Abstract
Immune checkpoint inhibitors have transformed cancer therapy. However, only a fraction of patients benefit from these treatments. The variability in patient responses remains a significant challenge due to the intricate nature of the tumor microenvironment. Here, we harness single-cell RNA-sequencing data and employ machine learning to predict patient responses while preserving interpretability and single-cell resolution. Using a dataset of melanoma-infiltrated immune cells, we applied XGBoost, achieving an initial AUC score of 0.84, which improved to 0.89 following Boruta feature selection. This analysis revealed an 11-gene signature predictive across various cancer types. SHAP value analysis of these genes uncovered diverse gene-pair interactions with non-linear and context-dependent effects. Finally, we developed a reinforcement learning model to identify the most informative single cells for predictivity. This approach highlights the power of advanced computational methods to deepen our understanding of cancer immunity and enhance the prediction of treatment outcomes.
Similar content being viewed by others
Introduction
Cancer immunotherapy, particularly immune checkpoint inhibitors (ICI), has revolutionized cancer treatment, offering durable responses in a subset of patients. ICI, such as anti-PD1 antibodies, work by reinvigorating exhausted T cells within the tumor microenvironment (TME), allowing the immune system to act effectively against tumor cells1. Despite their success, the majority of patients fail to respond to these therapies2, underscoring the need for reliable predictive biomarkers to identify patients who will likely benefit from treatment. Many efforts aim to decipher the behavior of the immune system against the tumor as a result of treatment3. In particular, characterizing the immune system in relation to different ICI responses and finding robust biomarkers to differentiate between responders and non-responders has become a major goal in the field of cancer immunity, often focusing on different cells in the TME4,5,6. Nevertheless, the complexity of immune interactions, and the context dependent behavior of the immune system to treatment are main obstacles in the endeavor for enhancing the efficacy of immunotherapy treatments.
Single-cell RNA sequencing (scRNA-seq) provides unparalleled insights into the cellular heterogeneity and dynamic states within the TME, capturing the complexity of immune responses at a granular level. However, despite the huge advances in the field of machine learning (ML), integrating this high-dimensional data with advanced models and techniques to predict therapeutic outcomes remains a challenge. Most existing approaches either oversimplify the data, losing valuable single-cell resolution7,8, or fail to provide interpretable results, limiting their clinical applicability9,10,11. Bridging this gap requires the development of innovative computational methods that retain data richness while offering clear insights into the biology underpinning the decision making of the models.
While machine learning in general has been widely adopted in the field of single-cell analysis12, relatively few studies use supervised learning to directly predict response to immunotherapy. For example, Dong et al. used cell-type proportion extracted from scRNA-seq data for training ML models to predict response to ICI8. In a different study7, Kang et al. used pseudo-bulk expression calculated from scRNA-seq. Prediction models with similar aims to ours have been developed, such as CloudPred13, which uses differentiable machine learning to predict lupus, and ScRAT14, which uses neural networks to predict COVID-19 phenotypes. However, these models have not been applied in the context of immunotherapy response.
Applying ML to scRNA-seq data raises a few challenges and open questions: for instance, should all single cells from a responding patient be labeled as ‘responders’, or should we use pre-existing knowledge on cell function to label cells as either “favorable’ or ’unfavorable’? How can we infer the patient class from the predicted classes of the associated single cells? Should we use all cells for prediction or perhaps a subset that is more predictive than others?
In this study we addressed these various questions by investigating scRNA-seq data of CD45+ cells from a cohort of ICI-treated melanoma patients15. We developed PRECISE (Predicting therapy Response through Extraction of Cells and genes from Immune Single-cell Expression data), a machine learning framework designed to predict immunotherapy response by and extract gene- and cell-level insights from scRNA-seq data. Using XGBoost (eXtreme Gradient Boosting) as the ML model16, our basic methodology involved labeling cells based on their sample’s response, training models in a leave-one-out manner, and then aggregating predictions to create a sample-level score. For feature selection (FS) we used Boruta17 which provided high performance in predicting patient response (AUC = 0.89). Following, we dissected the contribution of selected genes using feature importance scores and the more advanced Shapley Additive exPlanations (SHAP) values18, identifying complex, non-linear contribution of single genes and gene pairs to treatment response. Considering the cells-axis, we identified specific cell-types that are more informative than others in predicting response. Moreover, we devised a novel reinforcement learning model designed to identify single cells that are either predictive or non-predictive of patient response19. Applying these gene- and cell-based signatures to different datasets, we managed to achieve high prediction power across different cancer types. Additionally, our model demonstrated strong performance when applied on an external dataset comprising of multiple cancers. Overall, our findings highlight the potential of advanced computational approaches in enhancing our understanding of cancer immunity and informing treatment.
Results
Base model feature selection and per cell type prediction
We first utilized our previously published15 single-cell RNA sequencing (scRNA-seq) dataset that consists of 16,291 CD45+ cells from 48 tumor biopsies taken from 32 stage IV metastatic melanoma patients treated with ICI. 11 patients had longitudinal biopsies, and 20 patients had one or two biopsies taken either at baseline or during treatment. Tumor samples were classified according to radiologic evaluations as either progression/non-responder (NR; n = 31, including stable disease (SD) and progressive disease (PD) samples) or regression/responder (R; n = 17, including complete response (CR) and partial response (PR) samples). After the removal of non-informative genes (Methods), we constructed an XGBoost-based machine-learning model and investigated its power in predicting the patients’ response to ICI (Fig. 1a).
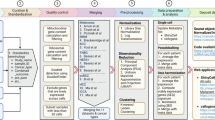
a Schematic workflow of the study: after preprocessing and quality control, the input data undergoes cell-type annotation to ensure a clean and well-annotated dataset (1). Each cell is then labeled according to its sample’s response status, and a classifier is trained at the single-cell level to differentiate responder cells from non-responder cells. The proportion of responder cells within a sample constitutes the sample score, serving as a prediction of likelihood to respond. This base model is then utilized in two main axes: (2 top, 3) interpretation of gene importance and behavior, using feature selection, feature importance analysis, and SHAP values; and (2 bottom, 4) analysis of cell importance, through cell-type prediction and a reinforcement learning framework for quantification of cell predictivity. (5) Finally, results from both axes are validated on independent datasets. b ROC curve for the base model predicting response to ICI using all cells and genes in the cohort. c XGBoost feature importance bar-plot showing the top 25 most important genes. d AUC scores indicating the prediction accuracy of the base model across different immune cell types. e Box plots comparing the scores produced by base XGBoost between responders (R) and non-responders (NR) across top most accurate cell subtypes, with significance values indicated by the Mann–Whitney U test. f ROC curve for the Boruta-selected model predicting response to ICI using all cells in the cohort. g AUC scores indicating the prediction accuracy of the Boruta-selected model across different immune cell types. h Bar-plot of the number of occurrences of each gene in the Boruta selection across the LOO folds, showing the top most robust genes. i Heatmap displaying the top genes selected by Boruta for different immune cell types, showing the number of occurrences of each gene for each cell type.
One of the main challenges in using supervised machine learning with single-cell data is the imbalance between the vast amounts of single-cells and the typically small number of samples from which these cells are taken. Naturally, the prediction target is often at the sample/patient level, raising the question of how best to assign labels to individual cells. To address this, we adopted a two-step approach: we trained the model and made predictions at the single-cell level, and then aggregated these predictions to devise a sample score. Each cell was labeled according to the response status of its sample of origin, such that cells from responding samples were classified as ‘responders’ and those from non-responding samples as ‘non-responders’. After predicting the labels for all cells in a sample, we calculated the proportion of cells predicted as ‘responders’, and this proportion constituted the sample score. The logic for this scoring could be described as a tug-of-war between ‘good’ and ‘bad’ cells. The more ‘good’ cells a sample contains, the more likely it is to be pulled towards a favorable response. Considering the small number of samples, training and testing was conducted in a leave-one-out (LOO) cross-validation manner, with each fold training the model on the cells from all samples except one, which was excluded for prediction. This approach also mitigates the stochastic effects often encountered in standard cross-validation. As our reference model, we included all 16,291 cells and all 10,082 genes that passed quality control (Methods). The ROC AUC score for this analysis was 0.84, showing reasonably high predictive accuracy (Fig. 1b, Supplementary Data 1). To further assess the reliability of LOO, we also performed stratified 5-, 7-, and 10-fold cross-validation with multiple random seeds. While AUC scores varied across seeds, the overall mean AUC over all k values and seeds remained consistent with LOO (0.84), confirming the robustness of our approach (Supplementary Data 2).
A slight refinement to this approach is to assign to each cell a probabilistic score rather than a binary classification for models that support probabilistic outputs (Methods). Currently, the final sample-level score reflects the proportion of ‘good’ cells. As a result, the key determinant of prediction is the fraction of responding cells and not their quality. The probabilistic approach allows for weighting each cell’s contribution to the overall sample response based on prediction confidence. This option was incorporated as a parameter in PRECISE, even leading to a slight improvement in prediction accuracy. However, for simplicity, interpretability, and generalizability, the binary classification approach was retained as the primary focus here.
To evaluate model performance, we compared XGBoost to several machine learning models, including LightGBM, Logistic Regression, Decision Trees, Random Forest, and simple multi-layered neural networks (Methods). XGBoost outperformed or performed comparably to most models (Supplementary Data 3). Interestingly, decision trees, despite their tendency to overfit, achieved high performance (AUC = 0.83–0.92, depending on depth). We speculate that the large number of cells relative to samples in this dataset makes decision trees more robust to noise than typically expected. Nevertheless, we focused our analysis on XGBoost, as it provides a balance between expressiveness and interpretability, making it the most suitable model for our approach. Decision trees, despite their strong performance here, may not generalize well in more complex datasets, as shown in later analyses (Discussion).
Additionally, we applied ScRAT14 to our dataset. Using its default parameters and adjusting it to a LOO framework, ScRAT achieved an AUC of 0.786, lower than our model’s AUC of 0.84 on the same dataset. Since ScRAT is based on transformer architectures, it inherently introduces greater complexity. While ScRAT’s attention mechanism provides some interpretability, our approach retains higher overall interpretability while achieving better predictive performance.
To identify genes that are important for predicting patient response to ICI, and can therefore serve as potential biomarkers, we used the feature importance metric, which measures the contribution of a feature to the prediction of the model based on the reduction in loss it achieves across the ensemble of trees. To quantify the importance of each gene in the cohort we extracted the importance score from each of the 48 folds in the LOO simulation, and averaged them across all folds (Fig. 1c). This analysis highlighted several genes as important for predicting patient response, including GAPDH, IFI6, LILRB4, GZMH, STAT1, LGALS1 and CD68 (Supplementary Data 4), all of which have already been found to relate to ICI and tumor immunity20,21,22,23,24,25. For example, CD68, a marker for macrophages, was found to be associated with poorer clinical outcomes in multiple cancer types in a pan-cancer analysis of expression data from TCGA and GTEx20. However, it is generally linked to M1 macrophages, which promote anti-tumor immunity26. STAT1 was found to promote immunotherapy response in melanoma21, and was found to be positively correlated with PD-L1 expression in ovarian cancer22. LILRB4, has been found to strongly suppress tumor immunity, with studies in murine tumor models demonstrating that its blockade can alleviate this suppression, resulting in antitumor efficacy23. Finally, GAPDH, receiving the highest feature importance score, has been shown to be involved in cancer progression and immune response regulation, particularly through its role in hypoxia-related pathways24,25.
Examining the sample scores revealed that while most samples received scores consistent with their response status, some samples were mis-scored. For example, sample Pre_P3, a non-responder, received a high score, whereas sample Pre_P28, a responder, received a low score (Supplementary Data 1). These discrepancies could be due to model performance, but they might also indicate specific traits of these samples that warrant further investigation. Specifically, sample Pre_P3 had a mutation in B2M limiting antigen presentation, demonstrating how factors not shown in the transcription are causing the tumors to respond or resist to treatment. In addition, sample Pre_P28 was unique in its high abundance of cells from the myeloid lineage, emphasizing the importance of the specific cells used for prediction. Following, and after establishing the prediction power of the base model that considers all genes and all cells, we investigated the model’s performance when using specific subsets of these entities.
Improving model performance by exploring cell and gene subsets
To improve model performance and study the contribution of different cell types and states, we first assigned individual cells to their respective cell type. We used the classification from the original paper which included 11 clusters determined by unsupervised clustering, and 13 cell types or states determined in a supervised manner using known gene markers15. We then applied the XGBoost model to each of the different cellular groups separately, using as input for each sample only cells from the given group. As expected, the per cell-type/cluster prediction resulted in different ROC AUC scores for each of the groups tested, testifying for their overall importance in predicting patient response. The results (Fig. 1d, e) show that T cells subsets achieve substantially higher scores in comparison to the rest of the cell-types (0.87–0.82), in concordant with their known role in immune response in general, and specifically in melanoma27. B cells (0.64), Macrophages (0.61) and other cell types were considerably less predictive. While the quantity of cells in each cellular groups is different and can potentially affect the accuracy of the model, this does not seem to be the dominant factor since there are T cell subsets that have a comparable number of cells to other cell-types, but produce much higher AUC scores (Fig. 1d). Intriguingly, the model’s accuracy was consistently higher for the supervised cell types compared to the unsupervised clusters (Supplementary Fig. 1), suggesting that the supervised clusters may capture more biologically meaningful signals and thus exhibit stronger predictive power.
We next moved to investigate if and to what extent feature selection would improve the base model predictions. To this end we explored 5 main methods for evaluating feature contribution - top highly variable genes (HVG), top differentially expressed, Lasso regression-based FS, simple top feature importance (Supplementary Data 5), and Boruta (Methods). Importantly, in all feature selection techniques applied, except for HVG method which is independent of response, we assured that selection was made independently from the predicted sample, thus avoiding any train-test leakage.
One of the main advantages of Boruta over other feature selection techniques is the automatic determination of the number of features17. While its hyperparameters can be tuned to adjust the number of selected features, the method works well without any tuning, providing a statistically justified feature selection rather than an arbitrary manual choice. Unlike simple top feature importance method, which can introduce variability, Boruta selects only the most consistently important genes, enhancing robustness. Therefore, although other feature selection techniques could offer slightly better accuracy for a specific choice of gene number or hyperparameters, Boruta remained the most reliable method for consistent and robust performance (Supplementary Data 5). The AUC scores produced by the other methods were mostly in the range 0.81-0.85, depending on the method and the hyperparameter, while Boruta produced a score of 0.89 (Fig. 1f), manifesting its superiority in this selection. Finally, to account for the observed variability in performance achieved for different cellular groups and gene subsets, we used Boruta feature selection to extract the top most contributing features on the base model and per cell-type, getting insight into the most important cell type-specific genes (Supplementary Data 6). To this end, we ran Boruta in the same manner as before, but this time focusing on each cell-type and cluster (Methods). As expected, the accuracy across most cell-types increased (Fig. 1d, g). Exploring the top genes in each group (Fig. 1h, i), we found genes with an established relation to cancer immunity and activation. For example, In NK cells, FCGR3B, known as CD16b, is involved in antibody-dependent cellular cytotoxicity (ADCC)28,29. It was suggested that NK cells mediate ADCC-driven cytotoxicity specifically against PD-L1-positive cells, demonstrating the role of ADCC in enhancing the effectiveness of immunotherapies, particularly those targeting PD-L130. In B cells, CXCR4 is expressed on the surface of mature B cells and interacts with CXCL12 to recruit regulatory B cells to the tumor site. These cells can inhibit T cell activity, contributing to an immunosuppressive tumor environment. HLA-DRB1 and HLA-DP1, both part of the MHC-II complex, were found in CD4+ helper T cells to be of the greatest importance. In addition, CD38+HLA-DR+ expressing CD4+ and CD8+ T cells were significantly expanded following ICI treatment in HNSCC (head and neck squamous cell carcinoma)31. GAPDH, CD38, STAT1 and IFI6 all have well established roles in T cells activation and immunotherapy, and therefore they appear robust in most T cells subtypes24,25,32,33,34,35. In addition to these general markers, more specific markers were extracted, such as PRF1 in cytotoxic CD8 T cell, a key component of the cytotoxic machinery of activated T cells, directly involved in killing tumor cells36.
SHAP value investigation reveals complex gene behaviors and interactions
While XGBoost’s feature importance provides some insight into the most contributing genes, its interpretability is limited. For instance, feature importance is non-directional, and thus does not indicate whether a gene is more associated positively or negatively with response. To address this, and considering that gene behavior may extend beyond simple linear or dichotomous relationships, we employed SHAP (SHapley Additive exPlanations) values18 (Methods). SHAP values, derived from cooperative game theory, measure both the direction and magnitude of a gene’s effect on the predicted response (Fig. 2a, b), allowing visualization of how gene expression influences it. Moreover, one of the strengths of SHAP values derives from its ability to capture context dependent contributions of each gene, such that the SHAP value for a gene can differ between two cells even if they share similar expression levels of that gene (Fig. 2c).
a 20 Top genes with highest absolute Shapley score identified in the model. b SHAP value summary plot depicting the impact of each of the most important genes on model output. Gene expression (shown by coloring) as a function of SHAP value (x-axis) showing the relation between expression pattern and model’s prediction. c Scatter plots showing the relationship between gene expression and SHAP values for key genes (GAPDH, STAT1, IFITM2, and HSPA1A). d Interaction plots illustrating the SHAP value dependencies between specific gene pairs: GAPDH & STAT1 (left), CD38 & CCL5 (middle), CCR7 & HLA-B (right). Expression of the first gene shown as x-axis position, and the expression of the interacting gene shown as coloring; SHAP value shown as y-axis position. Decision trees beneath the plots show the simplified relation of the gene-pairs conditional expression with response to ICI. e Interaction plots comparing SHAP value dependencies between gene pairs trained on all cells vs T cells: CCL5 & CD38 (left), CCL4 & HLA-B (right). Expression of the first gene shown as x-axis position, and the expression of the interacting gene shown as coloring; SHAP value shown as y-axis position. f Waterfall plots showing SHAP value contributions to individual model predictions (sample prediction). Four examples of patients predicted as responders or non-responders, with differences in gene expression patterns contributing to the outcome predictions. Pre_P35 – Responder predicted correctly, Pre_P2 – Non-Responder predicted correctly, Pre_P28 – Responder predicted as Non-Responder, Pre_P3 – Non-Responder predicted as Responder.
After training an XGBoost model over all of the cells in the cohort, SHAP values were calculated from that model, quantifying the contribution of each gene for each cell in the cohort. For example, as illustrated in Fig. 2c, GAPDH, and STAT1’s expressions are associated with non-response. The directionality of the SHAP value is such that high SHAP values are associated with response, and low SHAP values with non-response. However, the relationships of the two genes differ: GAPDH exhibits a continuous relationship with a non-zero threshold separating responders from non-responders, whereas STAT1 shows an on-off behavior, with zero expression linked to response and non-zero expression linked to non-response. Conversely, other genes such as HLA-G (Supplementary Fig. 2a) were associated with response. Additionally, SHAP reveals other, non-monotonic, gene behaviors. For example, HSPA1A (Fig. 2c) shows a complex pattern where both very low and very high expression are associated with response, while the middle range is more related to non-response. Another interesting example is of IFITM2 where zero expression is related to favorable response, the middle range has less effect on the prediction and high IFITM2 pushes the model towards non-response.
Going beyond the effects of single genes, we next investigated SHAP values for gene interactions. Focusing on the top most important genes that were found in the FS section, we examined their most interactive partners in predicting immunotherapy response (Fig. 2d). Notable pairs, showing a prominent conditional behavior in relation to SHAP, include GAPDH-STAT1, CD38-CCL5, and CCR7-HLA-B (additional pairs are shown in Supplementary Fig. 2b). To further simplify and enhance interpretability, we created decision trees for each gene pair. These trees align well with interaction plots, quantifying expression thresholds that best differentiate responses. As shown in Fig. 2d, the interaction between GAPDH and STAT1 highlights that cells with low GAPDH (<11.1) and high STAT1 (>2.45) are associated with non-response, whereas low GAPDH with low STAT1 is linked to response. High GAPDH is associated with response regardless of STAT1 expression. As previously mentioned, GAPDH plays a broad role in the immune microenvironment through hypoxia-related pathways, glycolysis-mediated acidification, and ferroptosis—all of which have been implicated in unfavorable immunotherapy responses24,25,37. Consistently, SHAP analysis highlights its interactions with multiple genes and their combined impact on response (Supplementary Fig. 3). The CCR7-HLA-B pair shows a different relation to response, where the separation by CCR7 is close to zero (<0.68), demonstrating an on-off conditionality, where low CCR7 is associated with non-response. In higher values of CCR7, HLA-B best separates between outcomes with expression threshold of 11.3, where lower HLA-B expression relates to response, and higher to non-response. Such interaction analyses constitute a great way of modeling conditional effects of genes - relationships that are challenging to find without advanced models like SHAP.
SHAP dependencies for the model that is trained on all cells, captures in part the cell-types and sub cell-types expression patterns. Given that cells in different clusters exhibit distinct expression profiles, a cluster associated with response or non-response is likely to reflect its unique expression pattern in the dependence plot. For example, cluster G1 (B cells, n = 1445), which is significantly more abundant in responders, shows a nearly 2.5-fold higher proportion of low GAPDH, low STAT1 cells compared to the entire cohort. In contrast, G6 and G11 (Exhaustion clusters, n = 2222 and n = 1129) are significantly more abundant in non-responders. Consistently, only a small fraction of these cells exhibits low GAPDH and STAT1 expression, with G6 showing a 3.3-fold lower abundance, and G11 a 20-fold lower abundance compared to its proportion in the cohort. To explore intra-cell-type dependencies, we reduce the influence of this effect by also focusing solely on T cells (Fig. 2e).
The T cell-specific analysis revealed that in certain gene pairs, interactions became more distinct. In particular, the separation of expression patterns was clearer in T cells for both CCL5-CD38 and CCL4-HLA-B (Fig. 2e), highlighting the importance of these interactions within this specific cell type. In the CCL4-HLA-B, the dependence is almost undetectable using the whole cohort, but with the T cells this is one of the strongest dependencies. In other gene-pairs, such as GAPDH-STAT1, the dependence is clearer in the whole cohort rather than in T cells, probably due to the effect of cell-type bound expressions described above.
Interestingly, all the chemokines in this analysis—CCL4, CCL5, and CCR7— have an on-off behavior. Non-zero expression, albeit low, of these chemokines is sufficient to dramatically alter the behavior of other genes in relation to the immune response (Fig. 2d, e). CCL4 and CCL5 can promote anti-melanoma immune response38, and are known to have a role in recruitment of immune cells39,40. They have also been found to promote tumor development and progression by recruiting regulatory T cells to the tumor site41. It is not surprising then, that their presence or absence can alter the effect of other genes on the immune response. This is especially true for HLA-B, which is critical for activating CD8+ T cells; the activation function can only be exerted following recruitment of these T cells, facilitated by chemokines like CCL4.
To further elucidate the behavior of the model, we leveraged SHAP values’ ability to deconvolve the contributing factors to each prediction, allowing us to understand the model’s choices. We analyzed the model’s performance to understand why certain samples were correctly or incorrectly predicted. We focused on four samples: Pre_P2 and Pre_P35 that were correctly predicted, and Pre_P3 and Pre_P28 that were misclassified. Using SHAP values, we trained the XGBoost model on all cells except those from the target sample, mimicking the actual prediction process. We then summed the contributions of all cells in each left-out-sample to obtain aggregated scores for each gene (Methods). These scores, sorted by absolute value, reveal the most critical genes for the model’s predictions (Fig. 2f, Supplementary Fig. 4). SHAP waterfall plots are commonly used for interpreting a single prediction, but here, since each sample contains many prediction targets (cells), the plots represent an aggregation of these values. Therefore, the scores for each sample depict the average contribution of each gene to the prediction of all the cells in that sample – showing a general trend rather than a single contribution value. For instance, Pre_P35 was classified correctly as a responder, primarily due to low GAPDH and HLA-B values, with smaller contributions from several other genes. Pre_P2 was correctly classified as a non-responder, mostly due to a high GAPDH expression, as well as a high HLA-B and IFITM2 expression and low HLA-G expression. Pre_P28 was misclassified as a non-responder due to the high HLA-C, TNFAIP3, and HLA-B expression, coupled with low HLA-G. Conversely, Pre_P3 was misclassified as a responder due to the average low expression of genes including GAPDH, HLA-B, HLA-C, FOS, STAT1, CD38, TSC22D3, and EPSTI1. Although a low HLA-G score pushed the model toward non-response, the cumulative effect of the rest of the genes was stronger.
Overall, the ability to interpret the model’s performance and decision-making is crucial for model improvement, gaining sample-level insights, and ensuring the model’s applicability in a clinical setting, which is the ultimate goal. Once the genes that affect the model’s prediction either positively or negatively are identified, it is possible to examine the distribution of these genes through the cohort and in the different cell-types.
Reinforcement learning enables labeling each cell’s predictivity
Our analysis revealed that certain subsets of T cells perform best in predicting patient response. However, it remains unclear whether every cell within this group is necessary to achieve that level of predictive accuracy, or if some cells may not contribute to the model’s prediction and even detract from its performance. Specifically, up until now we considered all the cells from a responding sample as responders and all the cells from a non-responding sample as non-responders. This approach represents a simplified model rather than a reflection of reality, as the TME is heterogeneous, and contains synergistic, contradictory, and inert factors. For example, while the proportion or exhausted T cells mostly associates with non-responding samples in baseline, they do exist also in responders42. To address that, we developed a novel reinforcement leaning (RL) framework that is built upon the basic model. Our goal was to quantify each cell’s effect on model prediction and create a cell-level score by rewarding cells with directional rewards when they are predicted correctly. By doing this, we move from a coarse, cluster-level measure, to a fine-grained, cell-level score.
In general lines, we aim to score each cell based on its directional contribution to the sample’s prediction. In order to achieve this, the RL framework should reward cells that are predicted correctly for their correct contribution to the model’s prediction, and penalize cells that are predicted incorrectly. Our methodology reflects this ambition by iteratively predicting the score of each cell and updating its value according to whether it was predicted correctly. The cells that are repeatedly predicted to the right direction—responders receiving positive scores and non-responders negative scores—accumulate higher absolute scores, reflecting their strong predictive value. Cells with the largest positive or negative scores are the most predictive cells, while those with scores near zero scores, are less predictive.
First, to get a continuous score, the XGBoost model was switched from a Classifier model to a Regressor model. To create the most accurate model possible, we only used the top genes from the FS section. This not only improved running time but also ensured a better distribution of the labels across samples. Initially, each cell from a responder sample is labeled 1, and each cell from a non-responder is labeled -1. Then, the model iteratively updates the continuous labels for each cell, ultimately assigning a score to each (Methods). Cells that are consistently predicted in the same direction will receive repeated rewards and therefore will be pushed to the edges of the distribution. In contrast, cells that are often predicted incorrectly will be iteratively normalized without any reward, resulting in scores with a low absolute value. These scores reflect the strength and direction of each cell’s contribution to the response of the sample (Fig. 3a, b). Large positive and negative scores indicate strong predictive power of positive and negative response, respectively, while close to zero scores indicate an overall weak association of these cells with patient response.
a tSNE plots showing RL prediction scores (left) and immune cell clusters (right). Clusters are colored by group, and the RL prediction scores range from highly predictive to non-predictive (lower limit is determined as -3 and upper limit as 6 for visualization purposes). b Histogram of RL prediction distribution. c Bar plots showing the proportion of non-predictive (top), non-responders predictive (middle), and responders predictive (bottom) cells by cluster. d Stacked bar plots representing the proportion of RL labels (R predictive, non-predictive, NR predictive) for each patient sample. Top 31 samples are Non-Responders and bottom 17 samples are Responders. e Dot plot of the top most differentially expressed genes between the three RL bins, showing log fold change of key genes in non-responders predictive (NR predictive), non-predictive, and responders predictive (R predictive) groups.
Examining the score distribution across clusters and cell types, we categorized each cell into three bins based on its score: ‘non-response predictive’ (< −0.5), ‘non-predictive’ (between −0.5 and 0.5), and ‘response predictive’ (>0.5). Figure 3a illustrates the distribution of these bins across the cohort. There is a clear association between the proportion and direction of predictive cells within each cluster and the relation of the cluster to response (Fig. 3c), as determined by the original study15. Specifically, clusters G3 (Monocytes/Macrophages), G6 (Exhausted CD8 + T-cells), and G11 (Lymphocytes exhausted/Cell cycle), which are prevalent in non-responders, also had the highest proportion of non-response predictive cells. Conversely, clusters G1 (B-cells) and G10 (Memory T-cells) that are associated with responders, were the top clusters in terms of response predictive cells. This analysis also highlights clusters G9 (Exhausted/HS CD8+ T-cells) and G7 (Regulatory T-cells), which have a relatively high prevalence of non-response predictive cells, and cluster G5 (Lymphocytes), which contains more response predictive cells, although with a lower proportion (see Supplementary Fig. 5 for cell-type RL distribution). This observation serves as a sanity check, reinforcing the validity of our model.
Notably, non-predictive cells are well distributed across most clusters and are prevalent throughout the cohort. This might result from the arbitrary threshold used to bin the cells, however, examining the distribution of the scores across the cohort shows that most of the non-predictive cells are indeed close to zero and are therefore not affected by the specific threshold used (Fig. 3b). In addition, it is important to note that these cells have passed rigorous quality control, so their non-predictive phenotype is not likely due to low-quality data but rather suggests a different level of participation in immunity. Finally, the spreading of non-predictive cells across different clusters and cell-types suggests that simply filtering cell-types or focusing on a single cluster is not sufficient, emphasizing the potential need for a finer method to filter cells and narrow the analysis to predictive cells.
Following, we examined the distribution of RL labels within each sample (Fig. 3d). As expected, the proportion of predictive cells in well-classified samples was much higher, while misclassified samples had a higher proportion of non-predictive cells. Although likely reflecting to some extent the accuracy of the model, this also highlights the heterogeneity of the TME both within individual samples and across different samples.
To characterize these cells and understand what makes a cell predictive or non-predictive of patient response, we performed differential expression analysis among the three bins, as shown in Fig. 3e. As expected, the genes used in the RL model appeared in the top differentiators list, but other significant genes also emerged, suggesting that the RL model manages to generalize and create biological groups. These include PKM, PGAM1 and ACTB43,44,45 in non-response predictive, TCF7, IL7R in non-predictive, and SELL, MS4A146 in response predictive cells. Although some biological genes were extracted as differentiators of non-predictive cells, this group does not seem to have strong markers differentiating it from the rest of the cells.
Quantifying individual cell participation in immune response provides novel, granular insights into the tumor microenvironment. This approach also offers an avenue for identifying and filtering out non-contributory cells, refining predictions related to immunotherapy response.
Gene- and cell-based signatures predict patient response to ICI
To evaluate the generalizability of our findings, we first constructed a gene signature differentiating between responders and non-responders (Fig. 4a). To this end, we extracted the genes with the highest feature importance scores from each fold of the LOO (Methods). Analyzing the intersection of genes from the 48 folds, we selected the top 11 genes that consistently intersected across the folds: GZMH, LGALS1, GBP5, HLA-DRB5, CCR7, IFI6, GAPDH, HLA-B, EPSTI1, CD38, and STAT1. Although somewhat arbitrary, the number of genes selected was based on the minimum number of top genes needed from each fold to form the intersection group: the top 11 genes required relatively few top genes from each fold, whereas those further down the list required a larger number of intersecting genes, making them less robust (Supplementary Fig. 6). Pathway enrichment analysis of the 11 genes (Methods)47 found pathways related to immune activation, interferon signaling, and T cell related pathways, including cytokine signaling in the immune system, and immune system regulation (Supplementary Data 7). By examining the SHAP distribution for these genes’ expression, we determined that all except CCR7 were negatively correlated with response. Following, the score we devised was an equally weighted combined score of the genes in the signature, with the weight of CCR7 specifically negatively adjusted (Methods).
a T Cell ROC curves showing the predictive power of the 11-gene signature score combined with RL-based filtration in distinguishing responders from non-responders across multiple cancer types. The blue curve represents the ROC curve before filtration, and the red curve represents the ROC curve after filtration. Box plots, separated into pre- and post-treatment samples (where available), depict the 11-gene signature scores before RL filtration. The datasets included are: Melanoma – Sade-Feldman et al.15, TNBC (Triple-Negative Breast Cancer) - Zhang et al.48, NSCLC (Non-Small Cell Lung Cancer) – Caushi et al.49, BCC (Basal Cell Carcinoma) – Yost et al.51, Glioblastoma – Mei et al.50, HER2+ & ER+ & TNBC - Bassez et al.52, Breast Cancer - Tietscher et al.54. b Kaplan-Meier survival analysis of the 11 genes in the signature using bulk RNA-seq data across various cancer types. The survival curves illustrate the association between high and low expression of each gene. Plots include survival for genes – GAPDH, CD38, CCR7, HLA-DRB5, STAT1, GZMH, LGALS1, IFI6, EPSTI1, HLA-G, GBP5.
To mitigate biases arising from differences in cell-type abundance and considering that some datasets contain only T cells, we focused exclusively on T cells for all of our validation datasets. First, we evaluated the scores on the training melanoma cohort, which produced a high AUC score of 0.93 (n = \(48\), p-value = \(6.16\times {10}^{-7}\)). Importantly, a significant separation was observed when examining baseline and post-therapy samples separately (n = \(19\) and \(29\), p-values of \(3.5* {10}^{-4}\) and \(3.9* {10}^{-4}\), respectively).
To establish the robustness of this score, we analyzed additional publicly available datasets having scRNA-seq data from patients treated with ICI (Fig. 4a). In a triple-negative breast cancer (TNBC) dataset, our predictor achieved an AUC of 0.95 (n = \(13\), p value = \(0.003\))48, and baseline samples were perfectly separated using the score receiving an AUC of 1 (n = \(8\), p-value = \(0.018\)). In a non-small cell lung cancer (NSCLC) dataset containing only post-treatment samples, our score metric achieved an AUC of 0.76 (n = \(57\), p-value = \(5.5* {10}^{-4}\))49. Two additional datasets showed a positive trend in association with the score, though they were not as significant with this simple score: a glioblastoma dataset and a basal-cell carcinoma (BCC) dataset both receiving an AUC score of 0.68 (n = \(11\), p-value = \(0.2\) and n = \(22,\) p-value = \(0.08\), respectively)50,51. The BCC dataset showed a relatively better separation using only baseline samples (AUC = \(0.77\)), though due to the smaller sample size, this separation remained insignificant (n = \(11\), p-value = \(0.09\)).
Next, we applied our predictor to a dataset of HER2 positive, ER positive and TNBC patients having annotations for clonal expansion phenotype52. The dataset consists of two cohorts, one of treatment naive samples, and the second of patients receiving neoadjuvant chemotherapy prior to anti-PD1. In both cohorts, the score was significantly associated with expansion, with the first cohort receiving an AUC of 0.81 and the second 0.77 (n = \(58\), p-value = \(8.1* {10}^{-5}\), and n = \(22\), p-value = \(0.03\)). Combined, these datasets achieved an AUC score of 0.79 (n = \(80\), p-value = \(2.8* {10}^{-5}\)). This trend remained significant when separating baseline and post-treatment samples. This finding suggests that samples with expanded clonal populations are more likely to be non-responsive. This also aligns with a study in melanoma patients, which showed that highly expanded clonotype families were predominantly distributed in cells with an exhausted phenotype, characterized by decreased diversity of T cell receptors (TCRs)53. Lastly, we applied our signature to a dataset of breast cancer patients having a classification for exhausted and non-exhausted TME54. This data predominantly contained samples from treatment-naive breast cancer patients. Our predictor achieved an AUC of 0.86 and significantly differentiated between these two TME states (n = \(14\), p value = \(0.01\)).
Following, we sought to validate the RL based filtration of non-predictive cells by constructing a cell-filtration score (Methods). This score guided the removal of cells from each dataset, after which we reapplied the 11-gene signature to the remaining cells and tested for prediction accuracy (Fig. 4a). While the genes identified in the differential expression analysis in the previous section effectively differentiate response-predictive from non-response-predictive cells, as aforementioned, non-predictive cells had less obvious distinguishing markers (Fig. 3e). Since we aim to apply the score to remove non-predictive cells in new datasets, a more sophisticated way of characterizing these cells is needed. To address this, we used a logistic regression model to predict who are the non-predictive cells. Although this classifier cannot be directly applied to other datasets due to differences in scale and potential batch effects, we can derive a more generalizable score from it that can be applied to new data. The filtration score was built according to the logistic regression classifier of non-predictive cells, using the coefficients of the top 100 most important genes (Supplementary Data 8). A weighted mean over all these genes created a separation between the different RL classes (Supplementary Fig. 7), and determined the RL-based filtration-score (Methods).
To balance the removal of non-predictive cells while minimizing the risk of filtering out predictive ones, we chose to remove 40% of the cells with the highest scores across all datasets. The results were robust to the threshold choice between 15-60%: as expected, less filtration led to smaller improvements, while more filtration resulted in greater improvements, up to a certain point (Supplementary Data 9). In the melanoma dataset that was used for devising the gene and cell scores15, the cell-filtration improved the AUC score to over 0.94 (n = \(48\), p-value = \(2.55\times {10}^{-7}\)). Applying this filtering to the other datasets used for validation, achieved an improvement in the prediction accuracy in five out of the six. Specifically, the glioblastoma dataset showed a substantial improvement in AUC, from 0.68 to over 0.9 (n = \(11\), p-value = \(0.03\)). Filtration in the BCC dataset produced an improved AUC of 0.7 (n = \(22\), p-value = \(0.06\)). The HER2 positive ER positive and TNBC dataset consisting of the two cohorts, saw an increase in AUC to 0.8 in relation to expansion (n = \(80\), p-value = \(1.3* {10}^{-5}\)). The AUC of the breast cancer cohort labeled with exhaustion increased to 0.88 (n = \(14\), p-value = \(0.009\)). In the TNBC48 the AUC was improved from 0.95 to 0.98 (n = \(13\), p value = \(0.0016\)). In the NSCLC dataset the AUC score was reduced to 0.73.
Gene signature predicts patients’ survival
Finally, we examined the specific genes that make up the 11-gene signature, using bulk RNA-seq datasets to assess their association with patient overall survival (Fig. 4b). It has been shown that immune activation is related to improved survival in different cancer types55,56,57. We expected, therefore, that higher immune responsiveness would be associated with better survival, hence the signature should differentiate survival rates in this direction. Of note, validation in bulk RNA sequencing is challenging due to the mixture of different cell types. However, since survival analyses look at outcomes across a cohort, they are less dependent on the precision of cell-type-specific resolution. What matters is whether the overall expression of these immune genes, in the context of the mixed population, correlates with better or worse outcomes for the patients. Moreover, strong immune-related signals can still emerge because immune gene expression often reflects the overall immune activity in the tumor microenvironment.
To examine this, we used the GEPIA2 online tool58, and assessed the relation between each of the 11 genes and survival. Using the median expression as a cutoff and using all cancer types available in GEPIA (Methods), we found that 7 out of the 11 genes, CCR7, STAT1, GBP5, LGALS1, EPSTI1, IFI6 and GAPDH, were significantly differentiating the overall survival time in the expected direction (after Bonferroni correction). The remaining 4 genes, HLA-DRB5, HLA-B, GZMH and CD38, showed no significant association with survival in either direction (Fig. 4b). The fact that this signal comes from all cancer types and shows that patients with high expression of these genes in their tumor exhibit an improved survival rate, strengthens our hypothesis that this could indeed be the immune related activation.
Discussion
In this study, we developed PRECISE, a machine learning framework designed to predict response to ICI from single-cell RNA-seq data. Our aim was to provide a roadmap for how single-cell data can be used in prediction tasks, such as treatment response, while maintaining model interpretability. Machine learning models stand out as the only technology able to capture the complexity level seen in the TME and reflected in single-cell data. The main advantage of these supervised learning models is that they are trained directly on the target and adapt to the characteristics of the data, instead of merely searching for patterns linked to the target.
The ability to work simultaneously on two levels of complexity—sample-level and cell-level—is crucial for any model designed for single-cell prediction and interpretation. Our model enables exactly that: initially, by assigning each cell a label according to its sample; next, by combining cell-level predictions into a sample response score; later, by aggregating the single-cell effects on the model using SHAP and grouping them into sample contributions; and lastly, through the reinforcement learning model, diffusing the sample responses back down to assign a predictivity score to each cell. This approach leverages the large amounts of single-cell data to effectively train machine learning models, which are known to require an abundance of data. At the same time, it ensures that meaningful conclusions can be drawn at the sample level, creating a robust and interpretable prediction framework.
While ROC AUC was chosen as the primary evaluation metric due to its threshold independence, practical applications require selecting an optimal threshold based on clinical or policy-driven objectives—such as minimizing false positives (FPR) or false negatives (FNR). A threshold can be optimized using cross-validation. Nonetheless, in the melanoma dataset used for training, the default threshold of 0.5 yielded the highest accuracy (0.833 for the base model). However, since the proportion of responder samples may influence predictions, aligning the threshold with this proportion could serve as an alternative baseline, offering an intuitive approach without requiring additional optimization.
We decided to focus on Boruta as our main feature selection method because it proved to be both reliable in terms of prediction accuracy and, just as importantly, did not require manually choosing the number of selected genes. For the base model and T cells, Boruta dramatically outperformed other methods, even when their parameters were optimized (0.89 with Boruta compared to 0.83–0.855 for the others). Lasso achieved slightly better AUCs in a few specific cell types for certain alpha values, but the differences were minor (Supplemental Data 5). Boruta proved to be a reliable choice when tree-based ML models are used.
Our model is highly flexible, both in terms of implementation and objective. Although we used XGBoost, many other algorithms with sufficient capacity could handle the data complexity equally effectively. Additionally, we opted to include all genes that passed preprocessing steps, capturing the strongest signals across the data. However, this approach may overlook substantial subsets of the data. Future analyses could focus on specific gene groups (e.g., metabolic genes) to explore different dimensions and reveal new insights.
Differences between cancer types, as well as the differences in prior treatments and medical backgrounds, make transferability of results from one dataset to another very challenging. Despite these challenges, the 11-gene signature showed a positive trend in all datasets, indicating the robustness and generalizability of the model’s results. The RL filtration score also showed a positive trend in most datasets, improving prediction accuracy and effectively removing noisy, non-predictive cells.
Focusing on the immune system is an excellent way to establish transferability between cancer types, since while the cancers can greatly differ, the immune system retains common features. This likely explains why our model’s results could be generalized across different cancer types. While it is encouraging to see that markers identified in a single dataset are predictive in several other datasets, it is likely that the resulting genes are not universally applicable. Moreover, the model is expected to perform poorly if applied directly on separate datasets without proper integration. In fact, large batch effects could be present even within a dataset between different samples, possibly affecting performance. To examine this we applied our approach on a dataset comprising of multiple cohorts from different cancer types59, including 139 samples of patients treated with ICIs. The data was normalized to create a standardized dataset while maintaining sparsity, which is an important factor in terms of computational efficiency (Methods). The prediction was performed in a 10-fold cross-validation manner, and several machine learning models and parameters were explored, including XGBoost, LightGBM, Logistic Regression, Decision Trees, and three simple multi-layered neural networks. These models achieved high predictive performance, with AUC scores ranging from 0.815 to 0.89 (Supplementary Fig. 8, and Supplementary Data 10). These results demonstrate the generalization ability of the model, and its potential applicability, especially after proper data integration. Interestingly, Logistic Regression outperformed all other models, while Decision Trees, which previously achieved the highest accuracy, performed the worst. Future work should aim to broaden the application of this model, possibly by utilizing foundation models to effectively integrate single-cell data into a unified embedding60,61 (interpretability should be kept in mind). Expanding the model’s input data should enhance model robustness and enable new possibilities, including adding a high-level model to aggregate scores from different cell types alongside sample-level features—such as age, gender, and cell-cell communication—to generate a more comprehensive and accurate overall sample score.
Another point to consider is the use of both baseline and post-treatment samples. While the ultimate objective of such models is the prediction of patient response before treatment, we included in our training data both pre- and post-treatment samples to maintain as much data as possible for the training step. Nonetheless, we found that the prediction accuracy was similar in the validation datasets when estimated on pre-treatment samples alone, and in some cases even produced better separation. Additionally, it should be noted that prediction on post-treatment samples has a value in cases of advanced line ICI re-challenge, for patients not responding to first line treatments. Predicting patient response to ICI in post-therapy samples is crucial for identifying dynamic biomarkers that reflect therapy-induced changes in the tumor microenvironment and immune system. Unlike pre-therapy markers, post-treatment signatures can provide real-time insights into treatment efficacy, resistance mechanisms, and patient stratification for further intervention. This approach enables early identification of non-responders, guiding adaptive treatment strategies and optimizing clinical outcomes. Moreover, it aids in refining therapeutic targets and improving the design of future immunotherapy trials.
This study demonstrates the potential of machine learning models trained on single-cell data to predict immunotherapy responses and provide deep biological insights. The two-step approach employed here enables the transition between sample- and cell-level predictions, providing granular interpretability unavailable in parallel methods. Moreover, this is the first application of such an approach in the context of ICI response prediction, making this work a novel contribution to immunotherapy research. Our approach of reinforcing well-predicted cells to assess their directional influence on response classification is, to our knowledge, introduced here for the first time. We believe that this demonstrates the potential of multi-level predictive modeling and encourages further innovation in this direction. Using PRECISE (Predicting therapy Response through Extraction of Cells and genes from Immune Single-cell Expression data), we offer a roadmap through which single-cell data can be used not only for sample predictions, but also for understanding complex biological systems such as the TME. Machine learning models in this context can go beyond mere prediction – we showed their utility in capturing gene-gene interactions, identifying non-linear expression patterns, and quantifying cell-type and single-cell participation in immune response. With sufficient data and further development, these models could become exceedingly powerful tools. Expanding training to larger, multi-dataset cohorts will be crucial for improving generalization.
Methods
Data collection and preprocessing
We utilized a publicly available scRNA-seq dataset containing immune cells from melanoma patients, generated using Smart-seq technology15,62. The dataset included 16,291 immune cells from 48 samples. Following initial quality control done by the authors, preprocessing primarily involved filtration of genes based on expression levels. Specifically, we excluded non-coding genes, mitochondrial genes, and ribosomal protein genes (genes starting with ‘MT-‘, ‘RPS’, ‘RPL’, ‘MRP’, and ‘MTRNR’). Additionally, only genes expressed in at least 3% of cells were retained, resulting in 10,082 genes. Cells were labeled according to their sample’s response to ICI therapy, with cells from responding samples labeled as responders and those from non-responding samples labeled as non-responders.
Additional datasets were collected and preprocessed for validation, including droplet-based datasets containing read counts48,49,50,51,52. In the droplet-based datasets, expression levels were first normalized to a standard sum of 10,000 reads per cell and then log-transformed. It is important to note that the per-cell read coverage in these datasets was often higher than 10,000. Only samples with defined clinical annotations (response or clonal expansion) were included in the analysis. We considered only patients treated with either immune checkpoint inhibitors (ICI) or a combination of ICI and chemotherapy. Additionally, a dataset of a non-ICI cohort containing mostly treatment-naive patients with annotations for exhausted and non-exhausted tumor microenvironment (TME) was included54.
Cell-type assignment and unsupervised clustering
In our previously published melanoma dataset15, we used the original classification of T cells that was based both on gene markers and manual curation. Eleven unsupervised clusters were identified using k-means clustering, and cell-type assignments were determined through supervised classification based on marker genes, as reported in the original article. In most other datasets used for external validation, T cells were either extracted using flow cytometry, or were identified in silico using computational approaches in the paper49,50,51,52,54 generating the data. The TNBC dataset48 was the only dataset without cell-type annotations. Identification of T cells in this dataset was done by unsupervised clustering, followed by differential expression on these clusters and determination of T cells according to known markers.
Base machine learning model
Training was conducted in a leave-one-out (LOO) cross-validation manner, where the model was trained on all samples except one, and predictions were made for the left-out sample. The model was trained and used for prediction at the single-cell level, learning to predict the label associated with the sample’s response. After predicting the labels of all cells in the cohort, we calculated the proportion of cells predicted as responders in each sample, forming the sample score. Accuracy of the prediction was evaluated using the Receiver Operating Characteristic (ROC) Area Under the Curve (AUC) score between the sample scores and the sample response labels, making the accuracy assessment robust to the threshold choice. From each model fold, feature importance scores were extracted for each gene using the model’s built-in ‘feature_importance_’ attribute, which quantifies each gene’s contribution to the model’s prediction.
Probabilistically weighted extension of the base model
For models that produce probabilistic outputs (e.g., XGBoost), we implemented an alternative approach using ‘predict_proba’ function, which assigns each cell a probability score between 0 and 1, reflecting its confidence in being classified as a responder. Instead of a binary classification, the final sample-level score is computed as the mean probability across all cells, effectively weighting each cell’s contribution based on prediction confidence. This approach has been incorporated as an optional parameter in models that support probabilistic predictions.
XGBoost classifier supervised predictive model
An XGBoost model, known for its balance between accuracy and interpretability, was trained to predict the response label of each cell16. The XGBoost model was built using the XGBoost Python package. The classifier was trained using the default parameters for XGBoost, except for a learning rate (LR) of 0.2, a max_depth of 7, and an alpha of 1. The model’s performance was not sensitive to these parameters. No hyperparameter tuning was required. The objective function was set to ‘binary:logistic’. An important parameter included was the ‘scale_pos_weight’, set to the ratio of the number of negative cells to positive cells, addressing potential bias towards responders or non-responders in the data. Although accuracy was high without it, including this parameter ensures better generalization for more biased data.
Feature importances metric for XGBoost
The feature importance parameter was set to ‘gain’, which is the default parameter. The ‘gain’ feature importance calculates the average reduction in loss that the feature causes.
-
1.
Per-Split Gain: Each time a feature is used in a split, the reduction in the objective function (cross-entropy loss in our case) is recorded as \({gain}(s)\).
-
2.
Total Gain for a Feature: Summing the gain across all splits where a feature g is used:
where \({S}_{g}\) are all splits using g.
Average Gain: The total gain is divided by the number of times the feature appears in splits:
Final Feature Importance: To normalize feature importances so they sum to 1, the average gain is divided by the sum of average gains across all features:
ensuring FI sums to 1.
The exact calculation can change in different implementations, but the measure is the same – how much a gene’s expression contributes to reducing loss and improving model accuracy.
Different models evaluation, and K-fold CV prediction
To evaluate the robustness and generalizability of our approach, we tested multiple machine-learning models beyond XGBoost. These included Logistic Regression, Decision Trees, Random Forest63, LightGBM64, and Neural Networks with varying architectures (Supplementary Data 3). For neural networks, we tested three architectures: NeuralNet_1 ([64, 32] nodes), NeuralNet_2 ([128, 64, 32] nodes), and NeuralNet_3 ([32, 16] nodes), using different learning rates and epoch numbers to optimize performance. The default settings included 10 epochs and a learning rate of 0.005, with additional parameter combinations explored: epochs [5, 20, 40] and learning rates [0.001, 0.01]. For Decision Trees, max depths of [4, 6, 8, 10, 12] were tested. All other models were set with default parameter values. Model performance was evaluated using ROC AUC scores derived from LOO cross-validation. Neural networks were implemented in PyTorch65 using the Adam optimizer and ReLU activation function for hidden layers66. LightGBM64 was executed using the LightGBM Python package, while all other models were run with scikit-learn63.
To assure the validity of the LOO CV, we performed K-fold cross-validation with stratified 5-, 7-, and 10-fold splits. Each fold maintained the responder/non-responder ratio, ensuring balanced class representation across training and validation sets, splitting by sample. Cross-validation was repeated across five random seeds (1–5), and mean AUC scores were computed to assess variability and robustness (Supplementary Data 2).
Per cluster/cell-type prediction
Cellular groups we’re extracted from the original paper15, and included 11 clusters determined by unsupervised clustering, and 13 cell types or states determined using known gene markers. To examine cluster- and cell type-specific predictions, we ran the classification model with LOO on each cellular group, and obtained an AUC score for each cluster independently. Samples were excluded from the leave-one-out process if they didn’t contain any cells from the specific cellular group, and the accuracy scores were adjusted accordingly.
Boruta feature selection
We used the boruta_py from the Python Boruta package. Boruta is a wrapper algorithm that iteratively removes features that are statistically less important than random probes (shadow features). We used Boruta to extract only the confirmed genes from its output in a leave-one-out fashion. We set alpha to 0.02, which produced a small, robust number of features. The chosen features were saved for interpretation and used for LOO prediction, ensuring no train-test leakage.
Lasso regression based feature selection
Lasso regression, a technique that performs both variable selection and regularization, was used for feature selection. Lasso zeroes the coefficients of non-contributing features. Using different values of the regularization parameter alpha, we performed LOO predictions with Lasso-based feature selection. Although results were reasonable, they were consistently lower than those obtained with Boruta and required handpicking of the alpha value. Feature selection was done after scaling the genes, using Lasso from Python’s sklearn package (with max_iter set to 500).
Simple feature selections
We also examined the performance of simple feature selection methods, such as top differentially expressed genes and the most highly variable genes. Differential expression analysis between responding and non-responding samples was conducted within the LOO using either Wilcoxon ranksum test, t-test (scanpy), or Fisher’s exact tests with significance values of 0.05, 0.01, and 0.001. Highly variable genes were extracted using scanpy’s function on the top 1000, 2000, 4000, and 8000 top highly variable genes. The Wilcoxon ranksum test produced the best results among these methods but still yielded consistently lower performance compared to Boruta and resulted in a much higher number of genes.
SHAP analysis
SHAP (SHapley Additive exPlanations) values were used to interpret the model’s predictions. SHAP values explain the impact of each feature (gene) on the model’s output, providing insights into gene importance and interactions. We applied SHAP using the SHAP Python package on the genes: GZMH, LGALS1, GBP5, HLA-DRB5, CCR7, IFI6, GAPDH, HLA-B, EPSTI1, CD38, STAT1. SHAP automatically identified gene interactions and the mean contributions of genes to a sample’s prediction. We averaged the SHAP scores for each gene across the sample to generate sample- level Waterfall plots (Fig. 2, Supplementary Fig. 4).
Decision trees for SHAP interaction analysis
To model the pairwise conditional behaviors of genes as extracted by SHAP analysis, we used decision trees. Each time, we insert the expression pattern of two genes extracted by SHAP to the decision tree model. We used the DecisionTreeClassifier from python’s sklearn with max_depth set to 2. This limit on the depth helped exhibit a simple relation between the two genes.
Reinforcement learning framework
To quantify the directional predictivity of each cell, we constructed a reinforcement learning framework based on XGBoost, but this time we used a regression model.
Initially, responder cells are assigned a score of 1 and non-responders -1. In each iteration, the model is trained on all but one sample (LOO) and then predicts scores for the cells in the excluded sample. Cell scores are updated based on whether they align with their sample’s response label—positively for responders and negatively for non-responders. If the prediction matches the true response, the score is reinforced by a small factor scaled by the prediction value; otherwise, it remains unchanged. For example, a cell from responder sample receiving a prediction of 5, will be updated by a reward of eta*5; a cell from a non-responder predicted the same will not be rewarded. At the end of each iteration, scores are normalized to maintain scale and penalize misclassified cells. This process runs for 200 iterations with an adjustable learning rate of 0.01, gradually amplifying the most predictive cells. The pseudocode below illustrates this process:
Initialization
-
1.
Initialize responder cells with label 1 and non-responder cells with label -1
-
2.
Set the list of chosen features to those extracted with feature selection
-
3.
Set the learning rate (eta = 0.01) and the number of iterations (200)
Iteration Process
For each iteration:
-
1.
For each sample:
Train the model on all samples except the current sample.
Predict the labels for the current sample.
for each cell in sample:
-
If the sample is a non-responder and the prediction value is negative, or if the sample is responder and the prediction value is positive:
-
Update the label: label = label +eta*prediction
-
Else:
-
Do not update the label.
-
2.
Normalize the absolute updated value of the labels to maintain their scale.
Return:
Return the updated labels.
The labels were then classified into ‘Responder Predictive’ for \({label} > 0.5\), ‘Non-Responder Predictive’ for \({label} < -0.5\), and ‘Non-Predictive’ for \(-0.5 < {label} < 0.5\). These groups were then characterized with differential expression analysis.
Application of 11-gene signature on external datasets
We created a pseudo-bulk expression matrix by averaging the expression of each gene across all T cells in each sample. To normalize the differences in the scale of expression among different genes, we divided the expression of each gene in a sample by the total expression of that gene across all samples. Formally,
Where P is the pseudo-bulk expression matrix with the samples as rows and genes as columns, and E is the new, scaled matrix. The normalization term in the denominator sums the expression of a given gene over all samples in the dataset.
After calculating the scaled pseudo-bulk, the score for each sample was calculated as follows:
This score was then used to differentiate between the different responses in the samples.
Pathway enrichment analysis of top features
Pathway enrichment analysis of the 11-gene signature was performed using the Reactome online tool47, following standard enrichment settings. Only human related pathways were included. The significance levels before and after multiple-hypothesis correction, number of intersecting genes per pathway and other results were recorded in Supplementary Data 7 to assess the contribution of the 11-gene set to the identified pathways.
Cell filtration score calculation and application
The filtration score was calculated based on a logistic regression model. The model was trained to differentiate non-predictive cells from the rest of the cells (response predictive and non-response predictive) using the python LogisticRegression algorithm from sklearn63, with the max_iter parameter increased to 500 to assure convergence. The 100 genes with the largest absolute coefficient values were extracted from the model and used to create a weighted score (Supplementary Data 8).
Given the vector of the weights extracted by the logistic regressor and the expression matrix E normalized for each gene’s expression:
The score for a specific cell is calculated by the dot product of these two vectors
The cells with the largest scores are most likely to be non-predictive. The cells with the top 40% scores were filtered out in all datasets.
Bulk RNA-seq survival analysis
GEPIA2, an online tool for analyzing RNA sequencing expression data, was used to examine the relation of the expression of the 11 genes to overall survival58. Using the median expression as a cutoff, this produced Kaplan-Meier survival curves for each gene, over all available cancer types in GEPIA2. These include: ACC, BLCA, BRCA, CESC, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LAML, LGG, LIHC, LUAD, LUSC, MESO, OV, PAAD, PCPG, PRAD, READ, SARC, SKCM, STAD, TGCT, THCA, THYM, UCEC, UCS and UVM.
Data Availability
All data used in this study is published and publicly available. The scRNA-seq data for the melanoma training cohort used in this paper is available through accession number GEO: GSE120575. TNBC scRNA-seq data was downloaded from GSE169246. NSCLC scRNA-seq data was downloaded from GSE176021. Glioblastoma scRNA-seq was downloaded from Figshare (https://doi.org/10.6084/m9.figshare.22434341). BCC was attained from GSE123813. HER2 positive, ER positive and TNBC scRNA-seq data was accessed from the Lambrecht’s lab website (https://lambrechtslab.sites.vib.be/en/single-cell). Breast cancer scRNA-seq data classified by exhaustion was obtained from ArrayExpress database at EMBL-EBI under E-MTAB-10607. The integrated dataset was downloaded from the CELLxGENE Discover database (https://cellxgene.cziscience.com/collections/61e422dd-c9cd-460e-9b91-72d9517348ef).The methods described in this manuscript have been implemented in a tool called PRECISE, which is freely available at the GitHub repository. The repository also includes a tutorial for its use: https://github.com/yizhak-lab-ccg/PRECISE-.
Code availability
The methods described in this manuscript have been implemented in a tool called PRECISE, which is freely available at the GitHub repository. The repository also includes a tutorial for its use: https://github.com/yizhak-lab-ccg/PRECISE-.
Abbreviations
- scRNA-seq :
-
Single-cell RNA sequencing
- TME :
-
Tumor microenvironment
- ICI :
-
Immune checkpoint inhibitors
- LOO :
-
Leave-one-out
- ROC :
-
Receiver Operating Characteristic
- AUC :
-
Area Under the Curve
- SHAP :
-
SHapley Additive exPlanations
- RL :
-
Reinforcement learning
- FS :
-
Feature selection
- HNSCC :
-
Head and neck squamous cell carcinoma
- BCC :
-
Basal-cell carcinoma
- NSCLC :
-
Non-small cell lung cancer
- TNBC :
-
Triple-negative breast cancer
- ADCC :
-
Antibody-dependent cellular cytotoxicity
- ML :
-
Machine learning
- NR :
-
Non-responder/Non-responsive
- R :
-
Responder/Responsive
- LR :
-
Learning rate
- HVG :
-
Highly variable genes
- TCRs :
-
T cell receptors
References
Pardoll, D. M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012).
Sharma, P., Hu-Lieskovan, S., Wargo, J. A. & Ribas, A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017).
Fares, C. M., Van Allen, E. M., Drake, C. G., Allison, J. P. & Hu-Lieskovan, S. Mechanisms of resistance to immune checkpoint blockade: why does checkpoint inhibitor immunotherapy not work for all patients? Am. Soc. Clin. Oncol. Educ. Book 39, 147–164 (2019).
Ganesan, S. & Mehnert, J. Biomarkers for response to immune checkpoint blockade. Annu. Rev. Cancer Biol. 4, 331–351 (2020).
Toor, S. M., Sasidharan Nair, V., Decock, J. & Elkord, E. Immune checkpoints in the tumor microenvironment. Semin. Cancer Biol. 65, 1–12 (2020).
Wu, T. & Dai, Y. Tumor microenvironment and therapeutic response. Cancer Lett. 387, 61–68 (2017).
Kang, Y., Vijay, S. & Gujral, T. S. Deep neural network modeling identifies biomarkers of response to immune-checkpoint therapy. iScience 25, 104228 (2022).
Dong, Y. et al. Prediction of immunotherapy responsiveness in melanoma through single-cell sequencing-based characterization of the tumor immune microenvironment. Transl. Oncol. 43, 101910 (2024).
Liu, R., Dollinger, E. & Nie, Q. Machine learning of single cell transcriptomic data from anti-PD-1 responders and non-responders reveals distinct resistance mechanisms in skin cancers and PDAC. Front. Genet. 12, 806457 (2022).
Erfanian, N. et al. Deep learning applications in single-cell genomics and transcriptomics data analysis. Biomed. Pharmacother. 165, 115077 (2023).
Ma, Q. & Xu, D. Deep learning shapes single-cell data analysis. Nat. Rev. Mol. Cell Biol. 23, 303–304 (2022).
Raimundo, F., Meng-Papaxanthos, L., Vallot, C. & Vert, J.-P. Machine learning for single-cell genomics data analysis. Curr. Opin. Syst. Biol. 26, 64–71 (2021).
He, B. et al. CloudPred: Predicting Patient Phenotypes From Single-cell RNA-seq. In Biocomputing 2022 337–348 (WORLD SCIENTIFIC, Kohala Coast, Hawaii, USA, 2021). https://doi.org/10.1142/9789811250477_0031.
Mao, Y. et al. Phenotype prediction from single-cell RNA-seq data using attention-based neural networks. Bioinformatics 40, btae067 (2024).
Sade-Feldman, M. et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013.e20 (2018).
Chen, T. & Guestrin, C. XGBoost: a scalable tree boosting system. In Proc. 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794 (ACM, San Francisco California USA, 2016). https://doi.org/10.1145/2939672.2939785.
Kursa, M. B., Jankowski, A. & Rudnicki, W. R. Boruta – A System for Feature Selection. Fundam. Informaticae 101, 271–285 (2010).
Lundberg, S. M. & Lee, S.-I. A Unified Approach to Interpreting Model Predictions. In Advances in Neural Information Processing Systems vol. 30 (Curran Associates, Inc., 2017).
Sutton, R. S. & Barto, A. G. Reinforcement Learning: An Introduction (MIT Press, 2018).
Zhang, J., Li, S., Liu, F. & Yang, K. Role of CD68 in tumor immunity and prognosis prediction in pan-cancer. Sci. Rep. 12, 7844 (2022).
Horowitch, B. et al. Subsets of interferon signaling predict response to immune checkpoint blockade in melanoma patients. Clin. Cancer Res. Off. 29, 2908–2918 (2023).
Liu, F. et al. Expression of STAT1 is positively correlated with PD-L1 in human ovarian cancer. Cancer Biol. Ther. 21, 963–971 (2020).
Sharma, N., Atolagbe, O. T., Ge, Z. & Allison, J. P. LILRB4 suppresses immunity in solid tumors and is a potential target for immunotherapy. J. Exp. Med. 218, e20201811 (2021).
GAPDH: a common housekeeping gene with an oncogenic role in pan-cancer - PMC. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10470192/.
Peng, C., Ye, H. & Yi, Z. GAPDH: unveiling its impact as a key hypoxia-related player in head and neck squamous cell carcinoma tumor progression, prognosis, and therapeutic potential. Am. J. Cancer Res. 13, 5846–5867 (2023).
Oshi, M. et al. M1 Macrophage and M1/M2 ratio defined by transcriptomic signatures resemble only part of their conventional clinical characteristics in breast cancer. Sci. Rep. 10, 16554 (2020).
Ahmed, H. et al. Role of T cells in cancer immunotherapy: Opportunities and challenges. Cancer Pathog. Ther. 1, 116–126 (2022).
Meng, F. et al. Leveraging CD16 fusion receptors to remodel the immune response for enhancing anti-tumor immunotherapy in iPSC-derived NK cells. J. Hematol. Oncol. 16, 62 (2023).
Aguilar, O. A. et al. The CD16 and CD32b Fc-gamma receptors regulate antibody-mediated responses in mouse natural killer cells. J. Leukoc. Biol. 113, 27–40 (2023).
Park, J.-E. et al. Anti-tumor effects of NK cells and anti-PD-L1 antibody with antibody-dependent cellular cytotoxicity in PD-L1-positive cancer cell lines. J. Immunother. Cancer 8, e000873 (2020).
Luoma, A. M. et al. Tissue-resident memory and circulating T cells are early responders to pre-surgical cancer immunotherapy. Cell 185, 2918–2935.e29 (2022).
Chen, L. et al. CD38-Mediated immunosuppression as a mechanism of tumor cell escape from PD-1/PD-L1 Blockade. Cancer Discov. 8, 1156–1175 (2018).
Revach, O.-Y. et al. Disrupting CD38-driven T cell dysfunction restores sensitivity to cancer immunotherapy. bioRxiv 2024.02.12.579184. https://doi.org/10.1101/2024.02.12.579184 (2024).
Nakayama, Y. et al. Phospho-STAT1 expression as a potential biomarker for anti-PD-1/anti-PD-L1 immunotherapy for breast cancer. Int. J. Oncol. 54, 2030–2038 (2019).
Liu, J. et al. PLEK2 and IFI6, representing mesenchymal and immune-suppressive microenvironment, predicts resistance to neoadjuvant immunotherapy in esophageal squamous cell carcinoma. Cancer Immunol. Immunother. CII 72, 881–893 (2022).
Osińska, I., Popko, K. & Demkow, U. Perforin: an important player in immune response. Cent. Eur. J. Immunol. 39, 109–115 (2014).
Caradec, J., Sirab, N., Revaud, D., Keumeugni, C. & Loric, S. Is GAPDH a relevant housekeeping gene for normalisation in colorectal cancer experiments? Br. J. Cancer 103, 1475–1476 (2010).
Vilgelm, A. E. & Richmond, A. Chemokines modulate immune surveillance in tumorigenesis, metastasis, and response to immunotherapy. Front. Immunol. 10, 333 (2019).
Griffith, J. W., Sokol, C. L. & Luster, A. D. Chemokines and Chemokine receptors: positioning cells for host defense and immunity. Annu. Rev. Immunol. 32, 659–702 (2014).
Honey, K. CCL3 and CCL4 actively recruit CD8+ T cells. Nat. Rev. Immunol. 6, 427–427 (2006).
Mukaida, N., Sasaki, S.-I. & Baba, T. CCL4 signaling in the tumor microenvironment. Adv. Exp. Med. Biol. 1231, 23–32 (2020).
Jiang, P. et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 24, 1550–1558 (2018).
Chen, M., Liu, H., Li, Z., Ming, A. L. & Chen, H. Mechanism of PKM2 affecting cancer immunity and metabolism in Tumor Microenvironment. J. Cancer 12, 3566 (2021).
Zheng, Y. et al. PGAM1 inhibition promotes HCC ferroptosis and synergizes with Anti-PD-1 immunotherapy. Adv. Sci. 10, 2301928 (2023).
Gu, Y. et al. A pan-cancer analysis of the prognostic and immunological role of β-actin (ACTB) in human cancers. Bioengineered 12, 6166–6185 (2021).
Mudd, T. W., Lu, C., Klement, J. D. & Liu, K. MS4A1 expression and function in T cells in the colorectal cancer tumor microenvironment. Cell. Immunol. 360, 104260 (2021).
Fabregat, A. et al. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinform. 18, 142 (2017).
Zhang, Y. et al. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell 39, 1578–1593.e8 (2021).
Caushi, J. X. et al. Transcriptional programs of neoantigen-specific TIL in anti-PD-1-treated lung cancers. Nature 596, 126–132 (2021).
Mei, Y. et al. Siglec-9 acts as an immune-checkpoint molecule on macrophages in glioblastoma, restricting T-cell priming and immunotherapy response. Nat. Cancer 4, 1273–1291 (2023).
Yost, K. E. et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 25, 1251–1259 (2019).
Bassez, A. et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat. Med. 27, 820–832 (2021).
Oliveira, G. et al. Phenotype, specificity and avidity of antitumour CD8+ T cells in melanoma. Nature 596, 119–125 (2021).
Tietscher, S. et al. A comprehensive single-cell map of T cell exhaustion-associated immune environments in human breast cancer. Nat. Commun. 14, 98 (2023).
Lu, L., Bai, Y. & Wang, Z. Elevated T cell activation score is associated with improved survival of breast cancer. Breast Cancer Res. Treat. 164, 689–696 (2017).
Duan, Z. et al. CD39 expression defines exhausted CD4+ T cells associated with poor survival and immune evasion in human gastric cancer. Clin. Transl. Immunol. 13, e1499 (2024).
Gentles, A. J. et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 21, 938–945 (2015).
Tang, Z., Kang, B., Li, C., Chen, T. & Zhang, Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47, W556–W560 (2019).
Gondal, M. N., Cieslik, M. & Chinnaiyan, A. M. Integrated cancer cell-specific single-cell RNA-seq datasets of immune checkpoint blockade-treated patients. Sci. Data 12, 139 (2025).
Cui, H. et al. scGPT: toward building a foundation model for single-cell multi-omics using generative AI. Nat. Methods 21, 1470–1480 (2024).
Szałata, A. et al. Transformers in single-cell omics: a review and new perspectives. Nat. Methods 21, 1430–1443 (2024).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Pedregosa, F. et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Ke, G. et al. LightGBM: a highly efficient gradient boosting decision tree. In Advances in Neural Information Processing Systems vol. 30 (Curran Associates, 2017).
Paszke, A. et al. PyTorch: an imperative style, high-performance deep learning library. In Proceedings of the 33rd International Conference on Neural Information Processing Systems 8026–8037 (Curran Associates Inc., Red Hook, NY, USA, 2019).
Agarap, A. F. Deep Learning using Rectified Linear Units (ReLU). arXiv.org https://arxiv.org/abs/1803.08375v2 (2018).
Acknowledgements
We would like to thank Prof. Gad Getz, Dr. Moshe Sade-Feldman, Dr. Liron Zisman, Elad Zisman, Ofir Shorer, and Sapir Levin for fruitful discussions and helpful comments on the manuscript. Figure 1A was created with BioRender.com using a paid license.This work was supported by the Ministry of Science and Technology (2032895), by the Israel Science Foundation (3614/19), and by the Israel Cancer Research Fund (2032965 23-204-RCDA). This work received additional support from the Ruth and Bruce Rappaport Technion Integrated Cancer Center (RTICC).
Author information
Authors and Affiliations
Contributions
K.Y. supervised the project and guided it throughout. K.Y. and A.P. jointly wrote the main manuscript text. A.P. prepared the figures and performed the analysis. Both authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Pinhasi, A., Yizhak, K. Uncovering gene and cellular signatures of immune checkpoint response via machine learning and single-cell RNA-seq. npj Precis. Onc. 9, 95 (2025). https://doi.org/10.1038/s41698-025-00883-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41698-025-00883-z