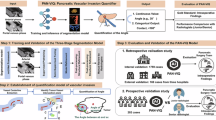
Abstract
Laparoscopic pancreatic surgery remains highly challenging due to the complexity of the pancreas and surrounding vascular structures, with risk of injuring critical blood vessels such as the Superior Mesenteric Vein (SMV)-Portal Vein (PV) axis and splenic vein. Here, we evaluated the High Resolution Network (HRNet)-Full Convolutional Network (FCN) model for its ability to accurately identify vascular contours and improve surgical safety. Using 12,694 images from 126 laparoscopic distal pancreatectomy (LDP) videos and 35,986 images from 138 Whipple procedure videos, the model demonstrated robust performance, achieving a mean Dice coefficient of 0.754, a recall of 85.00%, and a precision of 91.10%. By combining datasets from LDP and Whipple procedures, the model showed strong generalization across different surgical contexts and achieved real-time processing speeds of 11 frames per second during surgery process. These findings highlight HRNet-FCN’s potential to recognize anatomical landmarks, enhance surgical precision, reduce complications, and improve laparoscopic pancreatic outcomes.
Similar content being viewed by others
Introduction
As an increasingly popular treatment for pancreatic diseases, laparoscopic pancreatic surgery offers a minimally invasive approach, reducing recovery time, postoperative pain and hospital stays1. However, laparoscopic pancreatic surgery remains highly challenging due to the complexity of the pancreas and surrounding structures. One of the main challenges for pancreatic surgery is the manipulation of critical anatomical vessels, especially the Superior Mesenteric Vein-Portal Vein (SMV-PV) axis and the splenic vein2. These vessels pose a significant challenge due to their susceptibility to intraoperative bleeding, exacerbated by the veins’ delicate nature3. SMV-PV axis is a crucial factor in determining the resectability of pancreatic tumors, especially when venous invasion occurs, requiring preservation or reconstruction for successful surgery4,5,6. Proper management of this axis during venous resection is essential to minimize complications and maintain splenic vein function, which significantly impacts surgical outcomes7. Therefore, accurate identification and careful handling of the SMV-PV axis and splenic vein is one of the most difficult complications in pancreatic surgery4. However, as the surgeon operating with the laparoscopy cannot use the “sense of touch” to identify blood8, it is important to enhance the visual identification of SMV-PV axis and splenic vein during laparoscopic pancreatic surgery.
In the field of medical imaging recognition, Deep Learning (DL) technology has catalyzed significant breakthroughs in previous medical imaging studies, like ultrasound9, PET-CT10, CT11, MR12 and Retinal Fundus Photographs13. Specifically, in the realm of intelligent surgery, DL has proven its merit in accurately identifying and segmenting critical arteries, such as renal14 and mesenteric arteries15, achieving impressive precision of 0.937. Over the past three years, several leading publications have reported the application of DL technology in identifying anatomical landmarks and safety assessments in surgical procedures16,17,18,19,20. However, these studies predominantly focused on cholecystectomy and endoscopic pituitary surgery, leaving other surgical scenarios, such as pancreatic surgery, relatively unexplored.
Here, we present two examples of SMV-PV axis and the splenic vein recognition in laparoscopic pancreatic surgery, including Laparoscopic Distal Pancreatectomy (LDP) and Pancreaticoduodenectomy (Whipple procedure). LDP and Whipple are both widely recognized as a standard procedure for both benign and malignant pancreatic diseases. LDP is particularly for lesions located in the pancreatic tail or body21,22,23,24, while the Whipple procedure is primarily performed for lesions located in the pancreatic head, as well as the duodenum, common bile duct, or surrounding areas25. However, both LDP and Whipple are complex surgical procedure that requires high level qualifications and trainings for surgeons: surgeons typically need to complete hundreds of laparoscopic procedures in other areas under to develop the expertise necessary for safe performance of LDP26 and Whipple27,28. Therefore, it is important to enhance the venous anatomical landmarks for LDP Whipple procedure, for assisting surgery and educational purpose.
In both processes, we constructed an annotated database of SMV-PV images from experienced surgeons. We then employed the High Resolution Network (HRNet)29 to train the model, which identified and delineated the SMV-PV axis and splenic vein in LDP, and explored the real time segmentation of anatomical landmarks. Our model allows for instant identification of the anatomical landmarks during pancreatic surgery, enhancing surgical precision. Overall, this work can enhance the safety, reduces the surgeon’s stress levels, and contribute to DL-based anatomical landmarks identification in pancreatic surgery.
Results
Dataset description
In this study, the dataset was divided into training and testing sets, as shown in Table 1. For the LDP group, 25 cases were included, resulting in 126 videos and 12,694 frames. The training set consisted of 10,434 frames from 19 patients, while the testing set included 2260 frames from 6 patients. In the Whipple group, 30 cases were included, yielding 138 videos and a total of 35,986 frames. The training set comprised 28,915 frames from 22 patients, and the testing set included 7071 frames from 8 patients (Supplementary Table 1). This distribution ensures a comprehensive evaluation of the model’s performance across varying surgical contexts.
Recognition of SMV-PV axis and the splenic vein separately
Firstly, we tried individual recognition of SMV-PV axis and the splenic vein in LDP. Here we used multiclass segmentation to classify each image pixel into one of three categories: non-vein, SMV-PV or splenic vein. To test the model performance in different image quality, we classified the surgery images into high and low difficulty groups (Supplementary Table 1, also see Methods and Supplementary Fig. 2). For the splenic vein, recall and precision are slightly lower in high-difficulty cases (0.740 and 0.809) compared to low-difficulty cases (0.771 and 0.822), resulting in a lower mean Dice score (0.452 vs. 0.545). Similarly, for the SMV-PV axis, recall and precision also decline in high-difficulty cases (0.754 and 0.682) compared to low-difficulty cases (0.795 and 0.646), with mean Dice scores of 0.463 and 0.466, respectively. Overall, both segmentation accuracy and efficiency remain underwhelming, with performance further degrading in more complex scenarios, underscoring the need for model improvement (Supplementary Table 2).
However, we noticed that the multiclass segmentation recognition results were suboptimal (Supplementary Table 2), as the model might struggle to differentiate between specific types of blood vessels (see more in discussion). Anatomically, the splenic vein merges with the SMV to form the PV, so we hypothesized that treating them as a single entity could improve model performance. By merging the two veins into one unified “venous anatomical landmark,” we aimed to enhance the model’s generalization ability and its applicability across different surgical procedures. As a result, the SMV-PV axis and splenic vein were classified as a single “combined vein” entity for all subsequent binary segmentation analyses of vein vs. non-vein.
Recognition of SMV-PV axis and the splenic vein as venous anatomical landmarks
We then performed binary segmentation analyses of anatomical landmarks in LDP and Whipple surgeries (Table 2 and Supplementary Table 3). In the LDP group, the model achieved a Recall of 84.10%, Precision of 76.30%, and Dice coefficient of 0.645 on low-difficulty cases, while performance on high-difficulty cases dropped to a Recall of 60.20%, Precision of 73.40%, and Dice of 0.465 (Table 2). In the Whipple group, results were similar, with a Recall of 82.50%, Precision of 86.60%, and Dice of 0.668 on low-difficulty cases, but lower performance on high-difficulty cases (Recall 61.90%, Precision 80.70%, Dice 0.512) (Table 2). Notably, using the combined dataset (All) yielded the highest performance on low-difficulty cases (Recall 92.40%, Precision 85.60%, Dice 0.738) and showed improved performance on high-difficulty cases (Recall 72.50%, Precision 80.50%, Dice 0.537) compared to either LDP or Whipple alone (Table 2). These results indicated strong model performance in the vein recognition. Moreover, our model, which requires only a A100 Tensor Core GPU, demonstrated strong real-time capability of 11 frames per second (fps) (Supplementary Video 1).
We then compared the model performance differences between HRNet-FCN and Dlink-Net +30, Deeplabv3++31, U-Net+++32 and UperNet_swin_tiny+33,34 in the binary segmentation task of the SMV-PV axis system (Supplementary Table 4), including key metrics of Giga Floating Point Operations per Second (GFLOPs), recall, precision, false negative rate, false positive rate, and mean Dice coefficient. Although different models had different advantages, we noticed that HRNet-FCN outperformed all other models with a relatively high GFLOPs (38.75), the highest Mean Dice coefficient (0.720) and Recall (0.946) in low-difficulty cases, and a competitive performance in high-difficulty cases with a lower computational cost. These results further support our model selection by demonstrating its efficiency and effectiveness in vascular segmentation tasks.
Recognition of venous anatomical landmarks with combine training set
To further evaluate the model’s generalization ability, we began by removing the boundary between difficulty levels (Table 3, Supplementary Table 5). In LDP group, although the high/low difficulty ratio was nearly 2:1 (Supplementary Table 1), the overall recall for uniform difficulty levels closely matched that of the low-difficulty level. Interestingly, both precision and Dice scores were improved when we combined the training sets. For example, the combined training set achieved a recall of 89.70% and precision of 93.50% on the LDP test set, resulting in a Dice score of 0.713 (Table 3). This suggests that combining different sub-training sets enhances the model’s overall performance. Consequently, we merged the LDP and Whipple training sets and examined the test set under both individual and uniform conditions. Specifically, the Whipple test set with the combined training achieved a recall of 83.80%, precision of 90.40%, and a Dice score of 0.767, slightly outperforming the LDP test set’s Dice score of 0.713 in the same combined training setup (Table 3). Additionally, when tested with both training and test sets combined, the model achieved intermediate performance, with a recall of 85.00%, precision of 91.10%, and Dice score of 0.754 (Table 3), sitting between the individual performances of LDP and Whipple.
Furthermore, the combined dataset enables more accurate and comprehensive recognition. Compared to the model trained solely on the LDP dataset, the model trained on the combined dataset can identify previously unrecognized regions (identification) or further refining the recognition of existing regions (completion) on both LDP and Whipple test set (Fig. 1). Our model can accurately identify the SMV-PV axis in real-time during pancreatic surgery, enhancing precision and supporting surgeons with clear, reliable visualization throughout the procedure. We performed real-time processing on an 11-fps video both in the laboratory and, importantly, in real-world operating room settings (Supplementary Video 1 and 3) and non-real-time processing on a 24-fps video (Supplementary Video 2). In summary, integrating the SMV-PV axis and the splenic vein, along with unifying difficulty levels, resulted in combined training sets that enhanced the performance across different test sets.
Overall, by integrating the veins, unifying difficulty levels, and combining training sets, our HRNet-FCN model demonstrated robust performance in the vein recognition task. The enhanced results achieved with the combined dataset indicate that the model generalizes well to different surgical contexts and can effectively support real-time vessel recognition, ensuring precision and safety in complex surgical operations.
Discussion
This study developed a machine learning approach for detecting the representative veins in laparoscopic pancreatic surgery, which is the first anatomical landmarks recognition for laparoscopic surgery to our best knowledge. It has now been experimentally deployed in real-world operating room settings.
Our HRNet-FCN model has the following advantages: 1) unlike traditional encoder-decoder architectures such as U-Net and DeepLabV3+, which downsample high-resolution inputs and later upsample them to recover fine details, HRNet maintains high-resolution representations by parallelly connecting multi-scale sub-networks and performing repeated multi-scale fusion. By allowing high-resolution features to be continuously refined with contextual information from lower-resolution branches, HRNet enhances segmentation accuracy, particularly for fine vascular structures. 2) We incorporated FCN to optimize feature representation for our specific surgical application, ensuring robust performance in real-time vascular segmentation. Here, we noticed that false positives were often caused by similar features, while the false negative cases were mainly caused by occlusion, followed by lack of distinction from surrounding tissues (Supplementary Fig. 4). Notably, we found that many small fragments were not eliminated during post-processing, leading to a high number of false positives. Therefore, we adjusted the convolutional kernel from 7 × 7 to 69 × 69, which resulted in a significant improvement in precision (Supplementary Fig. 3, Supplementary Table 6). Furthermore, experienced surgeons prefer a clearer field of view during surgery, these small targets offer limited assistance, making our streamlined approach highly justified.
However, when returning to multiclass segmentation task, the performance was less satisfactory. Previous attempts by Nakamura35 and Tokuyasu36, using the YoloV3 algorithm for multi-object detection during gallbladder resection without relying on pixel-level segmentation resulted in accuracy ranging only from 0.07 to 0.3235. Despite subsequent enhancements to the algorithm elevating the mean Dice coefficient to 0.72, 0.49, 0.46, and 0.6636, respectively, the results still felt short of expectations. Multiclass segmentation in anatomical landmarks involves categorizing different objects based on pixel-level segmentation. The multiclass segmentation of the SMV-PV axis and splenic vein obtained a mean Dice coefficient of around 0.5, indicating the challenges due to: 1) The indistinct visual contrast between those anatomical landmarks contributed to lower recognition accuracy, whereas the mean Dice coefficient for identification of gallbladder triangle is notably higher; 2) Obstructions of tissues or instruments often segmented the boundary of anatomic landmark structures, dividing them into several smaller segments, thereby complicating the recognition; 3) Simultaneous objects in multiclass semantic segmentation tasks may lead to a significant decrease in recognition accuracy37. Therefore, we merged the anatomical landmarks and applied binary segmentation for better model performance. Future iterations may adopt instance segmentation to identify critical vessels, including the dorsal pancreatic and first jejunal veins38. Also, ICG fluorescence might further improve vascular recognition and warrants future investigation39.
Despite the overall progress medical imaging, there still remains a data scarcity across diverse medical scenarios40,41, wherein some certain investigations have been limited into merely a single case42,43. This data deficiency represents a critical bottleneck for the ongoing research in DL-powered medical applications. Our study aims to address this gap by supplementing data for venous anatomical landmarks in laparoscopic pancreatic surgery. Here, we found that a larger, more diverse training set significantly improved performance (see improvement from Tables 2 to 3), highlighting the importance of data diversity for generalization and the model’s adaptability for minimal fine-tuning across datasets, so it is crucial to gather more diverse and extensive datasets from a wide range of medical centers and surgeons. Also, future work might use active learning to prioritize high-difficulty cases for annotation and retraining, while leveraging pre-trained networks from large-scale medical datasets to minimize manual effort and improve performance.
Our dataset currently lacks cases with venous anomalies, partly due to the limited sample size, which should be included in future work. Additionally, advanced tumor cases involving vascular structures, such as those with vascular involvement or borderline resectable pancreatic cancer (BRPC), are rarely performed laparoscopically at our institution due to clinical practices and guidelines and were therefore excluded44. Future studies could explore more datasets in these venous anomalies.
Although our model runs at 11-fps, human vision perceives delays under 100 ms as seamless45, ensuring a smooth experience. We implemented a dual-screen setup, with one displaying raw footage and the other showing AI-enhanced visualization (Supplementary Video 3), facilitating surgical decision-making while minimizing patient risk. However, some misidentifications, particularly false positives, still occur during the surgery (Supplementary Video 4), indicating an important area for future improvement. A key challenge is that certain structures, such as portions of the liver, share similar imaging characteristics with the SMV-PV complex and splenic vein due to their venous blood composition. To address this, future work could explore larger Transformer-based models with built-in spatial awareness46, which may enhance feature differentiation and reduce false positives. Additionally, our current algorithm only recognizes single image frames, lacking the contextual information under surgical logic. Current research used contextual information was not optimal, achieving a mean Dice coefficient of 0.718 and a recall rate of 0.507 for renal artery identification43, indicating further improvement required.
Sometimes bleeding might happen beyond instant visual recognition, so it is possible to integrate vessel and instrument recognition to predict bleeding caused by device contact, retrospectively tracing the source after hemorrhage, minimizing risks and adverse outcomes. Future work may incorporate multimodal techniques to enhance predictive accuracy and intraoperative decision-making. Furthermore, our works lays the foundation for AR-assisted surgical navigation, which could enhance intraoperative precision and safety through interactive overlays and 3D reconstructions47,48,49.
In summary, we conducted image segmentation in laparoscopic pancreatic surgery, identifying the contours of the SMV-PV axis and splenic vein, achieving commendable recognition results in operating room. Our study supplemented the research on automatic identification based on AI technology in pancreatic scenarios, confirming the effective identification of anatomical landmarks in pancreatic surgery. The model may further combine with laparoscopic and robotic surgical systems to enhance patients’ safety in pancreatic surgery.
Methods
Dataset generation
Patients who underwent laparoscopic distal pancreatectomy at Peking Union Medical College Hospital between January 2021 and June 2022 were included. Inclusion criteria were: 1) Videos should contain the whole procedures of surgery, 2) Procedures should be laparoscopic distal pancreatectomy or Whipple, and 3) Appearance of the SMV-PV axis in the video for at least ≥2 min. Exclusion criteria were: 1) Any interruptions to laparoscopic surgery, 2) Any history of abdominal or pelvic surgeries, 3) Cases with vascular anatomical variations, and 4) Tumors with vascular invasion or borderline resectable pancreatic cancer (BRPC). All patients had signed the informed consent form for the collection of surgical videos. Ethical approvals were both obtained from The Institutional Review Board (IRB) of Peking Union Medical College Hospital (PUMCH). Approval I-25PJ0451 authorized the collection of surgical videos for dataset inclusion, while I-25PJ0643 permitted the clinical deployment of the AI model in the operating room.
To generate our data set, video data from those patients were randomly assigned in a 5:1 ratio to the training and testing sets. Then, one senior surgeon selected all videos with the SMV-PV axis and splenic veins and further extracted them at 1 frame per second to generate the image set. Four junior surgeons delineated the contours of the SMV-PV axis and splenic vein from these images, and classified them into high and low difficulty groups (Supplementary Fig. 2). Any image meeting either of the following criteria was classified into the high difficulty group: 1) The external rectangular area of the target ≤64 × 64 pixels; 2) Target vessels were unclear for any reasons such as blurred, poor exposure, overexposure, blood immersion, or fascial coverage (Supplementary Fig. 2).
Model training
All images were scaled down to 960 × 540 by a 4 × 4 convolutional kernel. Data augmentation, including random flipping, brightness enhancement, contrast enhancement, and saturation enhancement were performed. Subsequently, all pixels in those images underwent z-score normalization to minimize the noise50. The images were then fed into the High Resolution Network (HRNet)-Full Convolutional Network (FCN), which was pre-trained on ImageNet dataset, for training.
HRNet maintained high resolution position-information throughout the whole process. There are two key features that could help HRNet to keep the information: 1. A parallel structure of feature maps with different resolutions; 2. An exchange of information between the high and low resolutions feature maps. In this study, HRNet is structured into 3 stages, and each stage comprises 3 different steps: 1. Downsampling all existing feature maps to generate a new feature map with 1/2 resolution level (e.g. 1/4 → 1/8); 2. Applying convolution to all maps to extract new features; 3. Fusing information through upsampling and downsampling. Finally, all feature maps were fused and sent to FCN to restore them to original resolution and generated the predictions. The overall schematic of the HRNet_FCN module is illustrated in Fig. 2. Please note that downsampling reduces the size of an image to decrease computation and capture essential features, while upsampling increases the image size to meet computational or network requirements. Training process is shown in Supplementary Fig. 1.
a Diagram of High-Resolu48tion Network (HRNet)-Full Convolutional Network (FCN). Images were scaled to 960 × 540 to input into HRNet_V2 Network for feature extraction. Feature maps at 1/4, 1/8, 1/16, and 1/32 levels were generated by HRNet_V2. After deconvolution, these feature maps were concatenated to form a fused feature map containing all information at different levels. Finally, the FCN performed convolution and deconvolution to obtain the result. b Details of the parallel structure. The original image underwent three phases to form feature maps at four different scaling levels. Parallel arrows represented convolution, downward arrows represented downsampling, and upward arrows represented upsampling.
Due to the significantly lower total number of pixels in the SMV-PV axis and splenic vein (positive samples) compared to the number of background pixels (negative samples) at a ratio of approximately 1:15, the indispensable balance of positive and negative samples was adjusted by a cross-entropy loss function based on their weighted ratio. For a given image, the expression of the loss function was:
where ytrue represented the label category, ypred represented the predicted label category of each pixel. n represented the total sum of all pixels in the image. The weight of each class (yclassweight) is generated as the reciprocal of the proportion of pixels in the entire image for that category.
Where num_yclass represented the total number of pixels of different classes in ground truth.
To eradicate spikes at the edges of the predicted areas and connect small predicted regions to achieve better recognition, morphological opening and closing were applied using the cv2.morphologyEx function from the Python library. The kernel size is 69 × 69 pixels.
Training parameters were set as follows: batch size of 16, SGD optimizer, Softmax classifier, an initial learning rate of 0.01 with polynomial decay to a minimum of 0.0001, spanning 80,000 epochs. The model was trained on NVIDIA A100 Tensor Core GPU, with Python 3.6 and PyTorch 0.4.1.
Evaluation
The Intersection over Union (IoU) was applied to assess the success predictions (Fig. 3). When the IoU ratio of the predicted box and the ground truth box was ≥a specific IoU threshold (in this study, usually 0.1 or 0.3), it was a true positive; otherwise, it was a false positive. Additionally, if a predicted box exists for a certain object but there is no corresponding ground truth box, it is called a false positive. Based on these results, precision and recall were calculated. The mean Dice coefficient was also used to evaluate the accuracy of model predictions. In terms of real-time performance, floating-point operations per second (FLOPs) were employed. The definitions of those parameters are in Supplement File1.
Recall (sensitivity) refers to the proportion of ground truth boxes correctly detected by the model out of all ground truth boxes. Precision refers to the proportion of true positives among all predicted boxes. The formulas for both are as follows:
Dice coefficient is a metric to evaluate the model’s detection performance. A higher mean Dice coefficient indicates better detection performance. The formula for calculating the mean Dice coefficient is as follows:
Disclosure of AI assistance
This manuscript used AI tools for language editing, with all generated content reviewed and approved by the authors. The use of AI is transparently disclosed in accordance with journal guidelines. No patient data were processed using AI, ensuring full compliance with privacy and ethical standards.
Data availability
The datasets used and analyzed during the current study available from the corresponding author on reasonable request.
Code availability
The underlying code for this study is not publicly available but may be made available to qualified researchers on reasonable request from the corresponding author.
References
Ammori, B. J. Pancreatic surgery in the laparoscopic era. Jop 4, 187–192 (2003).
Nagakawa, Y. et al. The Straightened Splenic Vessels Method Improves Surgical Outcomes of Laparoscopic Distal Pancreatectomy. Dig. Surg. 34, 289–297 (2017).
Liang, S., Hameed, U. & Jayaraman, S. Laparoscopic pancreatectomy: indications and outcomes. World J. Gastroenterol. 20, 14246–14254 (2014).
Kang, C. M. et al. Laparoscopic distal pancreatectomy with division of the pancreatic neck for benign and borderline malignant tumor in the proximal body of the pancreas. J. Laparoendosc. Adv. Surg. Tech. A 20, 581–586 (2010).
Hellman, P. et al. Surgical strategy for large or malignant endocrine pancreatic tumors. World J. Surg. 24, 1353–1360 (2000).
Pedrazzoli, S. Surgical Treatment of Pancreatic Cancer: Currently Debated Topics on Vascular Resection. Cancer Control 30, 10732748231153094 (2023).
Addeo, P. et al. Management of the splenic vein during a pancreaticoduodenectomy with venous resection for malignancy. Updates Surg. 68, 241–246 (2016).
Bari, H., Wadhwani, S. & Dasari, B. V. M. Role of artificial intelligence in hepatobiliary and pancreatic surgery. World J. Gastrointest. Surg. 13, 7–18 (2021).
Yao, Z. et al. Preoperative diagnosis and prediction of hepatocellular carcinoma: Radiomics analysis based on multi-modal ultrasound images. BMC Cancer 18, 1089 (2018).
van Helden, E. J. et al. Radiomics analysis of pre-treatment [(18)F]FDG PET/CT for patients with metastatic colorectal cancer undergoing palliative systemic treatment. Eur. J. Nucl. Med. Mol. Imaging 45, 2307–2317 (2018).
Hosny, A., Parmar, C., Quackenbush, J., Schwartz, L. H. & Aerts, H. Artificial intelligence in radiology. Nat. Rev. Cancer 18, 500–510 (2018).
Hoang, U. N. et al. Assessment of multiphasic contrast-enhanced MR textures in differentiating small renal mass subtypes. Abdom. Radio. 43, 3400–3409 (2018).
Gulshan, V. et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 316, 2402–2410 (2016).
Casella, A. et al. NephCNN: A deep-learning framework for vessel segmentation in nephrectomy laparoscopic videos. In Proc. 2020 25th International Conference on Pattern Recognition (ICPR), 6144–6149 (2021).
Kitaguchi, D. et al. Real-time vascular anatomical image navigation for laparoscopic surgery: experimental study. Surg. Endosc. 36, 6105–6112 (2022).
Mascagni, P. et al. Artificial Intelligence for Surgical Safety: Automatic Assessment of the Critical View of Safety in Laparoscopic Cholecystectomy Using Deep Learning. Ann. Surg. 275, 955–961 (2022).
Madani, A. et al. Artificial Intelligence for Intraoperative Guidance: Using Semantic Segmentation to Identify Surgical Anatomy During Laparoscopic Cholecystectomy. Ann. Surg. 276, 363–369 (2022).
Wu, S. et al. SurgSmart: an artificial intelligent system for quality control in laparoscopic cholecystectomy: an observational study. Int. J. Surg. 109, 1105–1114 (2023).
Khan, D. Z. et al. Artificial intelligence assisted operative anatomy recognition in endoscopic pituitary surgery. npj Digital Med. 7, 314 (2024).
Cheng, K. et al. Artificial intelligence-based automated laparoscopic cholecystectomy surgical phase recognition and analysis. Surg. Endosc. 36, 3160–3168 (2022).
Kudsi, O. Y., Gagner, M. & Jones, D. B. Laparoscopic distal pancreatectomy. Surg. Oncol. Clin. N. Am. 22, 59–73 (2013). vi.
Chung, J. C., Kim, H. C. & Song, O. P. Laparoscopic distal pancreatectomy for benign or borderline malignant pancreatic tumors. Turk. J. Gastroenterol. 25, 162–166 (2014). Suppl 1.
Cai, H., Feng, L. & Peng, B. Laparoscopic pancreatectomy for benign or low-grade malignant pancreatic tumors: outcomes in a single high-volume institution. BMC Surg. 21, 412 (2021).
Groot, V. P. et al. Patterns, Timing, and Predictors of Recurrence Following Pancreatectomy for Pancreatic Ductal Adenocarcinoma. Ann. Surg. 267, 936–945 (2018).
Gagner, M. & Palermo, M. Laparoscopic Whipple procedure: review of the literature. J. Hepatobiliary Pancreat. Surg. 16, 726–730 (2009).
Liao, C. H. et al. The feasibility of laparoscopic pancreaticoduodenectomy-a stepwise procedure and learning curve. Langenbecks Arch. Surg. 402, 853–861 (2017).
Sahakyan, M. A. et al. Implementation and training with laparoscopic distal pancreatectomy: 23-year experience from a high-volume center. Surg. Endosc. 36, 468–479 (2022).
van Ramshorst, T. M. E. et al. Learning curves in laparoscopic distal pancreatectomy: a different experience for each generation. Int. J. Surg. 109, 1648–1655 (2023).
ke, S. et al. High-Resolution Representations for Labeling Pixels and Regions (2019).
Zhou, L., Zhang, C. & Wu, M. D-LinkNet: LinkNet with Pretrained Encoder and Dilated Convolution for High Resolution Satellite Imagery Road Extraction. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW) 192–1924 (2018).
Chen, L.-C., Papandreou, G., Schroff, F. & Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv, http://arxiv.org/abs/1706.05587 (2017).
Zhou, Z., Rahman Siddiquee, M. M., Tajbakhsh, N. & Liang, J. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4. 3-11 (Springer, 2018).
Xiao, T., Liu, Y., Zhou, B., Jiang, Y. & Sun, J. Unified Perceptual Parsing for Scene Understanding. In Proc. Computer Vision – ECCV 2018: 15th European Conference, Munich, Germany, September 8–14, 2018, Proceedings, Part V 432–448 (Springer-Verlag, Berlin, Heidelberg, 2018).
Liu, Z. et al. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. In Proc. IEEE/CVF International Conference on Computer Vision (ICCV) 9992–10002 (2021).
Tokuyasu, T. et al. Development of an artificial intelligence system using deep learning to indicate anatomical landmarks during laparoscopic cholecystectomy. Surg. Endosc. 35, 1651–1658 (2021).
Nakanuma, H. et al. An intraoperative artificial intelligence system identifying anatomical landmarks for laparoscopic cholecystectomy: a prospective clinical feasibility trial (J-SUMMIT-C-01). Surg. Endosc. 37, 1933–1942 (2023).
Roß, T. et al. Comparative validation of multi-instance instrument segmentation in endoscopy: Results of the ROBUST-MIS 2019 challenge. Med Image Anal. 70, 101920 (2021).
Logarajah, S. I. et al. Whipple pancreatoduodenectomy: A technical illustration. Surg. Open Sci. 7, 62–67 (2022).
Ishizawa, T. “Bon mariage” of artificial intelligence and intraoperative fluorescence imaging for safer surgery. Artif. Intell. Surg. 3, 163–165 (2023).
Loukas, C., Gazis, A. & Schizas, D. Multiple instance convolutional neural network for gallbladder assessment from laparoscopic images. Int. J. Med. Robot. 18, e2445 (2022).
Leibetseder, A., Schoeffmann, K., Keckstein, J. & Keckstein, S. Post-surgical Endometriosis Segmentation in Laparoscopic Videos. In Proc. International Conference on Content-Based Multimedia Indexing (CBMI) 1–4 (2021).
Sonsilphong, S., Sonsilphong, A., Hormdee, D. & Khampitak, K. In 2022 International Electrical Engineering Congress (iEECON). 1–4 (IEEE, 2022).
Caballas, K., Bolingot, H. J., Libatique, N. & Tangonan, G. Development of a Visual Guidance System for Laparoscopic Surgical Palpation using Computer Vision, (2021).
Tempero, M. A. et al. Pancreatic Adenocarcinoma, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl Compr. Canc Netw. 19, 439–457 (2021).
Nielsen, J. Usability Engineering (Morgan Kaufmann Publishers Inc., 1994).
Chen, J. et al. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. ArXiv, http://arxiv.org/abs/2102.04306 (2021).
Kasai, M., Aihara, T. & Yamanaka, N. Enhancing liver surgery and transplantation: the role of 3D printing and virtual reality. Artif. Intell. Surg., 4, 180–186 (2024).
Ping, L. et al. Application and evaluation of surgical tool and tool tip recognition based on Convolutional Neural Network in multiple endoscopic surgical scenarios. Surgical Endosc. 37, 7376–7384 (2023).
Hua, S. et al. Automatic bleeding detection in laparoscopic surgery based on a faster region-based convolutional neural network. Ann. Transl. Med. 10, 546 (2021).
Shorten, C. & Khoshgoftaar, T. M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 6, 60 (2019).
Acknowledgements
This work was supported by National High Level Hospital Clinical Research Funding, No. 2022-PUMCH-A-052 and No. 2022-PUMCH-B-003. The funder played no role in study design, data collection, analysis and interpretation of data, or the writing of this manuscript.
Author information
Authors and Affiliations
Contributions
Data Collection, Z.W., J.G., C.F., X.H., R.C.; Parameter and Model Adjustment, Z.W., Q.Y.; Original Draft Preparation, Z.W., J.S., Q.Y., R.C., H.Z., L.P., S.H.; Review and Editing, Z.W., J.S., R.C., H.Z., S.H.; Supervision, S.H., W.W., J.S. and R.C. contributed equally to this work and should be considered co-first authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shi, J., Cui, R., Wang, Z. et al. Deep learning HRNet FCN for blood vessel identification in laparoscopic pancreatic surgery. npj Digit. Med. 8, 235 (2025). https://doi.org/10.1038/s41746-025-01663-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-01663-6