Abstract
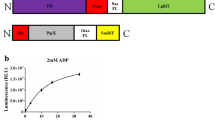
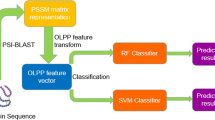
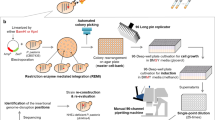
Deciphering protein–protein interactions (PPIs) is crucial for a comprehensive understanding of biological processes, yet current methodologies often provide incomplete interactome maps. Here, we present a sensitive bimolecular NanoBiT-based protein complementation assay platform for robust PPI detection in the fission yeast Schizosaccharomyces pombe. Our platform utilizes two NanoLuc moieties, SmBiT and LgBiT, fused to interacting protein partners, generating a quantifiable luminescent signal upon interaction. To maximize the chances of detection and mitigate potential issues arising from tag position-dependent inactivation of bait proteins, our system enables simultaneous expression of two distinct bait constructs within a single cell: one with the LgBiT-tag fused at its N-terminus and another at its C-terminus. For the prey, we constructed a comprehensive ORFeome library of fission yeast proteins, each fused with SmBiT at its C-terminus, leveraging homologous recombination tools. We established efficient high-throughput methods for cloning and selection of single-copy integrants, enabling the rapid construction of the prey library and reliable identification of true yeast transformants. Validating the platform, high-throughput screening using the general transcription elongation factor Tfs1 successfully identified previously undetectable interactors. This versatile platform not only significantly expands the scope of interactome discovery but also offers a powerful tool for future protein-compound interaction studies.
Similar content being viewed by others
Data availability
The authors confirm that all data required to support the conclusions of this article are included within the text, figures, and tables. Protein–protein interaction screening data generated using the NanoBiT system in this study have been submitted to the IMEx (http://www.imexconsortium.org) consortium through IntAct [X]44 and assigned the identifier IM-30540. Strains and plasmids are available upon request or from the Yeast Genetic Resource Center (YGRC/NBRP) of Japan.
Abbreviations
- NanoBiT:
-
Nanoluciferase binary technology
- ORF:
-
Open reading frame
- PCA:
-
Protein complementation assay
- PCR:
-
Polymerase chain reaction
- PPI:
-
Protein–protein interaction
- Y2H:
-
Yeast-two-hybrid
References
De Las Rivas, J. & Fontanillo, C. Protein-protein interaction networks: Unraveling the wiring of molecular machines within the cell. Brief. Funct. Genomics 11, 489–496 (2012).
Tsai, C. J., Ma, B. & Nussinov, R. Protein-protein interaction networks: how can a hub protein bind so many different partners?. Trends Biochem. Sci. 34, 594–600 (2009).
Titeca, K., Lemmens, I., Tavernier, J. & Eyckerman, S. Discovering cellular protein-protein interactions: Technological strategies and opportunities. Mass Spectrom. Rev. 38, 79–111 (2019).
Fields, S. & Song, O. K. A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 (1989).
Tebo, A. G. & Gautier, A. A split fluorescent reporter with rapid and reversible complementation. Nat. Commun. 10, 2822 (2019).
Pelletier, J. N., Campbell-Valois, F. X. & Michnick, S. W. Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc. Natl. Acad. Sci. U. S. A. 95, 12141–12146 (1998).
Algar, W. R., Hildebrandt, N., Vogel, S. S. & Medintz, I. L. FRET as a biomolecular research tool—Understanding its potential while avoiding pitfalls. Nat. Methods 16, 815–829 (2019).
Xu, Y., Piston, D. W. & Johnson, C. H. A bioluminescence resonance energy transfer (BRET) system: Application to interacting circadian clock proteins. Proc. Natl. Acad. Sci. U. S. A. 96, 151–156 (1999).
Ito, T. et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. U. S. A. 98, 4569–4574 (2001).
Yu, H. et al. High-quality binary protein interaction map of the yeast interactome network. Sci. AAAs 322, 104–110 (2008).
Hofmann, J. L., Maheshwari, A. J., Sunol, A. M., Endy, D. & Zia, R. N. Ultra-weak protein-protein interactions can modulate proteome-wide searching and binding. bioRxiv https://doi.org/10.1101/2022.09.30.510365 (2022).
Dixon, A. S. et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 11, 400–408 (2016).
Vyas, A., Freitas, A. V., Ralston, Z. A. & Tang, Z. Fission yeast Schizosaccharomyces pombe: A unicellular, “Micromammal” model organism. Curr. Protoc. 1, e150 (2021).
Matsuyama, A. et al. pDUAL, a multipurpose, multicopy vector capable of chromosomal integration in fission yeast. Yeast 21, 1289–1305 (2004).
Matsuyama, A., Shirai, A. & Yoshida, M. A novel series of vectors for chromosomal integration in fission yeast. Biochem. Biophys. Res. Commun. 374, 315–319 (2008).
Matsuyama, A., Hashimoto, A., Nishimura, S. & Yoshida, M. A set of vectors and strains for chromosomal integration in fission yeast. Sci. Rep. 13, 315–319 (2023).
Shirai, A. et al. Global analysis of gel mobility of proteins and its use in target identification. J. Biol. Chem. 283, 10745–10752 (2008).
Kim, D. U. et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 28, 617–623 (2010).
Nishimura, S. et al. Marine antifungal theonellamides target 3β-2-hydroxysterol to activate Rho1 signaling. Nat. Chem. Biol. 6, 519–526 (2010).
Bauer, F. et al. Translational control of cell division by Elongator. Cell Rep. 1, 424–433 (2012).
Matsuyama, A. et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 24, 841–847 (2006).
Vo, T. V. et al. A proteome-wide fission yeast interactome reveals network evolution principles from yeasts to human. Cell 164, 310–323 (2016).
Ohsawa, S. et al. Nitrogen signaling factor triggers a respiration-like gene expression program in fission yeast. EMBO J. 43, 4604–4624 (2024).
Forsburg, S. L. & Rhind, N. Basic methods for fission yeast. Yeast 23, 173–183 (2006).
Petersen, J. & Russell, P. Growth and the environment of Schizosaccharomyces pombe. Cold Spring Harb. Protoc. 2016, pdb-top079764 (2016).
Matsuyama, A., Yabana, N., Watanabe, Y. & Yamamoto, M. Schizosaccharomyces pombe Ste7p is required for both promotion and withholding of the entry to meiosis. Genetics 155, 539 (2000).
Shirai, A., Matsuyama, A., Yashiroda, Y., Arai, R. & Yoshida, M. Rapid and reliable preparation of total cell lysates from fission yeast. Protoc. Exch. (2006).
Chiu, P. C. et al. Ferrichrome, a fungal-type siderophore, confers high ammonium tolerance to fission yeast. Sci. Rep. 12, 17411 (2022).
Matsuyama, A., Hashimoto, A., Arioka, M. & Y. M.,. Improvement of targeting efficiency by promoter replacement of markers in integration vectors. Genes Cells 30, e70013 (2025).
Hoffman, C. S. Preparation of Yeast DNA. Curr. Protoc. Mol. Biol. 39, 13–11 (1997).
Jacobus, A. P. & Gross, J. Optimal cloning of PCR fragments by homologous recombination in Escherichia coli. PLoS ONE 10, e0119221 (2015).
Keeney, J. B. & Boeke, J. D. Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136, 849–856 (1994).
Bernard, P. & Couturier, M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 226, 735–745 (1992).
Reaume, S. E. & Tatum, E. L. Spontaneous and nitrogen mustard-induced nutritional deficiencies in. Arch. Biochem. 22, 331–338 (1949).
Arita, Y. et al. Microarray-based target identification using drug hypersensitive fission yeast expressing ORFeome. Mol. Biosyst. 7, 1463–1472 (2011).
Nagao, K. et al. bfr1+ a novel gene of Schizosaccharomyces pombe which confers brefeldin A resistance, is structurally related to the ATP-binding cassette superfamily. J. Bacteriol. 177, 1536–1543 (1995).
Nishi, K. et al. A leptomycin B resistance gene of Schizosaccharomyces pombe encodes a protein similar to the mammalian P-glycoproteins. Mol. Microbiol. 6, 761–769 (1992).
Papadopoulos, D. et al. MYCN recruits the nuclear exosome complex to RNA polymerase II to prevent transcription-replication conflicts. Mol. Cell 82, 159-176.e12 (2022).
Zatreanu, D. et al. Elongation factor TFIIS prevents transcription stress and R-loop accumulation to maintain genome stability. Mol. Cell 76, 57-69.e9 (2019).
Siametis, A. et al. Transcription stress at telomeres leads to cytosolic DNA release and paracrine senescence. Nat. Commun. 15, 4061 (2024).
Cho, W. K. et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Sci. AAAs 361, 412–415 (2018).
Göös, H. et al. Human transcription factor protein interaction networks. Nat. Commun. 13, 766 (2022).
Ohana, R. F. et al. HaloTag7: A genetically engineered tag that enhances bacterial expression of soluble proteins and improves protein purification. Protein Expr. Purif. 68, 110–120 (2009).
del Toro, N. et al. The IntAct database: Efficient access to fine-grained molecular interaction data. Nucleic Acids Res. 50, D648–D653 (2022).
Acknowledgements
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers 22K05361 and 23H05473 and by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) KAKENHI Grant Number 23H04882 for Transformative Research Areas (A). Grammar and linguistic refinement of this manuscript were kindly assisted by Gemini 1.5 Pro (Google Cloud AI Studio).
Funding
This work was supported by the Japan Society for the Promotion of Science (Grant Number: 22K05361 and 23H05473), and the Ministry of Education, Culture, Sports, Science and Technology (Grant Number: 23H04882). The authors declare that no other funding was received for this study.
Author information
Authors and Affiliations
Contributions
This work was conceptualized by A.M. and M.Y. All experiments were done by A.M., A.H., and F.A. A.M. and F.A. prepared the original draft. S.N., M.A. and M.Y. edited the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Azadeh, F., Hashimoto, A., Nishimura, S. et al. Development of a global screening system for detecting protein–protein interactions by luminescence complementation in fission yeast. Sci Rep (2026). https://doi.org/10.1038/s41598-026-35430-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-026-35430-8