Abstract
Squamous transformation of prostate adenocarcinoma is a rare resistance mechanism in patients with advanced prostate cancer that impacts both prognosis and treatment. Herein, we profile circulating chromatin in serial plasma samples collected from a patient with metastatic prostate cancer that experienced squamous transformation. We detect dynamic changes in gene regulation from circulating chromatin reflecting the emergence of squamous differentiation, enabling non-invasive diagnosis and monitoring of this resistance phenotype, with potential therapeutic implications.
Similar content being viewed by others
Introduction
Prostate cancer is the most common non-cutaneous cancer in men in the United States and the second leading cause of cancer mortality1. In men with advanced prostate cancer, disruption of androgen receptor (AR) signaling with androgen deprivation therapy (ADT) is the backbone of systemic therapy. Later in the disease course, men with advanced prostate cancer often develop resistance to ADT, termed castration-resistant prostate cancer (CRPC)2. Lineage plasticity is a well-established mechanism of resistance in metastatic (m)CRPC. This event most commonly occurs as neuroendocrine transformation, molecularly characterized by deleterious genomic alterations in RB1 and TP53 and widespread epigenomic reprogramming, allowing prostate cancers to shed androgen dependence3,4,5,6,7,8,9. Squamous transformation is another form of lineage plasticity that is less common and less well understood, with an incidence of at least 1%10. While squamous cell prostate cancer (SqCPC) can develop de novo, about 50% of cases emerge through treatment-induced trans-differentiation of prostate adenocarcinoma11. In such cases, the squamous component is believed to originate from squamous metaplasia of acinar and ductal elements11. Treatment-emergent SqCPC has important prognostic and therapeutic implications. Patients with SqCPC have a poor prognosis12. Moreover, conventional prostate cancer therapies have limited efficacy in SqCPC13, and histology-directed chemotherapy may be more effective11,13,14,15. There are no tractable clinical tests to monitor for squamous transformation in men with mCRPC. Histologic analysis of tumor tissue is the current gold-standard. However, this approach has major shortcomings. Intra-patient tumor heterogeneity is well-established in mCRPC; consequently, analyzing a single metastatic biopsy is subject to sampling error and may miss treatment-emergent histologic transformation. Further, the invasive nature of tumor tissue biopsies prohibits serial testing to monitor for treatment-emergent resistance mechanisms.
Analysis of circulating tumor (ct)DNA is a promising means to detect histologic transformation from patient plasma that overcomes these challenges16,17,18. Cell-free (cf)DNA is wrapped around histone proteins, which bear methylation and acetylation modifications that mark active gene regulatory elements. These modifications change dynamically as tumor cells evolve, thus profiling cfDNA fragments bound to histones in the blood provides a real-time readout of transcriptional programs that can enable non-invasive cancer subtyping19,20,21. Recent studies have demonstrated a role for liquid biopsy epigenomic profiling to non-invasively detect AR alterations and neuroendocrine differentiation arising in prostate cancer18,19,22,23,24,25. In this study, we report the case of a patient with metastatic hormone-sensitive prostate adenocarcinoma that underwent treatment-associated squamous transformation. We identify epigenomic signatures of squamous differentiation in plasma samples collected from the patient over time. Our approach provides a dynamic assessment of squamous differentiation that expands the capability of liquid biopsy technologies for longitudinal monitoring of lineage plasticity.
Results
Clinical case
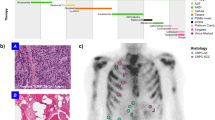
A 55-year-old man was diagnosed with de novo low-volume metastatic hormone-sensitive prostate cancer. At the time of diagnosis, transrectal ultrasound-guided biopsy of the prostate showed prostate adenocarcinoma. Initial treatment (as part of a clinical trial)26 comprised neoadjuvant leuprolide, apalutamide, abiraterone and prednisone for 6 months followed by radical prostatectomy, then maintenance leuprolide monotherapy (Fig. 1a). Ten months after surgery, his PSA rose to 3.22 ng/ml despite suppressed testosterone, indicating castration resistance, at which time abiraterone and prednisone were restarted. After 16 months of treatment, he developed progressive disease with scans showing new osseous metastases in the cervical, thoracic and lumbar spine as well as pathologically enlarged mediastinal and left hilar lymph nodes. Despite progressive disease on imaging, his PSA was unusually low at 0.04 ng/ml. The patient next received 177Lu-PSMA-617. After four cycles, bone scan showed mixed response in the osseous metastases, and CT scan showed further growth of the previously enlarged mediastinal and left hilar lymph nodes.
a Clinical course of the patient. ADT: Androgen deprivation therapy, mCRPC: metastatic castration resistant prostate cancer, FDG: Fluorodeoxyglucose, PSMA: Prostate specific membrane antigen, ICI: Immune checkpoint inhibitor, SBRT: Stereotactic body radiation therapy. b Comparative PSMA-PET and FDG-PET imaging (I) PSMA-PET scan with low uptake in the same two hilar lymph nodes. (II) FDG-PET scan with high metabolic activity in two left hilar lymph nodes. (III) PSMA-PET scan with avid uptake at the same T10 metastasis. (IV) FDG-PET scan with avid uptake at the T10 metastasis. c Histologic and immunohistochemical findings of mediastinal lymph nodes biopsies (I) Hematoxylin and eosin (H&E) stain showing squamous morphology. (II) Positive staining for p40. (III) Negative staining for NKX3-1. (IV) Weak staining for ERG. (V) H&E stain highlighting focal tumor clusters without squamous differentiation, (VI) which show NKX3-1 positivity. (VII) H&E stain of radical prostatectomy specimen (with treatment effect): Residual intraductal (left; with surrounding basal cells) and invasive (middle/right; without basal cells) carcinoma was present (b represents a higher magnification of boxed area in a). The tumor cells had pale cytoplasm and small round nuclei with prominent nucleoli. c Focally, in less than 5% of the tumor, squamous differentiation (sqd) was seen in the intraductal carcinoma; squamous differentiation was not seen in the invasive component. d Genomic profiling of the patient’s primary tumor from the radical prostatectomy specimen and a metastatic lesion from an enlarged mediastinal lymph node with low PSMA expression on PSMA PET scan (tumor purity 30% and 50%, respectively).
Imaging
Prostate-specific membrane antigen (PSMA) is an AR-regulated transmembrane protein that is highly expressed on the surface of most prostate cancer cells and has been leveraged as the target for FDA approved molecular PET scans to monitor therapeutic response in men with mCRPC27,28,29,30,31,32. This patient underwent a PSMA-PET scan following the CT scan above, which showed low PSMA tracer uptake in the left hilar lymph nodes (Fig. 1b. I), which can be observed in poorly differentiated, AR indifferent, and/or neuroendocrine prostate tumors. An FDG-PET scan was obtained, which confirmed the enlarged mediastinal and hilar lymph nodes were FDG avid (SUVmax 8.8) (Fig. 1b. II). In contrast, another lesion in the T10 vertebral body exhibited strong avidity for both PSMA (Fig. 1b. III) and FDG (Fig. 1b. IV), the typical pattern from a hypermetabolic metastatic prostate adenocarcinoma lesion.
Pathology
The patient underwent bronchoscopy and tissue biopsy of a mediastinal lymph node, and pathology revealed metastatic carcinoma with predominantly squamous features (review by expert genitourinary pathologist M.S.H.) in five of five cores. Immunohistochemistry demonstrated positive immunoreactivity for p40 (Fig. 1c. I-II) – a marker of squamous differentiation33. However, p16 staining was negative, arguing against an HPV-associated cancer of oropharyngeal origin34,35,36,37. The tumor cells were negative for TTF-1, a marker of lung and/or thyroid origin (data not shown). The cells were also negative for NKX3-1 (Fig. 1c. III), a highly specific marker of prostatic origin (Fig. 1c. IV), however, demonstrated a member of the TMPRSS2::ERG gene fusion, a pathognomonic genomic hallmark of prostate cancer. One of the biopsy cores contained a small focus of NKX3-1 positive acinar adenocarcinoma (Fig. 1c. V-VI), raising suspicion for treatment-emergent SqCPC. Notably, post hoc review of the radical prostatectomy specimen identified a small focus of intraductal squamous differentiation (approximately 1% of the overall tumor) (Fig. 1c. VII). Although there was no evidence of invasive SqCPC at the time, this suggests that the treatment-emergent metastatic SqCPC tumor may have been an outgrowth of a resistant subclone present at initial diagnosis rather than a result of trans-differentiation.
Next-generation sequencing
Next-generation sequencing-based genomic profiling was performed on the metastatic SqCPC tumor38. Comparison of the somatic profiles between the metastatic SqCPC tumor and the previously profiled predominantly acinar adenocarcinoma from the primary resection (prostatectomy) specimen (generated using the same assay) supported a common origin (Fig. 1d). First, an identical TMPRSS2::ERG fusion with matching breakpoints was identified in both primary and metastatic tumors, supporting a prostatic origin for the squamous lesion39. Further, a shared mutational profile (including presumed SNPs) was present in both primary tumor and SqCPC metastasis, including two pathogenic and five benign mutations (Fig. 1d; Supplementary Data 1). The metastatic SqCPC tumor had 9 additional pathogenic alterations not present in the primary tumor, including biallelic RB1 alterations consisting of a non-synonymous single-nucleotide variant predicted to be possibly damaging40 and a single copy number loss. RB1 biallelic loss is commonly found in treatment-emergent neuroendocrine prostate cancers41. To our knowledge, this is the first report of genomic profiling of a treatment-emergent SqCPC and suggests that RB1 inactivation may, more broadly, enable lineage plasticity in mCRPC.
Epigenomic profiling of plasma
We sought to evaluate whether we could leverage novel epigenomic liquid biopsy tools to non-invasively detect the squamous transformation event in this patient. We collected plasma samples at four timepoints throughout the patient’s disease course – two pre- and two post-SqCPC histologic transformation – and performed cfChIP-seq for two post-translational histone modifications: H3K4me3, which is enriched at active gene promoters42, and H3K27ac, which marks active promoters and enhancers43. Low-pass whole genome sequencing on cfDNA was also performed, and ctDNA fraction was estimated using ichorCNA44 (Fig. 2a). To look for evidence of squamous transformation we measured histone modifications at genes associated with squamous cell lineage. We observed an increased H3K4me3 signal at the promoter of DSC3 – a specific marker of squamous cell carcinoma (SCC)45 – in the plasma collected following the squamous transformation compared to the baseline sample. Similarly, we noted increased H3K4me3 signal at other SCC-associated genes such as SOX246, ETV447 and S100A1348,49. Conversely, the signal at the NKX3-1 promoter, a specific marker for prostate adenocarcinoma, remained similar across both timepoints (Fig. 2b). This finding indicates that activity of genes encoding squamous-specific markers can indeed be detected from circulating chromatin.
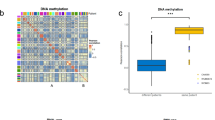
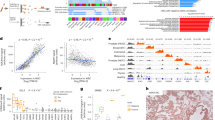
a Overview of the experimental workflow to detect and monitor squamous transformation from circulating chromatin. b Normalized signal from H3K4me3 cell-free ChIP-seq profiles at representative SCC-associated (DSC3, SOX2, ETV4, S100A13) and PRAD-associated (NKX3-1) loci in baseline and post-squamous transformation plasma samples. c Genomic Regions Enrichment of Annotations Tool analysis (GREAT) of TCGA squamous ATAC peaks (n = 7353). d Longitudinal tracking of the squamous (purple) and adenocarcinoma (blue) plasma epigenomic scores. H3K4me3 signal at housekeeping genes activity (red) and ctDNA fraction (green) included for comparison. All values were rescaled based on the mean signal in healthy plasma at each set of sites. e Correlation of the aggregated H3K27ac signal across adenocarcinoma and squamous sites with the estimated ctDNA fraction in plasma samples from patients with prostate adenocarcinoma.
To evaluate whether we could track the emergence of SqCPC in this patient, we assessed a set of sites with increased chromatin accessibility across several squamous cancer types50, representing regulatory elements whose activation is associated with squamous cell lineage (Supplementary Data 2). This set of ATAC-seq peaks was enriched for biologically relevant pathways associated with squamous differentiation51 (Fig. 2c). AR binding sites derived from cistrome DB were utilized to trace the adenocarcinoma component (Supplementary Data 2). We aggregated the H3K27ac signal at the squamous-specific accessible sites and AR binding sites to calculate prostate squamous and adenocarcinoma signals, respectively; signal at a set of housekeeping genes (Supplementary Data 2) was included as a control. Strikingly, we observed an increased H3K27ac signal at squamous-specific accessible sites in plasma drawn at timepoints 2 and 3, aligning with the timing of pathologic diagnosis of squamous transformation (Fig. 2d). This signal subsequently decreased after the administration of squamous histology-directed chemo-immunotherapy, corresponding to a reduction in size of the mediastinal and hilar lymph nodes with otherwise stable disease. These trends were observed despite the ctDNA fraction being consistently low (<0.05) throughout the patient’s disease course. Signal at housekeeping genes was consistent across the different timepoints. In addition to cancer subtyping, these results suggest that cfChIP-seq can serve as a tool for monitoring the response of distinct histologic components.
To validate that the circulating biomarkers used to detect squamous features are independent of the adenocarcinoma component and not simply a result of variations in cfDNA tumor content, we analyzed the association between the H3K27ac signal at the squamous-specific and adenocarcinoma-specific sites, respectively, with the ctDNA fraction in a cohort of previously profiled cfChIP-seq libraries from patients with metastatic castration-resistant prostate adenocarcinoma19. As expected, the H3K27ac signal at adenocarcinoma-specific sites significantly correlated with ctDNA fraction (R = 0.63; p < 0.0001; Fig. 2e). In contrast, no correlation was observed between the H3K27ac signal at squamous-specific sites and ctDNA fraction (R = 0.02; p = 0.92; Fig. 2e). These results suggest that levels of histone modifications at squamous-associated regulatory elements specifically reflect squamous-derived chromatin in plasma.
Discussion
In this manuscript, we report a patient with treatment-emergent squamous cell prostate cancer at the time of progression on 177Lu-PSMA-617 (Lutetium-PSMA) and demonstrate the ability to non-invasively detect the emergence of this resistant subclone through cfChIP-seq. This is the first application of liquid biopsy to detect treatment-emergent SqCPC and builds upon previous work, highlighting the potential of cfChIP-seq to detect and monitor an expanded array of clinically actionable resistance phenotypes arising from lineage plasticity20,25.
The patient initially presented with de novo oligometastatic hormone-sensitive PRAD, but at the time of progression on Lutetium-PSMA was found to have treatment-emergent SqCPC. Given the absence of established guidelines for SqCPC treatment, a tumor board convened, and histology-directed therapy was initiated with carboplatin, paclitaxel, and pembrolizumab. This regimen was extrapolated from standard-of-care squamous non-small cell lung cancer treatment based on the KEYNOTE-407 trial and the NCCN guidelines52,53. The patient responded for five months before progressing, highlighting the need for further data on the treatment of SqCPC and the importance of clinical trials focused on prostate cancer variant histologies.
In this case, SqCPC was diagnosed through tissue biopsy, immunohistochemistry, and next-generation sequencing. The diagnosis was initially suspected based on the immunohistochemical detection of NKX3-1 and p40 in one of the biopsy cores and was confirmed through next-generation sequencing by the presence of a TMPRSS2::ERG fusion, along with other alterations shared between the primary tumor and metastasis13,15,54. Notably, although the presence of the TMPRSS2::ERG fusion by genomics confirmed the prostatic origin of the squamous carcinoma, ERG IHC was weak. This finding is consistent with the loss of AR signaling, which drives transcription of the TMPRSS2::ERG fusion, in androgen-indifferent prostate cancer subtypes39,55,56. Moreover, this observation of treatment-emergent SqCPC at the time of resistance to Lutetium-PSMA highlights the need to further characterize the spectrum of resistance mechanisms to novel mCRPC therapies. Given the challenges associated with tumor tissue acquisition in late-stage prostate cancer patients, this molecular case report highlights the potential value of emerging liquid biopsy tools to provide non-invasive insights into clinically actionable tumor biology.
Molecular imaging has revolutionized our ability to gain non-invasive, dynamic insights into prostate cancer biology. For example, serial monitoring of mCRPC patients with paired FDG- and PSMA-PET scans facilitates characterization of distinct disease compartments. Specifically, FDG-avid PSMA-low/absent lesions are often indicative of AR-indifferent prostate cancer, such as poorly differentiated, neuroendocrine, or squamous subtypes. However, beyond quantifying tumor PSMA levels, these imaging techniques cannot characterize the specific tumor subtype, which is critical for optimal treatment selection. While subsequent tumor biopsies can provide this information, their invasive nature limits longitudinal, real-time assessment of tumor status. Herein, we demonstrate that cfChIP-seq can overcome these limitations to detect treatment-emergent SqCPC, building upon our previous work showing that plasma epigenomic profiling can non-invasively detect neuroendocrine prostate cancer18,19,22. Intriguingly, the plasma SqCPC signature preceded clinical diagnosis of squamous transformation by more than 9 months, suggesting a potential role for earlier detection of treatment-emergent variant histologies and delivery of histology-directed treatment than the current clinical paradigm enables.
In conclusion, our study highlights the growing utility of cfChIP-seq for monitoring mCRPC to generate non-invasive insights into tumor evolution and precise characterization of AR-indifferent subtypes. A noteworthy limitation of this study is that it involves a single patient, attributable to the rare occurrence of treatment-emergent squamous cell prostate cancer, who end up getting a tissue-based biopsy. However, this molecular case report highlights the growing potential of plasma epigenomic profiling for real-time, longitudinal assessment of heterogeneous tumor components, with important prognostic and therapeutic implications.
Methods
Study oversight and sample acquisition
This study adheres to all applicable ethical regulations. Informed consent for both participation and publication was obtained from the patient. Deidentified plasma samples were collected longitudinally from one patient at the Dana-Farber/Harvard Cancer Center. Plasma samples were collected under protocol number 01-045 approved by the Dana-Farber/Harvard Cancer Center (DF/HCC).
Regarding the cohort of patients with prostate adenocarcinoma, plasma samples and informed consents were collected under the same protocol. All samples were then de-identified and subsequently similarly processed.
Immunohistochemistry
Immunohistochemistry was performed on 4 µm-thick whole tissue sections from formalin-fixed paraffin-embedded (FFPE) tissue blocks, using the following antibodies: anti-NKX3-1 (Athena ES; Rabbit polyclonal 1:200); anti-p40 (Biocare; BC28 1:150); anti-ERG (Abcam; EPR3864 1:1800); and anti-TTF-1 (Agilent; 8G7G3/1 1:800) primary antibodies. Heat-induced antigen retrieval was performed in a pressure cooker with citrate buffer (pH 6). Signals were detected using the Dako EnVision Plus System (Agilent, Santa Clara, CA, USA), following the manufacturer’s recommendations.
Next-generation sequencing
Tumor profiling was performed by a hybrid capture-based targeted DNA sequencing panel (OncoPanel), on formalin-fixed, paraffin-embedded (FFPE) tissue sections as described previously38. In brief, H&E-stained slides were reviewed by a molecular pathologist to ensure tumor content of at least 20%. Indexed sequencing libraries were prepared from a 50-ng DNA sample using Illumina TruSeq LT reagents (Illumina). Custom solution-based hybrid capture (Agilent SureSelect; Agilent Technologies) was utilized to enrich for targeted exons and selected introns from 447 cancer genes (OncoPanel V3.1). Massively parallel sequencing was performed using HiSeq2500 or NovaSeq6000 (Illumina) to achieve a minimum mean target coverage of 50X and an average mean target coverage of 150X to 300X. Data were analyzed on an internally developed bioinformatics pipeline. Single-nucleotide variants, copy number variants, and structural variants were reviewed by a board-certified molecular pathologist.
Plasma cell-free ChIP-seq
H3K27ac and H3K4me3 cell-free ChIP-seq were performed serially on plasma samples using previously published methods19. The following antibodies were used: H3K27ac, Abcam # ab4729; H3K4me3, Thermo Fisher # PA5-27029.
Cell-free DNA extraction and low-pass whole-genome sequencing
Cell-free DNA extraction and LPWGS were performed on plasma samples using previously published methods19.
Squamous signature
The squamous feature set was a group of 7353 sites with chromatin accessibility that is consistently higher in squamous tumors of multiple lineages (Supplementary Data 2).
Analysis of cfChIP-seq data
H3K4me3/H3K27ac cf-ChIP-seq reads were aligned to the hg19 human genome build using Burrows–Wheeler Aligner version 0.7.174057 (RRID: SCR_010910). Non-uniquely mapping and redundant reads were discarded. MACS version 2.1.1 (RRID: SCR_013291)58 was used for ChIP-seq peak calling with a q value (false discovery rate (FDR)) threshold of 0.01. Fragment locations were converted to BED files using BEDTools59 (version 2.29.2) using the bamtobed command with the -bedpe flag set. For analyses involving overlaps with genomic regions, fragments were imported as GRanges objects and collapsed to 1 bp at the center of the fragment location to ensure that a fragment could map to only one site.
Quantification of peaks and transcription factor binding sites in plasma
We inferred transcriptional activity at sites of interest based on H3K27ac and H3K4me3, as described previously19. Briefly, peaks were resized to a 3-kb interval centered on the original peak, then binned into 40-bp windows. Fragment counts were aggregated across each 40-bp window for all peaks to obtain aggregate profiles for each sample. To account for variation in background signal across samples, we performed a ‘shoulder normalization’ step previously described19. We also normalized the signal in each bin to the aggregated signal at the common 10,000 DNAse hypersensitivity sites that are expected to be active across most tissue types and defined across the largest number of samples in ref. 60. For signal monitoring (Fig. 2d), the signal at each time point was rescaled based on the mean signal in a set of previously profiled healthy plasma samples. This was achieved by dividing the aggregated signal of each time point by the mean value in the healthy plasma for each of the squamous ATAC sites, AR binding sites, and housekeeping genes19.
Estimation of the circulating tumor DNA fraction in plasma
The ichorCNA R package (RRID:SCR_024768) was used to infer copy-number profiles and cfDNA tumor content from read abundance across bins spanning the genome using default parameters.
Data availability
Raw data (FASTQ files) and processed data (BED and BIGWIGS files) are available through GEO under accession numbers GSE291318 (prostate cancer samples) and GSE243474 (healthy samples).
Code availability
Scripts to reproduce analyses from this study are available at https://github.com/Baca-Lab/SCPC_manuscript.
References
Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 74, 12–49 (2024).
Watson, P. A., Arora, V. K. & Sawyers, C. L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 15, 701–711 (2015).
Takeda, D. Y. et al. A somatically acquired enhancer of the androgen receptor is a noncoding driver in advanced prostate cancer. Cell 174, 422–432.e413 (2018).
Taplin, M. E. et al. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N. Engl. J. Med. 332, 1393–1398 (1995).
Beltran, H. et al. The role of lineage plasticity in prostate cancer therapy resistance. Clin. Cancer Res 25, 6916–6924 (2019).
Beltran, H. et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med 22, 298–305 (2016).
Bluemn, E. G. et al. Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 32, 474–489.e476 (2017).
Ku, S. Y. et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355, 78–83 (2017).
Mu, P. et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 355, 84–88 (2017).
Malik, R. D. et al. Squamous cell carcinoma of the prostate. Rev. Urol. 13, 56–60 (2011).
Arva, N. C. & Das, K. Diagnostic dilemmas of squamous differentiation in prostate carcinoma case report and review of the literature. Diagn. Pathol. 6, 46 (2011).
Li, J. & Wang, Z. The pathology of unusual subtypes of prostate cancer. Chin. J. Cancer Res 28, 130–143 (2016).
Dizman, N. et al. Squamous transformation of prostate adenocarcinoma: a report of two cases with genomic profiling. Clin. Genitourin. Cancer 18, e289–e292 (2020).
Hanna, K. et al. Primary prostatic squamous cell carcinoma. Urol. Case Rep. 34, 101478 (2021).
Lau, H. D. & Clark, M. Metastatic squamous cell carcinoma transformed from prostatic adenocarcinoma following androgen deprivation therapy: A case report with clinicopathologic and molecular findings. Diagn. Cytopathol. 48, E14–e17 (2020).
Beltran, H. et al. Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J. Clin. Invest 130, 1653–1668 (2020).
De Sarkar, N. et al. Nucleosome patterns in circulating tumor DNA reveal transcriptional regulation of advanced prostate cancer phenotypes. Cancer Discov. 13, 632–653 (2023).
Franceschini, G. M. et al. Noninvasive detection of neuroendocrine prostate cancer through targeted cell-free DNA methylation. Cancer Discov. 14, 424–445 (2024).
Baca, S. C. et al. Liquid biopsy epigenomic profiling for cancer subtyping. Nat. Med 29, 2737–2741 (2023).
El Zarif, T. et al. Detecting small cell transformation in patients with advanced EGFR mutant lung adenocarcinoma through epigenomic cfDNA profiling. Clin. Cancer Res 30, 3798–3811 (2024).
El Zarif, T. et al. Epigenomic signatures of sarcomatoid differentiation to guide the treatment of renal cell carcinoma. Cell Rep. 43, 114350 (2024).
Berchuck, J. E. et al. Detecting neuroendocrine prostate cancer through tissue-informed cell-free DNA methylation analysis. Clin. Cancer Res 28, 928–938 (2022).
Chauhan, P. S. et al. Genomic and epigenomic analysis of plasma cell-free DNA identifies stemness features associated with worse survival in lethal prostate cancer. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.Ccr-24-1658 (2024).
Nawfal, R., El Hajj Chehade, R. & Berchuck, J. E. Unearthing a prostate cancer cfDNA signature that “stems” from AR alterations. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.Ccr-24-2849 (2024).
Sipola, J. et al. Plasma cell-free DNA Chromatin immunoprecipitation profiling depicts phenotypic and clinical heterogeneity in advanced prostate cancer. Cancer Res 85, 791–807 (2025).
Teo, M. Y. et al. Metacure: Multi-arm multimodality therapy for very high risk localized and low volume metastatic prostatic adenocarcinoma. J. Clin. Oncol. 37, TPS349–TPS349 (2019).
Bakht, M. K. et al. Neuroendocrine differentiation of prostate cancer leads to PSMA suppression. Endocr. Relat. Cancer 26, 131–146 (2018).
Bostwick, D. G., Pacelli, A., Blute, M., Roche, P. & Murphy, G. P. Prostate specific membrane antigen expression in prostatic intraepithelial neoplasia and adenocarcinoma: a study of 184 cases. Cancer 82, 2256–2261 (1998).
Chakraborty, P. S. et al. Metastatic poorly differentiated prostatic carcinoma with neuroendocrine differentiation: negative on 68Ga-PSMA PET/CT. Clin. Nucl. Med. 40, e163–e166 (2015).
Parida, G. K. et al. Adenocarcinoma prostate with neuroendocrine differentiation: potential utility of 18F-FDG PET/CT and 68Ga-DOTANOC PET/CT Over 68Ga-PSMA PET/CT. Clin. Nucl. Med 43, 248–249 (2018).
Tosoian, J. J. et al. Correlation of PSMA-targeted (18)F-DCFPyL PET/CT findings with immunohistochemical and genomic data in a patient with metastatic neuroendocrine prostate cancer. Clin. Genitourin. Cancer 15, e65–e68 (2017).
Usmani, S. et al. Molecular imaging in neuroendocrine differentiation of prostate cancer: 68Ga-PSMA versus 68Ga-DOTA NOC PET-CT. Clin. Nucl. Med 42, 410–413 (2017).
Bishop, J. A. et al. p40 (ΔNp63) is superior to p63 for the diagnosis of pulmonary squamous cell carcinoma. Mod. Pathol. 25, 405–415 (2012).
Esteve, A. et al. Low frequency of p16/CDKN2 gene mutations in esophageal carcinomas. Int J. Cancer 66, 301–304 (1996).
Hayashi, K. et al. High frequency of simultaneous loss of p16 and p16beta gene expression in squamous cell carcinoma of the esophagus but not in adenocarcinoma of the esophagus or stomach. Oncogene 15, 1481–1488 (1997).
Tokugawa, T., Sugihara, H., Tani, T. & Hattori, T. Modes of silencing of p16 in development of esophageal squamous cell carcinoma. Cancer Res. 62, 4938–4944 (2002).
Cuschieri, K. & Wentzensen, N. Human papillomavirus mRNA and p16 detection as biomarkers for the improved diagnosis of cervical neoplasia. Cancer Epidemiol. Biomark. Prev. 17, 2536–2545 (2008).
Garcia, E. P. et al. Validation of OncoPanel: A targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch. Pathol. Lab Med 141, 751–758 (2017).
Tomlins, S. A. et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 310, 644–648 (2005).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Aggarwal, R. et al. Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: a multi-institutional prospective study. J. Clin. Oncol. 36, 2492–2503 (2018).
Creyghton, M. P. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107, 21931–21936 (2010).
O’Geen, H., Echipare, L. & Farnham, P. J. Using ChIP-seq technology to generate high-resolution profiles of histone modifications. Methods Mol. Biol. 791, 265–286 (2011).
Adalsteinsson, V. A. et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 8, 1324 (2017).
Monica, V. et al. Desmocollin-3: a new marker of squamous differentiation in undifferentiated large-cell carcinoma of the lung. Mod. Pathol. 22, 709–717 (2009).
Boumahdi, S. et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature 511, 246–250 (2014).
Tang, Y. et al. Identification of ETV4 as a prognostic biomarker and correlates with immune cell infiltration in head and neck squamous cell carcinoma. Sci. Rep. 15, 7044 (2025).
Hu, Y., Han, Y., He, M., Zhang, Y. & Zou, X. S100 proteins in head and neck squamous cell carcinoma (Review). Oncol. Lett. 26, 362 (2023).
Li, R. et al. Systematic screening identifies a TEAD4-S100A13 axis modulating cisplatin sensitivity of oral squamous cell carcinoma cells. J. Oral. Pathol. Med. 50, 882–890 (2021).
Corces, M. R. et al. The chromatin accessibility landscape of primary human cancers. Science 362 https://doi.org/10.1126/science.aav1898 (2018).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Paz-Ares, L. et al. Pembrolizumab plus Chemotherapy for squamous non-small-cell lung cancer. N. Engl. J. Med 379, 2040–2051 (2018).
Riely, G. J. et al. Non-Small Cell Lung Cancer, Version 4.2024, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc Netw. 22, 249–274 (2024).
Autio, K. & McBride, S. Oligometastatic squamous cell transformation from metastatic prostate adenocarcinoma treated with systemic and focal therapy: a case report. J. Immunother. Precis Oncol. 5, 79–83 (2022).
Chen, Y. & Sawyers, C. L. Coordinate transcriptional regulation by ERG and androgen receptor in fusion-positive prostate cancers. Cancer Cell 17, 415–416 (2010).
Berchuck, J. E., Viscuse, P. V., Beltran, H. & Aparicio, A. Clinical considerations for the management of androgen indifferent prostate cancer. Prostate Cancer Prostat. Dis. 24, 623–637 (2021).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Meuleman, W. et al. Index and biological spectrum of human DNase I hypersensitive sites. Nature 584, 244–251 (2020).
Acknowledgements
S.C.B. is supported by the US Department of Defense award W81XWH-21-1-0358, the National Institutes of Health / National Cancer Institute U01 CA296432, the Damon Runyon Cancer Research Foundation, and the Fund for Innovation in Cancer Informatics. J.E.B. is supported by the US Department of Defense (W81XWH-20-1-0118, HT9425-23-1-0048).
Author information
Authors and Affiliations
Contributions
K.S.: Formal analysis, software, investigation, methodology, writing–original draft. R.N.: Data curation, investigation, writing–original draft. J.C.: Investigation. H.S.: Investigation. J.-H.S.: Investigation. S.E.S.: Investigation, writing–review, and editing. S.J.W.: Investigation, writing–review, and editing. J.K.R.: Investigation, writing–review, and editing. G.-S.M.L.: Investigation. R.H.C.: Investigation. Z.Z.: Investigation. G.S.G. Investigation. N.P.: Investigation. G.G.L.: Investigation. C.A.F.: Investigation, writing–review, and editing. M.S.H.: Investigation, writing–review, and editing. H.A.J.: Investigation, writing–review, and editing. A.D.C.: Resources, writing–review, and editing. T.K.C.: Resources, writing–review, and editing. M.L.F.: Resources, investigation, methodology, supervision, writing–review, and editing. S.C.B.: Conceptualization, resources, formal analysis, software, methodology, supervision, writing–review, and editing. J.E.B.: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, methodology, project administration, writing–review, and editing.
Corresponding authors
Ethics declarations
Competing interests
M.L.F. is a co-founder and shareholder of Precede Biosciences. S.C.B. is a co-founder and shareholder of Precede Biosciences. J.E.B. is an advisor/consultant to Genome Medical, Oncotect, Precede Biosciences, Tracer Biotechnologies, and Musculo, has equity in Cityblock Health, Genome Medical, Oncotect, Precede Biosciences, Tracer Biotechnologies, and Musculo, and has received speaker honoraria from Guardant Health.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Semaan, K., Nawfal, R., Canniff, J. et al. Plasma epigenomic profiling reveals treatment-emergent squamous transformation in prostate cancer. npj Precis. Onc. 9, 233 (2025). https://doi.org/10.1038/s41698-025-01031-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41698-025-01031-3