Abstract
Ex vivo lung perfusion (EVLP) enables advanced assessment of human lungs for transplant suitability. We developed a convolutional neural network (CNN)-based approach to analyze the largest cohort of isolated lung radiographs to date. CNNs were trained to process 1300 longitudinal radiographs from n = 650 clinical EVLP cases. Latent features were transformed into principal components (PC) and correlated with known radiographic findings. PCs were combined with physiological data to classify clinical outcomes: (1) recipient time to extubation of <72 h, (2) ≥ 72 h, and (3) lungs unsuitable for transplantation. The top PC was significantly correlated with infiltration (Spearman R: 0·72, p < 0·0001), and adding radiographic PCs significantly improved the discrimination for clinical outcomes (Accuracy: 73 vs 78%, p = 0·014). CNN-derived radiographic lung features therefore add substantial value to the current assessments. This approach can be adopted by EVLP centers worldwide to harness radiographic information without requiring real-time radiological expertise.
Similar content being viewed by others
Introduction
Precision medicine in transplantation is a growing field that emphasizes the study of candidate organs on an individualized basis1. Radiographic images of human lungs outside of the thoracic cavity represent an opportunity to study lung architecture in unprecedented detail. This unique radiographic dataset is derived from the ex vivo lung perfusion (EVLP) platform, which was developed to assess and treat isolated donor lungs prior to transplantation. EVLP is a breakthrough technology that has facilitated precision diagnostics, increased donor lung availability, and improved recipient outcomes2,3.
Currently, during EVLP, lungs are perfused at 37 °C and ventilated on a closed circuit for up to 6 h, and lung function is assessed to determine the suitability for transplantation4. Since the development of EVLP, there has been a surge of available isolated human lung data. The evaluation of lungs on EVLP enables an abundant flow of de-noised data free from the confounding factors present in complex physiological systems. Longitudinal measurements capture changes in lung function over time, providing key insights for surgical decision-making on organ suitability for transplantation. Significant efforts have been dedicated to investigating biomarkers for donor lung injury5,6,7, and recent studies have shown that combining physiological and biochemical parameters can improve the diagnostic value of individual EVLP features8,9. We have demonstrated in a machine learning model termed InsighTx, that combining a wide array of lung functional data obtained during EVLP can predict post-transplant outcomes10. However, EVLP-derived radiographs represent an important source of lung data that is easily obtainable but has been largely underutilized and understudied.
Lung radiography is an important aspect of EVLP diagnostics that provides unique, rich, and holistic information about donor lungs. In contrast to conventional chest radiographs, EVLP radiographs offer pristine, isolated images of donor lungs. Without obstruction from the heart, ribs, and chest wall tissue, ex vivo lung radiographs exhibit enhanced contrast that highlights the anatomy and abnormal findings against the well-displayed underlying structure. In a previous study, we used a cohort of EVLP X-ray images manually reviewed and scored by a radiologist to demonstrate that EVLP radiographic findings were associated with both the presence of physiological donor lung injury and lung transplant outcomes11. However, implementing this process is logistically challenging given that most EVLP centers do not have access to radiological expertize 24/7 to review X-ray images in real-time in the clinical operating room.
Over the past decade, powerful computing capabilities and large datasets have been the backbone driving the artificial intelligence revolution. Extensive studies have been published using machine learning methods to process and interpret chest radiographs12,13,14. As the field of deep learning has made significant strides, there has been a growing interest in applying computer vision methods to emerging fields such as EVLP. Among various techniques, convolutional neural networks (CNN) process image features by sliding filters across input images, a process that has found wide application in the field of medical imaging. Moreover, machine learning studies12,13,14 have shown that an unsupervised approach to radiographic images can be superior to conventional, manually derived labels. Taken together, a computer-vision approach to EVLP radiographs would fill a critical unmet need in facilitating advanced donor lung assessment for transplantation.
To harness the full potential of ex vivo lung radiographs in the absence of radiologists, herein we leverage computer vision to analyze this unique imaging source to quantify donor lung injury. The study objectives were to build an algorithm for automated interpretation of ex vivo lung radiographs, serving as a “virtual radiologist” within a clinical lung transplant team, and to develop a multi-modal approach to isolated lung evaluation. We first established an image processing pipeline based on CNN to analyze longitudinal X-ray images taken during EVLP. Classification performance of the CNN model was compared to manual radiographic labels. The interpretability of the CNN model was investigated using principal components (PCs) in the latent feature space. Finally, the prognostic value of radiographic analysis was combined with InsighTx10, a machine-learning model based on physiological and biological lung data.
Results
EVLP radiographs can be more accurately analyzed using computer vision methods
The baseline and pretrained performances of ResNet-50, ResNeXt-50, RexNet-100, EfficientNet-B2, EfficientNet-B3, and DenseNet-121 to classify donor lung outcomes are shown in Supplementary Table 1. These CNNs were trained to process temporal ex vivo lung radiographs before performing one classification for a given EVLP case. In the validation set, the CADLab-pretrained ResNet-50 model best predicted donor lung outcome with an accuracy of 66·9% (AUROC = 78·4%).
Our previous study reported the XGBoost performance of manually labeled radiographic images to predict donor lung outcomes, which resulted in an accuracy of 56 ± 4% and AUROC of 76 ± 2%15. When compared to the manual radiographic labels, the CNNs trained to classify outcomes using automatically extracted image features showed a significantly higher (p = 0·0007) mean accuracy of 63 ± 2%, and an equivalent AUROC of 76 ± 1% (p = 0·37) (Table 1).
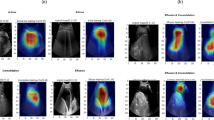
GradCAM maps in Fig. 1 revealed that the ResNet-50 model learned relevant clinical information. When the lungs were declined, the activated regions (highlighted in blue) were focused around the middle and lower lung lobes. Comparatively, when donor lungs were suitable for transplantation and good recipient outcomes occurred, the relevant model regions tended to be more diffuse throughout the image (i.e., not focused on any particular region). These observations were supported by cases with either high or low probability of the three predicted outcomes (Supplementary Fig. 1).
A CNN-based approach to processing EVLP radiographs extracts clinically relevant features from the ex vivo images
A one-dimensional vector comprising n = 4096 latent image features before the classification layer was extracted from the ResNet-50 model. Feature space transformation was performed using PC analysis to reduce the number of vectors describing the latent features. The graph in Supplementary Fig. 2 shows that the top ten PCs explained 98·7% of the variance in the feature space, where PC #1 alone explained 67%.
To investigate potential interpretation and meaning behind ResNet-50 latent features, the top ten PCs were correlated with manually labeled radiographic scores, donor data, and EVLP functional parameters. Fig. 2 indicates that high PC #1 values were strongly correlated with increased consolidation (Spearman R: 0·68, p < 0·0001), infiltration (Spearman R: 0·72, p < 0·0001), nodules (Spearman R: 0·25, p = 0·031), and interstitial lines (Spearman R: 0·24, p = 0·033). Other statistically significant correlations ranging from 0·23 to 0·38 were also found between the remaining PCs and radiographic labels (Fig. 2). PC #1 was strongly correlated with physiological and biological lung functions; additional statistically significant correlations were also observed with the other PCs (Supplementary Fig. 3A–C).
Features analyzed by computer vision are consistent with manually labeled radiographic abnormalities. The shaded bar on the right represents the Spearman correlation coefficients from +1.0 to −1.0. The shade and size of each circle in the grid indicate the extent to which each PC correlated with each radiographic abnormality. (PC principal component, *p < 0.05, **p < 0.01, ***p < 0.001).
A multi-modal fusion approach significantly improved the classification of transplant outcomes
Lastly, we evaluated the impact of a multi-modal approach to EVLP decision-making that combined imaging, physiological, and biochemical assessments. When trained to predict donor lung outcomes using physiological and biological EVLP data alone, the XGBoost-based InsighTx model achieved an accuracy of 73 ± 5% and AUROC of 88 ± 2%. Upon addition of the ten imaging PCs from the ResNet-50 latent features, the accuracy and AUROC significantly increased to 78 ± 4% (p = 0·014) and 94 ± 2% (p < 0·0001), as shown in Table 2. Importantly, both precision (73% vs. 64%, p = 0·02) and recall (79% vs. 72%, p = 0·009) increased significantly when PCs from X-ray image analysis were added to the InsighTx model. An example confusion matrix is provided in Supplementary Table 2. The prevalence of the three endpoints was consistent across cohorts and reflected the overall population prevalence of 38%, 23%, and 39% for transplanted lungs with recipients extubated within 72 h, more than 72 h, or unsuitable lungs for transplant, respectively. The Shapley Additive explanation values showed that PC #1 from ResNet-50 is the most impactful feature for classifying transplant outcomes, and PC #9 was also among the ten most important features (Supplementary Table 3). PC #9 did not correlate significantly with absolute values nor longitudinal changes of radiographic abnormalities (Fig. 2). Upon close inspection of radiographs with high and low values of PC #9, no distinct trends in visible features were identified. Considering that PC #9 accounts for less than 1% of the total model output (Supplementary Fig. 2), it is likely that it represents a complex feature not readily discernible by visual inspection.
Discussion
In this study, convolutional neural networks (CNN) were trained to classify lung transplant outcomes using longitudinally acquired ex vivo lung radiographs. The radiographic features automatically extracted from a CNN predicted clinical outcomes to the level of using manual radiographic scores. Moreover, we have demonstrated that latent features from a neural network can be extracted and used in downstream analyses. Upon investigation of the PCs of ResNet-50 latent features, the top PC, which accounted for two-thirds of the neural network output, was found to be strongly correlated with the clinical findings of radiographic consolidation, infiltration, and many other physiological injuries. When incorporated into the existing donor lung diagnostics using functional parameters, PCs of latent features not only significantly improved outcome predictions, but were also top ranked features for their contribution to the classification. This result provided quantitative evidence that incorporating radiographic analysis significantly improved donor lung prognostics.
This study represents a step forward in the application of machine learning methods for lung transplantation. Ex vivo lung radiographs provide isolated X-ray images of donor lungs with improved signal to noise compared to conventional chest radiographs. These images provide rich, qualitative, and semantic information about the injury status within regions of donor lungs. An image alone can depict various underlying issues of differing severities in distinct parts of donor lungs. Although EVLP has been widely adopted clinically as an invaluable technology for treating and assessing donor lungs, radiographs taken during EVLP are a unique and meaningful medium for donor lung prognostication that have previously been underused and understudied. Recently, we have reported the prevalence and diagnostic value of radiographic abnormalities in EVLP X-ray images15. To our knowledge, the current study is the first to apply deep learning to truly operationalize and leverage the prognostic potential of ex vivo lung radiographs.
The design of the CNN image processing pipeline was to mirror the surgeon’s decision-making process in EVLP, looking at X-ray images taken at different timepoints and examining the changes in donor lung condition over time. In contrast to many medical image classification studies12,13, pretraining models using large-scale chest X-ray images did not result in substantial improvements in classification performances. This is likely due to the distribution shifts in pixel intensities16 between chest X-ray and EVLP X-ray datasets. Nevertheless, the radiographic features automatically extracted from a CNN predicted clinical outcomes to the level of using manual radiographic scores. Importantly, the computer vision approach provides significant advantages in terms of clinical utility of automated image analysis versus training a separate model to analyze manually annotated radiographic assessments.
In future clinical implementations, the current tabular version of the InsighTx model can be combined with image analysis outputs, with PC #1 representing the level of radiographic consolidation and infiltration. This aligns with the observation that consolidation and infiltration are important radiographic abnormalities that should affect clinical judgment to transplant the lungs. The other neural network outputs that impact prognostic ability, PCs #2 to #10, were moderately correlated with other radiographic findings and functional parameters. These findings demonstrate that the ResNet-50 model learned radiographic patterns in donor lung injury relevant to the physiological function, and importantly, there is additional variance attributed to radiographic analysis that doesn’t already exist in the current functional assessments. Previous work by our group has demonstrated that the InsighTx model can increase the likelihood of transplanting suitable donor lungs and avoid transplantation of unsuitable lungs10. Herein we show that the addition of radiographic features to the model would significantly improve InsighTx performance and, as such, is likely to build on the impact of the model in surgical decision-making. However, prospective validation is necessary to fully validate these findings. Adding to the interpretability of the model, the class activation maps in this study showed that the neural network learned to increase the likelihood of declining donor lungs in response to changes in pixels in the middle and lower lung lobes. This is an important finding consistent with clinical observations that radiographic abnormalities were most prevalent in the middle and lower lobes17. When donor lungs were linked with good post-operative outcomes, the model tended to scan across the image and not be affected by a particular region, which is also aligned with clinical practice18,19.
There are limitations to this study. First, the interpretation of neural networks in image analysis remains a major hurdle in the field. Current saliency mapping methods in computer vision cannot reveal why or how the neural networks make decisions. The partial derivatives and averaging computations over many layers of a neural network also result in significant loss in precision and resolution of the saliencies. In our study, we applied GradCAM along with latent feature analysis to demonstrate that our model has learned information relevant to donor lung injury. Further clinical interpretation of GradCAM visualizations beyond these observations must be approached with caution. Second, the performance of radiographic analysis in classifying transplant outcomes was anticipated from our experience with other EVLP machine learning modeling. While decisions to transplant or decline lungs mainly depend on donor lung data, recipient outcomes are acknowledged to be significantly influenced by recipient factors. Because the CNNs here were agnostic to recipient information, it was a challenging task predicting recipient ventilation outcomes only using donor lung radiographs. However, similar to how clinicians evaluate donor lungs, radiographic analysis would only be considered along with other functional assessments. We therefore emphasize that incorporating X-ray features in the overall modeling indeed improved diagnostic performance. Third, the current image processing pipeline simultaneously analyzes temporal images from a given EVLP case. If only one image was available, a separate pipeline would be required. Single image analyses are however simpler and can be developed using the approach described herein.
In this study, we leveraged a unique dataset of isolated ex vivo human lung radiographs to obtain important prognostic information for transplant patients using computer vision. We have successfully achieved a multi-modal approach to EVLP decision making that includes radiographic analysis and lung function data. Deep learning models trained to interpret EVLP X-ray images were found to contribute considerably to the current donor lung evaluation techniques that use tabular functional assessments. Outputs of the adopted model can be largely interpreted as an assessment of clinical consolidation and infiltration. The resulting algorithm can be applied real-time in clinical EVLP; work is underway to store source code and data to accommodate the expanding database and to develop a user-friendly interface. Because there are no radiologists involved in EVLP procedures, a deep learning approach that automatically processes and analyzes EVLP radiographs can be considered a “virtual radiologist” for the transplant team. Implementing artificial intelligence in EVLP will ultimately add substantial value to the current donor lung diagnostics, serve as a step towards precision medicine, inform better clinical decisions, and improve the outcomes and quality of life for lung transplant recipients.
Methods
Study cohort
Clinical EVLP cases performed from 2008 to 2022 at Toronto General Hospital were considered in this study. All bilateral EVLP cases that had both 1 h and 3 h radiographs were included, yielding a total of n = 650 EVLP cases and n = 1300 radiographs. The cohort was split in an 80:20 ratio temporally into the training set (n = 520) and the validation set (n = 130), where the training set included cases performed between 2008 and 2020 and the validation set included cases from 2020 to 2022.
Inclusion and ethics
This study included all EVLP cases performed at our institution from 2008 to 2022. In accordance with the Declaration of Helsinki, University Health Network (UHN) Research Ethics Board (REB) and institutional approval was obtained for the collection, storage, and analyses of the biospecimens and data used in this study (UHN REB#12-5488-13 and UHN REB#11-0170-AE); informed patient consent was obtained from study participants.
Data collection and storage
The Toronto EVLP protocol has been previously reported4. Physiological and biological assessments were conducted using an ICU-grade ventilator, perfusate samples from the EVLP circuit, and pressure monitors. Radiographs were taken every 2 h15. Tabular data was stored within the Toronto Lung Transplant Program database, and radiographs on the UHN Picture Archiving and Communication System (PACS).
Image processing pipeline
The computational pipeline consisted of two main stages – pretraining and finetuning. Established CNN architectures including ResNet-5020, ResNeXt-5021, RexNet-10022, EfficientNet-B223, EfficientNet-B323, and DenseNet-12124 were used in both stages. The pretrain stage was performed on the PyTorch Image Models library25 available on GitHub. The CNN architectures were pretrained separately using two public chest X-ray datasets from the National Institutes of Health: one from the entire ChestX-ray14 dataset (n = 112,120) expanded from the ChestX-ray8 dataset26, and the other one that is a subset of the ChestX-ray14 dataset with cleaner labels (n = 10,000), published by the CADLab researchers27.
The finetune stage was developed using the PyTorch and Lightning libraries on Python (Version 3·10), which is composed of four modules: data loader, model training, on-the-fly validation, as well as inference. The model weights from the pretrained CNNs were used to initialize and further finetune the models using EVLP radiographs. The models were separately trained to classify donor lung outcomes as three classes: (1) transplanted lungs with recipient mechanical ventilation <72 h, (2) transplanted lungs with recipient mechanical ventilation ≥72 h, and (3) lungs deemed unsuitable for transplant.
The model performance was evaluated on the validation set using accuracy and area under the receiver operating characteristic (AUROC) curve. In the finetuning stage, the pipeline was designed to process 1 h and 3 h images separately from a given EVLP case, and then perform one single classification using the concatenated latent features from both images. An overview of this pipeline is shown in Fig. 3.
Class activation mapping
As a method for interpreting CNN classifications, gradient-weighted class activation (GradCAM)28 maps of the ResNet-50 model were generated using the pytorch-grad-cam library29 available on GitHub, as well as PyTorch Image Library and Seaborn packages. The last convolution layer in the third block of the ResNet-50 architecture was used to generate saliency maps. The activation saliencies were shown on the original images in full resolution.
Manual scoring of radiographs
As described in the previous study15, the manual labeling of the X-ray images was derived from scoring radiographic consolidation, infiltrate, atelectasis, nodule, and interstitial line findings across six lung regions (right upper lobe, right middle lobe, right lower lobe, left upper lobe, lingula and left lower lobe). In this dataset, infiltrate was defined similar to ground glass opacity, as an area of abnormal increased density through which the underlying lung markings can still be observed. The total score of each finding across all lung regions was used in subsequent analyses.
Principal component analysis
ResNet-50 and 115 cases in the validation cohort were used for the following analysis. A set of latent features that described the input images were extracted from the second to the last layer in the CNN, just before the classifier. PC analysis was performed to explain the latent features using ten vectors. The ten PCs from each EVLP case were correlated with donor information and EVLP parameters using Pearson correlations. Within the 115 cases, PCs from 38 cases were correlated with manually-derived radiographic labels from our previous study15 using Spearman correlations. The resulting heatmap was generated using the Corrplot package (version 0·92) in R.
Extreme gradient boosting model
The extreme gradient boosting (XGBoost) model30 is a state-of-the-art machine learning method for analyzing tabular data. The mechanism involves building an ensemble of decision trees and improving their overall performance by correcting errors from previous trees. In this study, two XGBoost were built using the validation cohort from the CNN image analysis (N = 115), which was then split 80:20 into training and validation sets. The first XGBoost model was trained in a similar fashion to InsighTx10, using EVLP physiological and biological parameters to classify the transplant endpoints described above. Another XGBoost model was trained on the same cohort with both EVLP tabular data and the ten PCs summarizing latent CNN features. The mean and standard deviation of the model performances were derived through bootstrapping. Statistical significance tests were conducted through one-tailed T-tests and Mann-Whitney U tests, using an alpha of 0.05. Model development and evaluation were performed through the Python Sci-kit Learn library.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Our study design did not include provisions to share the de-identified individual participant data, given historical concerns from our institution’s Research Ethics Board on the inherent risk of potentially identifying a participant using a combination of de-identified data fields. Thus, individual patient data from this study will not be made available in publicly accessible databases. However, the authors will take reasonable best efforts to grant access for requests from researchers affiliated to accredited research institutions wherever possible and with appropriate signed data sharing agreements.
Code availability
The study design approved by our institution did not include provisions to share source model weights from this study and it is not available in publicly accessible databases. However, the authors will take reasonable best efforts to grant access to related documents (e.g., study protocol, analysis plan) for requests from researchers affiliated to accredited research institutions wherever possible and with appropriate signed data sharing agreements. The image analysis pipeline can be found via GitHub (https://github.com/bowang-lab).
References
Ali, A. & Cypel, M. Ex-vivo lung perfusion and ventilation: where to from here? Curr. Opin. Organ Transplant. 24, 297–304 (2019).
Tian, D. et al. Outcomes of marginal donors for lung transplantation after ex vivo lung perfusion: a systematic review and meta-analysis. J. Thorac. Cardiovasc. Surg. 159, 720–730.e6 (2020).
Keshavjee, S. Human organ repair centers: Fact or fiction? JTCVS Open 3, 164–168 (2020).
Cypel, M. et al. Normothermic ex vivo lung perfusion in clinical lung transplantation. N. Engl. J. Med. 10, 1431–1440 (2011).
Yeung, J. C. et al. Physiologic assessment of the ex vivo donor lung for transplantation. J. Heart Lung Transplant. 31, 1120–1126 (2012).
Ferdinand, J. R. et al. Transcriptional analysis identifies potential novel biomarkers associated with successful ex‐vivo perfusion of human donor lungs. Clin. Transplant. 36, e14570 (2022).
Machuca, T. N. et al. Protein expression profiling predicts graft performance in clinical ex vivo lung perfusion. Ann. Surg. 261, 591–597 (2015).
Sage, A. T. et al. Prediction of donor related lung injury in clinical lung transplantation using a validated ex vivo lung perfusion inflammation score. J. Heart Lung Transplant. S1053249821022191 (2021) https://doi.org/10.1016/j.healun.2021.03.002.
Di Nardo, M. et al. Predicting donor lung acceptance for transplant during ex vivo lung perfusion: the EX vivo lung PerfusIon pREdiction (EXPIRE). Am. J. Transplant. 21, 3704–3713 (2021).
Sage, A. T. et al. A machine-learning approach to human ex vivo lung perfusion predicts transplantation outcomes and promotes organ utilization. Nat. Commun. 14, 4810 (2023).
Chao, B. T. et al. A radiographic score for human donor lungs on ex vivo lung perfusion predicts transplant outcomes. J. Heart Lung Transplant. Off. Publ. Int. Soc. Heart Transplant. S1053-2498 (24) 00004–4 (2024) https://doi.org/10.1016/j.healun.2024.01.004.
Akhter, Y., Singh, R. & Vatsa, M. AI-based radiodiagnosis using chest X-rays: a review. Front. Big Data 6, 1120989 (2023).
Moses, D. A. Deep learning applied to automatic disease detection using chest X‐rays. J. Med. Imaging Radiat. Oncol. 65, 498–517 (2021).
Jones, C. M. et al. Chest radiographs and machine learning – Past, present and future. J. Med. Imaging Radiat. Oncol. 1754-9485.13274 (2021) https://doi.org/10.1111/1754-9485.13274.
Chao, B. T. et al. Standardized radiographic evaluation of human donor lungs during ex vivo lung perfusion predicts lung injury and lung transplant outcomes. J. Heart Lung Transplant. 41, S13–S14 (2022).
Sailunaz, K., Özyer, T., Rokne, J. & Alhajj, R. A survey of machine learning-based methods for COVID-19 medical image analysis. Med. Biol. Eng. Comput. 61, 1257–1297 (2023).
Sakota, D. et al. Optical oxygen saturation imaging in cellular ex vivo lung perfusion to assess lobular pulmonary function. Biomed. Opt. Express 13, 328 (2022).
Shome, D. et al. COVID-transformer: interpretable COVID-19 detection using vision transformer for healthcare. Int. J. Environ. Res. Public. Health 18, 11086 (2021).
Lv, D. et al. A Cascade‐SEME network for COVID‐19 detection in chest x‐ray images. Med. Phys. 48, 2337–2353 (2021).
He, K., Zhang, X., Ren, S., Sun, J. Deep Residual Learning for Image Recognition. in 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 770–778 (IEEE, Las Vegas, NV, USA, 2016) https://doi.org/10.1109/CVPR.2016.90.
Liu, Z. et al. A ConvNet for the 2020s. in 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 11966–11976 (IEEE, New Orleans, LA, USA, 2022). https://doi.org/10.1109/CVPR52688.2022.01167.
Han, D., Yun, S., Heo, B. & Yoo, Y. Rethinking channel dimensions for efficient model design. (2020) https://doi.org/10.48550/ARXIV.2007.00992.
Tan, M. & Le, Q. V. EfficientNet: rethinking model scaling for convolutional neural networks. (2019) https://doi.org/10.48550/ARXIV.1905.11946.
Huang, G., Liu, Z., van der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. (2016) https://doi.org/10.48550/ARXIV.1608.06993.
Wightman, R. et al. Rwightman/pytorch-image-models: (2019) https://doi.org/10.5281/ZENODO.4414861.
Wang, X, et al. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. Proceedings of the IEEE conference on computer vision and pattern recognition. 2097–2106 (2017).
Tang, Y.-X. et al. Automated abnormality classification of chest radiographs using deep convolutional neural networks. Npj Digit. Med. 3, 70 (2020).
Selvaraju, R. R. et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 128, 336–359 (2020).
Gildenblat, J. et al. PyTorch library for CAM methods. GitHub. (2021) https://github.com/jacobgil/pytorch-grad-cam.
Chen, T. & Guestrin, C. XGBoost: A scalable tree boosting system. in Proc. of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 785–794 (ACM, San Francisco California USA, 2016) https://doi.org/10.1145/2939672.2939785.
Acknowledgements
This study was supported by the UHN Foundation and T-CAIREM (Temerty Center for AI Research and Education in Medicine) at the University of Toronto via an Innovation Grant for AI in Medicine. The funders of this study had no role in study design, data collection, analysis, data interpretation, or writing of this paper. The authors wish to thank the Biobank teams at UHN (TLTP Biobank) for their efforts to collect and process the samples used in this study. Additional thanks to the EVLP Team including all organ perfusion specialists and fellows, as well as Rasheed Ghany for their help with EVLP sampling and clinical database used in this study, respectively.
Author information
Authors and Affiliations
Contributions
B.T.C., A.T.S., and S.K. contributed to and verified all aspects of the work. Additionally: conceptualization: B.T.C., A.T.S., S.K.; data curation: B.T.C., S.K., TLTP Biobank, UHN EVLP Team; formal analysis: B.T.C., A.T.S., M.C.M., M.G.V.I.; funding acquisition: A.T.S., J.V., B.W., S.K.; investigation: B.T.C., A.T.S., B.W., S.K.; methodology: B.T.C., A.T.S., J.M., X.Z., B.W., S.K.; project administration: J.V., S.K.; resources: M.C., M.L., B.W., S.K.; software: B.T.C., J.M., B.W.; supervision: A.T.S., M.C., M.L., B.W., S.K.; validation: B.T.C., A.T.S., S.K.; visualization: B.T.C., M.G.V.I.; writing—original draft: B.T.C., A.T.S.; writing—review & editing: all authors. All authors had full access to all the data.
Corresponding author
Ethics declarations
Competing interests
Keshavjee serves as Chief Medical Officer of Traferox Technologies and reports personal fees from Lung Bioengineering, outside the submitted work. A.T.S., M.C.M., M.C., B.W., and S.K. declare ongoing patent applications with University Health Network (No.US63/314,930 & No. US63/315,042) related to machine learning models for ex vivo perfusion used in this study. The investigators fully adhere to policies at University Health Network that ensure academic integrity and management of potential conflicts of interest. B.T.C., J.M., M.G.V.I., X.Z., J.V., M.L. declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chao, B.T., Sage, A.T., McInnis, M.C. et al. Improving prognostic accuracy in lung transplantation using unique features of isolated human lung radiographs. npj Digit. Med. 7, 272 (2024). https://doi.org/10.1038/s41746-024-01260-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-024-01260-z