doi.org/10.1007/s10654-012-9720-8 (2012).
Billan, S., Kaidar-Person, O. & Gil, Z. Treatment after progression in the era of immunotherapy. Lancet Oncol. 21, e463–e476. https://doi.org/10.1016/S1470-2045(20)30328-4 (2020).
Chen, F. et al. Clinical progress of PD-1/L1 inhibitors in breast cancer immunotherapy. Front. Oncol. 11, 724424. https://doi.org/10.3389/fonc.2021.724424 (2021).
Chen, X., Zhang, W., Yang, W., Zhou, M. & Liu, F. Acquired resistance for immune checkpoint inhibitors in cancer immunotherapy: challenges and prospects. Aging (Albany NY) 14, 1048–1064. https://doi.org/10.18632/aging.203833 (2022).
Alipour, M. Molecular mechanism of Helicobacter pylori-induced gastric cancer. J. Gastrointest. Cancer 52, 23–30. https://doi.org/10.1007/s12029-020-00518-5 (2021).
Rojas-Rengifo, D. F. et al. Intramolecular energies of the cytotoxic protein CagA of Helicobacter pylori as a possible descriptor of strains’ pathogenicity level. Comput. Biol. Chem. 76, 17–22. https://doi.org/10.1016/j.compbiolchem.2018.05.016 (2018).
Baj, J. et al. Mechanisms of the epithelial-mesenchymal transition and tumor microenvironment in Helicobacter pylori-induced gastric cancer. Cells 9, 74. https://doi.org/10.3390/cells9041055 (2020).
Chen, S. Y., Zhang, R. G. & Duan, G. C. Pathogenic mechanisms of the oncoprotein CagA in H. pylori-induced gastric cancer (Review). Oncol. Rep. 36, 3087–3094, https://doi.org/10.3892/or.2016.5145 (2016).
Feng, S. et al. Syk and JNK signaling pathways are involved in inflammasome activation in macrophages infected with Streptococcus pneumoniae Biochem. Biophys. Res. Commun. 507, 217–222. https://doi.org/10.1016/j.bbrc.2018.11.011 (2018).
Maren, N. A. et al. Stepwise optimization of real-time RT-PCR analysis. Methods Mol. Biol. 2653, 317–332. https://doi.org/10.1007/978-1-0716-3131-7_20 (2023).
Hu, Q., Zhao, F., Guo, F., Wang, C. & Fu, Z. Polymeric nanoparticles induce NLRP3 inflammasome activation and promote breast cancer metastasis. Macromol. Biosci. 17, 147. https://doi.org/10.1002/mabi.201700273 (2017).
Wani, K., AlHarthi, H., Alghamdi, A., Sabico, S. & Al-Daghri, N. M. Role of NLRP3 inflammasome activation in obesity-mediated metabolic disorders. Int. J. Environ. Res. Public Health 18, 145. https://doi.org/10.3390/ijerph18020511 (2021).
Brahmer, J. R. et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 366, 2455–2465. https://doi.org/10.1056/NEJMoa1200694 (2012).
Delmonte, O. M. & Fleisher, T. A. Flow cytometry: Surface markers and beyond. J. Allergy Clin. Immunol. 143, 528–537. https://doi.org/10.1016/j.jaci.2018.08.011 (2019).
Chen, Z. W. et al. Helicobacter pylori CagA protein induces gastric cancer stem cell-like properties through the Akt/FOXO3a axis. J. Cell Biochem. 125, e30527. https://doi.org/10.1002/jcb.30527 (2024).
Zhang, X. et al. H. pylori CagA activates the NLRP3 inflammasome to promote gastric cancer cell migration and invasion. Inflamm. Res. 71, 141–155. https://doi.org/10.1007/s00011-021-01522-6 (2022).
Pachathundikandi, S. K. & Backert, S. Helicobacter pylori controls NLRP3 expression by regulating hsa-miR-223-3p and IL-10 in cultured and primary human immune cells. Innate Immun. 24, 11–23. https://doi.org/10.1177/1753425917738043 (2018).
Kim, S. E. et al. Gastroprotective effect of phytoncide extract from Pinus koraiensis pinecone in Helicobacter pylori infection. Sci. Rep. 10, 9547. https://doi.org/10.1038/s41598-020-66603-8 (2020).
Ta, N. et al. miRNA-1290 promotes aggressiveness in pancreatic ductal adenocarcinoma by targeting IKK1. Cell Physiol. Biochem. 51, 711–728. https://doi.org/10.1159/000495328 (2018).
Soudeh, G.-F. A review on the role of miR-1290 in cell proliferation, apoptosis and invasion. Front. Mol. Biosci. 8, 763338 (2021).
Alrashed, M. M., Alharbi, H., Alshehry, A. S., Ahmad, M. & Aloahd, M. S. MiR-624-5p enhances NLRP3 augmented gemcitabine resistance via EMT/IL-1beta/Wnt/beta-catenin signaling pathway in ovarian cancer. J. Reprod. Immunol. 150, 103488. https://doi.org/10.1016/j.jri.2022.103488 (2022).
Galland, S. et al. Tumor-derived mesenchymal stem cells use distinct mechanisms to block the activity of natural killer cell subsets. Cell Rep. 20, 2891–2905. https://doi.org/10.1016/j.celrep.2017.08.089 (2017).
Ni, B. et al. Tumor-associated macrophage-derived GDNF promotes gastric cancer liver metastasis via a GFRA1-modulated autophagy flux. Cell Oncol. (Dordr.) 46, 315–330. https://doi.org/10.1007/s13402-022-00751-z (2023).
Wei, C. et al. Characterization of gastric cancer stem-like molecular features, immune and pharmacogenomic landscapes. Brief Bioinform. 23, 1457. https://doi.org/10.1093/bib/bbab386 (2022).
Lu, F. et al. NLRP3 inflammasome upregulates PD-L1 expression and contributes to immune suppression in lymphoma. Cancer Lett. 497, 178–189. https://doi.org/10.1016/j.canlet.2020.10.024 (2021).
He, R. et al. L-Fucose ameliorates DSS-induced acute colitis via inhibiting macrophage M1 polarization and inhibiting NLRP3 inflammasome and NF-kB activation. Int. Immunopharmacol. 73, 379–388. https://doi.org/10.1016/j.intimp.2019.05.013 (2019).
Liu, H. et al. FNDC5 induces M2 macrophage polarization and promotes hepatocellular carcinoma cell growth by affecting the PPARgamma/NF-kappaB/NLRP3 pathway. Biochem. Biophys. Res. Commun. 582, 77–85. https://doi.org/10.1016/j.bbrc.2021.10.041 (2021).
Shadab, A. et al. Divergent functions of NLRP3 inflammasomes in cancer: a review. Cell Commun. Signal 21, 232. https://doi.org/10.1186/s12964-023-01235-9 (2023).
Kwon, H. J. et al. Genetic and immune microenvironment characterization of HER2-positive gastric cancer: Their association with response to trastuzumab-based treatment. Cancer Med. 12, 10371–10384. https://doi.org/10.1002/cam4.5769 (2023).
Qu, Y. et al. The effects of TNF-alpha/TNFR2 in regulatory T cells on the microenvironment and progression of gastric cancer. Int. J. Cancer 150, 1373–1391. https://doi.org/10.1002/ijc.33873 (2022).
Yi, J. et al. EGCG targeting STAT3 transcriptionally represses PLXNC1 to inhibit M2 polarization mediated by gastric cancer cell-derived exosomal miR-92b-5p. Phytomedicine 135, 156137. https://doi.org/10.1016/j.phymed.2024.156137 (2024).
Kiser, C., Gonul, C. P. & Genc, S. Nrf2 activator diethyl maleate attenuates ROS mediated NLRP3 inflammasome activation in murine microglia. Cytotechnology 76, 197–208. https://doi.org/10.1007/s10616-023-00609-8 (2024).
Verona, F. et al. Cancer stem cells and tumor-associated macrophages as mates in tumor progression: mechanisms of crosstalk and advanced bioinformatic tools to dissect their phenotypes and interaction. Front. Immunol. 16, 1529847. https://doi.org/10.3389/fimmu.2025.1529847 (2025).
Acknowledgements
The authors are grateful to the editor, the associate editor, and the reviewer.
Funding
This study was supported by grants from Medical research support project of Changshu Health Committee (No. CSWS202106), Changshu Science and Technology Program Project (No.CS202018), Basic Research Program-Medical application basic Research Project (No. CY202330) and Suzhou Science and Technology Bureau Clinical Trial Institution Capacity Improvement Project (SLT2023007).
Author information
Authors and Affiliations
Contributions
CW, YX and YG conceived the project. PW, YZ and XC collected specimens and data. CW, YX and PW conducted the experiments. YX, PW and YZ analyzed the data. CW and PW drafted the manuscript. PW, YX and YG confirm the authenticity of all the data. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of Affiliated Changshu Hospital of Nantong University.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wan, C., Wang, P., Xu, Y. et al. Mechanism and role of H. pylori CagA-induced NLRP3 inflammasome in gastric cancer immune cell infiltration. Sci Rep 15, 14335 (2025). https://doi.org/10.1038/s41598-025-98301-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-98301-8