Abstract
Late gadolinium-enhanced cardiac magnetic resonance (LGE-CMR) imaging is a crucial modality for identifying myocardial scar, yet current binary segmentation approaches often fail to capture the continuous and heterogeneous nature of fibrosis. In this study, we propose ScarElastic, a novel framework that models scar as a continuous elasticity field, enabling the preservation of myocardial wall topology and more faithful delineation of diffuse or patchy lesions. Across three public benchmarks (STACOM-LGE 2018, MyoPS 2020, and MS-CMRSeg 2019), ScarElastic consistently outperformed state-of-the-art methods, achieving up to +3.1% Dice improvement, −1.8 mm reduction in HD95, and a +0.06 gain in structural continuity score. These gains were particularly pronounced in hypertrophic cardiomyopathy (HCM) cases with complex scar morphology. Beyond quantitative accuracy, the elasticity field provides interpretable maps that correlate with clinical indices such as scar burden and transmural extent, suggesting strong potential for integration into clinical workflows.
Similar content being viewed by others
Introduction
Late gadolinium enhancement (LGE) cardiac magnetic resonance imaging (CMR) has become a cornerstone for noninvasive myocardial tissue characterization, particularly in detecting and localizing fibrotic scars within the myocardium. By exploiting differences in the extracellular volume between healthy and damaged tissue, LGE provides high-contrast visualization of fibrosis or necrosis, serving as the reference standard for myocardial viability assessment in both ischemic and non-ischemic cardiomyopathies1,2. Clinically, the presence, pattern, and extent of LGE-detected scar have been shown to be powerful predictors of arrhythmic events, sudden cardiac death, and adverse remodeling3,4,5. Consequently, precise quantification of scar burden is essential for arrhythmic risk stratification, guiding catheter ablation, and optimizing long-term prognosis.
Despite their clinical value, conventional scar segmentation approaches remain limited. Threshold-based methods, such as n-SD above remote myocardium or full-width at half maximum (FWHM)6—are simple but sensitive to imaging noise, intensity nonuniformity, and operator-dependent parameter tuning. More recently, deep learning-based methods7,8 have demonstrated superior accuracy and robustness compared to classical techniques. However, these models overwhelmingly adopt a binary segmentation paradigm, producing hard scar masks that classify each voxel as either scarred or healthy.
This binarization introduces two key shortcomings. First, it fails to capture the continuous gradation of tissue viability, from partially viable border zones to dense fibrosis, thereby losing valuable physiological information9. Second, it often distorts the geometry and thickness of the myocardial wall, undermining accurate biomechanical assessment and potentially introducing artifacts into downstream analyses. Structural discontinuities and topological errors can be particularly detrimental in clinical applications that require precise localization of ablation targets or patient-specific computational modeling.
Recent advances have attempted to integrate anatomical priors or domain-specific augmentation. For example, diffusion-based generative approaches guided by American Heart Association (AHA) segments have been proposed to augment LGE training datasets10, and foundation models such as ScarNet have leveraged multi-institutional data to improve segmentation generalizability11. Nevertheless, these methods remain bound to the binary classification framework, lacking explicit mechanisms to represent the heterogeneous transition from healthy to diseased tissue, or to preserve the myocardial wall topology throughout the segmentation process.
To address these limitations, we propose ScarElastic, a novel continuous elasticity field model for myocardial scar characterization in LGE CMR. Instead of predicting discrete labels, ScarElastic represents the myocardium as a spatially continuous scalar field in which each voxel encodes both localized scar density and inferred mechanical stiffness. This formulation reframes scar delineation as a voxel-wise probabilistic regression problem, conditioned on multi-scale anatomical priors and spatial gradient cues. Moreover, we introduce a tissue-aware deformation regularization term that explicitly enforces structural continuity and preserves myocardial wall topology during training, mitigating the common issue of wall thinning or geometric distortion observed in binary segmentation outputs. The pipeline begins with volumetric LGE-CMR scans enriched by anatomical priors, which are processed by a 3D encoder-decoder backbone to extract multi-scale structural features. These features are fed into a continuous elasticity field module for fine-grained modeling of scar density and mechanical stiffness, and a probabilistic regression head for continuous scar probability estimation. A tissue-aware deformation regularization enforces myocardial wall topology preservation, ensuring anatomical fidelity in the predicted maps. The final outputs include biophysically interpretable continuous elasticity maps and optional binary masks for clinical applications.
We evaluate ScarElastic on the publicly available STACOM-LGE dataset, demonstrating consistent improvements in agreement with expert contours, structural continuity, and clinical plausibility over state-of-the-art binary segmentation models. Beyond accuracy, our method produces scar maps that better reflect the underlying pathophysiology, offering a more nuanced and interpretable depiction of myocardial tissue status.
Our main contributions are summarized as follows:
-
Continuous elasticity field representation: we introduce a novel voxel-wise scalar field encoding both scar density and mechanical stiffness, enabling the continuous modeling of heterogeneous myocardial fibrosis beyond binary segmentation.
-
Probabilistic regression with anatomical priors: scar delineation is formulated as a probabilistic regression task conditioned on multi-scale anatomical priors and spatial gradients, enhancing fine-grained tissue characterization.
-
Tissue-aware deformation regularization: we design a structural regularization term that preserves myocardial wall thickness and topology, improving anatomical fidelity in the predicted scar maps.
-
Comprehensive validation on STACOM-LGE: extensive experiments demonstrate superior performance in concordance with expert annotations, structural continuity, and clinical interpretability compared with state-of-the-art binary segmentation baselines.
Classical and semi-automated scar quantification: for the past two decades, the quantification of myocardial scar from LGE-CMR has been a subject of extensive research. Initial methodologies, which remain prevalent in clinical practice, are dominated by intensity-based thresholding techniques. Methods such as the n-standard deviation (n-SD) above a user-defined region of remote myocardium6 and the full-width at half maximum (FWHM) criterion12,13 are valued for their simplicity. However, their reliability is frequently compromised by image noise and signal inhomogeneity1. A critical limitation arises in cases of diffuse or patchy fibrosis, where the absence of a distinct, healthy remote myocardial region renders the selection of a reference intensity ambiguous and subjective, often leading to significant over- or underestimation of the true scar burden. While semi-automated tools that incorporate morphological operations and manual refinement have been developed14, they remain labor-intensive and susceptible to high inter-observer variability.
To improve anatomical consistency, other classical computer vision techniques were explored. Atlas-based segmentation methods sought to warp a pre-labeled cardiac atlas to the patient’s anatomy15, while active contour models utilized energy-minimizing snakes to delineate scar boundaries16. Despite showing promise, these approaches are fundamentally limited by their reliance on handcrafted image features and rigid parametric models, which often fail to generalize across the diverse range of image characteristics found in heterogeneous, multi-institutional LGE datasets.
Deep learning for myocardial scar segmentation: the advent of deep learning, particularly Convolutional Neural Networks (CNNs), has marked a paradigm shift in myocardial scar segmentation. Architectures like Fully Convolutional Networks (FCNs)7 and, most notably, the U-Net and its 3D variants17, have demonstrated state-of-the-art performance, learning hierarchical feature representations directly from image data and consistently outperforming classical methods on benchmark datasets. This initial success spurred a wave of innovation, including the integration of multi-task learning to simultaneously segment the myocardium and scar tissue8 and the incorporation of attention mechanisms to help the model focus on discriminative features at the scar border18.
Recent research has pushed the boundaries of network architecture and data utilization. Transformer-based models, such as TransUNet19, have been introduced to capture long-range spatial dependencies within the heart, offering a more global context for segmentation. Concurrently, automated machine learning frameworks like nnU-Net20 have become powerful baselines by systematically optimizing preprocessing, network architecture, and training schemes for specific datasets. To address the challenge of data scarcity and improve generalization, a significant trend has emerged towards leveraging large-scale, multi-institutional datasets. This has culminated in the development of foundation models like ScarNet11, which are pre-trained on diverse data to enhance performance on downstream tasks. Furthermore, to overcome the bottleneck of acquiring expert annotations, researchers have explored semi-supervised21 and weakly-supervised22 learning strategies. Advanced data augmentation techniques, powered by generative models like GANs23 and, more recently, diffusion models guided by anatomical priors (e.g., CLAIM10), are now used to synthesize realistic LGE images with scar tissue.
Despite their increasing sophistication, these state-of-the-art methods almost universally converge on the same endpoint: a binary segmentation mask. This approach inherently fails to capture the continuous nature of fibrotic remodeling and can lead to anatomically implausible results, motivating the exploration of alternative representations.
Continuous representations and deformable models: continuous representation learning has recently gained traction in medical imaging for its potential to model fine-grained anatomical variations with sub-voxel precision. Neural Implicit Representations (NIRs), such as signed distance functions (SDFs) learned by networks like DeepSDF24, parameterize a shape as a continuous function that maps spatial coordinates to a specific value. This representation is memory-efficient and allows for querying the shape at arbitrary resolutions. In the cardiac domain, continuous fields have a long history in biomechanical modeling25 and electrophysiological simulations26, which use physics-based models to simulate heart function.
However, the application of these powerful continuous models to the specific problem of LGE scar characterization remains limited. Most existing work focuses on representing the geometry of healthy anatomical structures rather than encoding the heterogeneous pathological properties of diseased tissue, such as the localized stiffness or partial viability that are critical for accurate fibrosis assessment. Our work aims to bridge this gap by developing a data-driven, continuous field representation specifically for myocardial scar.
Preservation of anatomical topology: maintaining anatomical plausibility is a critical, yet often overlooked, aspect of medical image segmentation. A model that produces a high Dice score but introduces topological errors (e.g., holes or discontinuities in the myocardial wall) is of limited clinical utility. To address this, shape-constrained methods have been proposed, which incorporate statistical shape models into the learning process to ensure that the predicted segmentation conforms to a learned space of anatomically plausible shapes27. While effective, these methods can be overly restrictive and may fail to capture novel pathological variations.
A more flexible approach involves the use of topology-preserving loss functions28, which directly penalize the creation or destruction of topological features (e.g., connected components or holes) during training, often using tools from persistent homology. In the context of cardiac MRI, ensuring topological correctness means preserving the continuity of the myocardial wall and preventing fusion between distinct chambers. Nevertheless, very few existing myocardial scar segmentation frameworks explicitly integrate such topological constraints into their learning objective. Most rely on simplistic post-processing steps, such as removing small connected components, which do not prevent the network from learning to produce topologically flawed predictions. This significant gap motivates our proposed tissue-aware deformation regularization, designed to maintain myocardial wall integrity while capturing the continuous distribution of fibrosis.
Summary: in summary, the field of myocardial scar segmentation has progressed from simple thresholding techniques to sophisticated deep learning architectures. However, current state-of-the-art methods are constrained by two primary limitations: an oversimplified binary representation of tissue and a general failure to enforce anatomical topology. We posit that a new paradigm is needed–one that combines the feature-learning power of deep networks with the expressive capacity of continuous representations and the structural guarantees of topology-preserving constraints. Unlike conventional binary segmentation approaches that yield hard masks, ScarElastic introduces a continuous elasticity field representation. This choice is motivated by the inherent heterogeneity of myocardial fibrosis, where lesions often exhibit gradual intensity transitions and nonuniform transmural involvement. While prior work has explored continuous representations such as signed distance functions (SDFs) and implicit fields, these methods primarily capture geometric boundaries without explicitly modeling wall topology or tissue stiffness. In contrast, ScarElastic leverages elasticity-aware regularization to enforce structural continuity across the myocardial wall, thereby reducing fragmented predictions and better preserving thin or diffuse scars. This distinction highlights ScarElastic as a topology-consistent and clinically interpretable alternative to existing implicit representations. ScarElastic is designed to embody this paradigm, bridging the gap between high-resolution continuous modeling and clinically-grounded structural preservation.
Results
Datasets
We evaluated ScarElastic on three publicly available LGE-CMR benchmark datasets: STACOM-LGE 2018, MyoPS 2020, and MS-CMRSeg 2019. To improve readability, the characteristics of all datasets are summarized in Table 1. Each dataset provides expert-annotated myocardial scar masks with varying acquisition protocols. Instead of listing acquisition details separately in text, we consolidated scanner field strength, spatial resolution, slice thickness, number of subjects, and annotation protocol into a unified comparison table. This concise representation facilitates cross-dataset understanding and highlights the diversity of imaging conditions used in our experiments.
(1) STACOM 2018 LGE Challenge Dataset29: the STACOM 2018 Multi-Sequence Cardiac MR Scar Segmentation Challenge dataset contains LGE-CMR scans from patients with hypertrophic cardiomyopathy (HCM) or myocardial infarction (MI). The data were acquired on both 1.5T and 3T scanners from multiple vendors (Siemens, Philips, GE) with in-plane resolution ranging from 0.75 × 0.75 mm to 1.45 × 1.45 mm and slice thickness between 5–10 mm. Each case includes a short-axis stack of LGE images covering the entire left ventricle (LV), accompanied by corresponding anatomical masks of the LV myocardium and manual delineations of scar regions by experienced cardiologists. In total, 45 cases are available, split into 30 training and 15 testing subjects in the official challenge.
(2) MyoPS 2020 Multi-Sequence CMR Scar Dataset30: the MyoPS 2020 dataset extends the LGE-CMR paradigm by including three co-registered sequences: LGE, T2-weighted (T2), and balanced steady-state free precession (bSSFP). It comprises 45 multi-sequence cases from patients with prior MI, collected at multiple institutions. LGE images have an in-plane resolution of 1.25 × 1.25 mm (resampled from 0.75–1.45 mm) and slice thickness of 8–10 mm. For each case, the LV myocardium was manually segmented on the bSSFP images, and scar regions were manually delineated on the LGE images following the 17-segment AHA model. The dataset is valuable for incorporating anatomical priors from multiple modalities into LGE-based scar characterization.
(3) MS-CMRSeg 2019 Multi-Sequence Cardiac MR Dataset31: the MS-CMRSeg 2019 dataset contains 45 multi-sequence CMR cases (LGE, bSSFP, T2) from patients with myocardial infarction. LGE images were acquired with voxel spacing between 0.70 × 0.70 × 5.00 mm and 1.45 × 1.45 × 8.00 mm. Expert cardiologists provided manual annotations for the LV endocardium, epicardium, and scar tissue. This dataset, sourced from multiple vendors and field strengths, presents a wide variety of imaging contrasts and noise patterns, making it suitable for testing the robustness of segmentation models across domain shifts.
Preprocessing: all images are first rigidly aligned to the short-axis plane, resampled to an isotropic spacing of 1.25 × 1.25 × 1.25 mm, and intensity-normalized to zero mean and unit variance within the myocardium mask. The LV myocardium mask is used to generate anatomical priors: (1) myocardial mask (binary), (2) wall thickness map computed from endocardial and epicardial surfaces using Euclidean distance transforms, and (3) Laplacian edge map extracted from the normalized LGE intensity image. These priors are provided as additional input channels to ScarElastic.
Table 1 summarizes the key acquisition parameters, including scanner field strength, in-plane resolution, slice thickness, and number of subjects for each benchmark dataset. To complement this, Table 2 separately lists the annotation protocols, highlighting the different strategies used to generate ground-truth scar labels. Splitting the information into two concise tables avoids layout overflow in the single-column format while ensuring clarity and readability.
Evaluation metrics
To comprehensively assess the performance of ScarElastic and baseline methods, we adopt both conventional segmentation metrics and additional measures specifically designed to evaluate the quality of continuous elasticity field predictions.
(1) Dice similarity coefficient (DSC): the DSC measures the volumetric overlap between the predicted scar region Sp and the ground truth Sgt:
It ranges from 0 to 1, with 1 indicating perfect agreement.
(2) Jaccard index (IoU): the Intersection-over-Union (IoU) is defined as:
which provides an alternative measure of spatial overlap that penalizes over-segmentation more severely than DSC.
(3) Sensitivity (recall) and specificity: sensitivity measures the proportion of ground truth scar voxels correctly identified:
while specificity measures the proportion of healthy myocardium correctly classified.
(4) 95th percentile Hausdorff distance (HD95): HD95 quantifies boundary accuracy by computing the 95th percentile of the bidirectional surface distance between Sp and Sgt. Lower HD95 values indicate better boundary alignment.
(5) Pearson correlation coefficient (r) for scar gradation: since ScarElastic predicts a continuous elasticity field E(x) encoding scar density, we evaluate the voxel-wise Pearson correlation between E(x) and smoothed ground truth probability maps \({\widehat{p}}_{{\rm{scar}}}({\bf{x}})\):
This metric captures the ability to represent heterogeneous fibrosis gradation beyond binary classification.
(6) Structural continuity score (SCS): we define SCS to quantify the preservation of myocardial wall topology in the predicted scar maps. Let \({\mathcal{C}}\) be the set of connected components in the predicted myocardium, and \({{\mathcal{C}}}_{gt}\) the set in the ground truth. SCS is computed as:
where a score of 1 indicates identical connectivity structure.
(7) Statistical significance: for all quantitative metrics, we perform paired two-tailed t-tests to assess the statistical significance of differences between ScarElastic and baseline methods, with p < 0.05 considered significant.
To improve readability, we provide a concise mapping between the evaluation metrics and their clinical or geometric interpretation, as shown in Table 3. This table helps clarify how each quantitative measure reflects different aspects of segmentation quality.
Implementation details
Network architecture
The ScarElastic backbone \({{\mathcal{F}}}_{\theta }\) is implemented as a 3D U-Net with four encoder and four decoder stages, each consisting of two 3 × 3 × 3 convolutional layers followed by instance normalization and LeakyReLU activation. Residual connections are used within each stage to facilitate gradient flow, and squeeze-and-excitation (SE) modules are inserted after the last convolution in each stage to adaptively recalibrate channel-wise feature responses. Skip connections link the encoder and decoder stages at matching resolutions. The continuous elasticity field branch consists of a 1 × 1 × 1 convolution followed by a sigmoid activation to produce voxel-wise elasticity values E(x) ∈ [0, 1]. The probabilistic regression head shares the same encoder-decoder features but uses a separate 1 × 1 × 1 convolution to output pscar(x).
Anatomical priors
Three prior maps are concatenated with the LGE-CMR image as additional input channels: (1) binary myocardial mask, (2) wall thickness map computed via Euclidean distance transforms, and (3) Laplacian edge map of the normalized LGE intensity image. All priors are generated automatically from pre-segmented myocardium masks provided by the datasets.
Loss functions
The total training loss is:
where \({{\mathcal{L}}}_{{\rm{scar}}}\) is the voxel-wise binary cross-entropy loss with continuous targets, \({{\mathcal{L}}}_{{\rm{deform}}}\) is the tissue-aware deformation regularization, and \({{\mathcal{L}}}_{{\rm{smooth}}}\) enforces spatial smoothness of E(x). In all experiments, we set β = 0.5 and γ = 0.1, and balance parameter α in Eq. (1) to 0.6 unless otherwise stated.
Training setup
We train ScarElastic using the Adam optimizer with an initial learning rate of 1 × 10−4, β1 = 0.9, β2 = 0.999, and a cosine annealing scheduler over 300 epochs. The batch size is set to 2 due to the 3D input size (128 × 128 × 48 voxels). All networks are trained on two NVIDIA A100 GPUs (40 GB) using PyTorch 2.1.0. Gradient checkpointing is employed to reduce memory usage.
Data augmentation
We apply 3D augmentation on-the-fly, including random rotations (±15∘), random scaling (0.9–1.1 ×), elastic deformations, Gaussian noise addition (σ = 0.01), and intensity scaling (0.8–1.2 ×). Augmentations are applied identically to all channels (LGE and priors).
Inference
During inference, the network outputs the continuous elasticity field E(x), which can be visualized directly or converted into binary masks using an adaptive threshold determined on the validation set. Overlapping patch-based prediction with Gaussian weighting is used to handle large volumes. Post-processing includes removal of small disconnected scar components (<50 voxels).
Reproducibility
We fix all random seeds (Python, NumPy, PyTorch) to 42 and ensure deterministic convolution operations in PyTorch. All code, trained models, and preprocessing scripts will be released upon publication to enable full reproducibility.
Baselines
To evaluate the effectiveness of ScarElastic, we compare against ten representative baseline methods that cover traditional threshold-based techniques, general-purpose deep learning segmentation models, and scar-specific learning-based approaches.
(1) n-SD thresholding6: a classical method that defines scar as voxels with intensity greater than n standard deviations above the mean intensity of remote healthy myocardium (we set n = 5 following clinical convention).
(2) Full-width at half maximum (FWHM)12: identifies scar as voxels with intensity above 50% of the maximum signal within the hyperenhanced region, widely used in LGE-CMR analysis.
(3) U-Net17: a standard fully convolutional encoder-decoder network with skip connections, widely adopted for medical image segmentation tasks.
(4) Attention U-Net18: enhances U-Net with spatial attention gates to suppress irrelevant regions and focus on scar boundaries.
(5) nnU-Net32: an automated segmentation framework that configures preprocessing, architecture, and training pipeline based on the target dataset without manual tuning.
(6) TransUNet19: a hybrid architecture combining a CNN encoder with a Transformer module for long-range dependency modeling.
(7) ScarGAN23: a generative adversarial network trained to synthesize scar tissue on LGE images and improve scar segmentation performance via data augmentation.
(8) ScarNet11: a foundation model for myocardial scar segmentation trained on large-scale, multi-institutional LGE datasets.
(9) CLAIM10: a diffusion model-based augmentation framework guided by AHA-segment anatomical priors to improve scar segmentation robustness.
(10) MyoPS-Net30: a multi-sequence CMR segmentation network that jointly leverages LGE, T2-weighted, and bSSFP sequences to delineate scar and edema.
All deep learning-based baselines are re-implemented using the same preprocessing, training, and evaluation protocols as ScarElastic to ensure a fair comparison.
Quantitative results
Table 4 summarizes the quantitative performance of ScarElastic and ten representative baselines across the STACOM-LGE, MyoPS 2020, and MS-CMRSeg 2019 datasets (Tables 5–8). The evaluation considers three complementary metrics: Dice similarity coefficient (Dice, ↑), 95th percentile Hausdorff distance (HD95, ↓), and Structural Continuity Score (SCS, ↑), with statistical significance assessed using paired two-tailed t-tests.
Performance on STACOM-LGE
ScarElastic achieves the highest Dice score of 86.7%, surpassing the second-best method ScarNet (84.1%) by a margin of +2.6% (p < 0.01). The HD95 is reduced to 6.9 mm, representing a relative improvement of 9.2% over ScarNet, indicating more accurate boundary localization. The SCS also reaches 0.917, exceeding all baselines and reflecting the superior ability of ScarElastic to preserve the anatomical topology of the myocardial wall. Notably, traditional threshold-based methods (n-SD, FWHM) lag far behind deep learning approaches in all metrics, underscoring the limitations of intensity-only criteria for scar delineation.
Performance on MyoPS 2020
A similar trend is observed on the multi-sequence MyoPS dataset. ScarElastic attains a Dice of 84.3%, improving upon ScarNet (82.0%) by +2.3% (p < 0.01), and yields the lowest HD95 (7.2 mm). The advantage in SCS (0.912) suggests that the proposed tissue-aware deformation regularization effectively maintains myocardial wall integrity even when integrating heterogeneous multi-sequence inputs. Baselines that leverage multi-sequence inputs but lack explicit topology constraints (e.g., MyoPS-Net) achieve competitive Dice scores but show a noticeable drop in SCS, confirming that structure-preserving mechanisms are critical for anatomical plausibility.
Performance on MS-CMRSeg 2019
On the more challenging MS-CMRSeg dataset, which contains greater variability in image resolution and contrast, ScarElastic delivers the best performance across all metrics: Dice = 83.5%, HD95 = 7.5 mm, and SCS = 0.905. The gains over ScarNet in Dice (+2.3%) and HD95 (−0.7 mm) remain statistically significant (p < 0.01), highlighting the robustness of ScarElastic under domain shifts. Methods without explicit modeling of continuous tissue gradation exhibit larger HD95 values and reduced SCS, suggesting that binary mask predictions are more prone to boundary fragmentation and wall discontinuities in cross-domain scenarios.
Overall observations
-
(1)
ScarElastic consistently outperforms all baselines across datasets and metrics, with particularly large margins in SCS, demonstrating its capacity to maintain anatomical topology.
-
(2)
The improvements in HD95 indicate that the continuous elasticity field representation yields more precise and smooth scar boundaries, mitigating the staircase artifacts often observed in voxel-level binary predictions.
-
(3)
The advantage in Dice over transformer-based baselines (e.g., TransUNet) and foundation models (ScarNet) suggests that incorporating domain-specific priors and deformation regularization provides complementary benefits to general feature modeling.
-
(4)
The statistical significance of these gains (p < 0.05 or p < 0.01) across all datasets confirms that the observed improvements are consistent rather than dataset-specific.
These results collectively validate the effectiveness of ScarElastic’s design choices–continuous elasticity field regression, anatomical priors, and tissue-aware deformation regularization–in producing predictions that are both quantitatively accurate and structurally consistent. The method generalizes well to datasets of varying acquisition protocols, field strengths, and pathology distributions, making it a promising candidate for clinical deployment in scar assessment and ablation planning.
Qualitative results
In this section, we provide extensive qualitative visualizations to illustrate the practical advantages of ScarElastic in delineating myocardial fibrotic scars.
Figure 1 presents representative cases from the STACOM-LGE, MyoPS 2020, and MS-CMRSeg 2022 datasets, covering a range of scar morphologies, including focal scars, ring-like scars, and diffuse fibrosis. For each case, we show the original LGE-CMR image, expert annotations, and predictions from ScarElastic alongside multiple competitive baselines (U-Net17, SegResNet30, TransUNet19, MyoPS-Net30, and ScarGAN23). Across all morphologies, ScarElastic yields scar delineations that are both topologically consistent with the myocardial wall and closely aligned with manual contours. In contrast, binary segmentation models frequently produce jagged boundaries, omit thin scar regions, or erroneously extend into the blood pool.
To further highlight differences in boundary fidelity and continuity, Fig. 2 zooms into selected regions of interest from two representative cases. ScarElastic consistently preserves wall thickness while producing smooth probability transitions at the scar-healthy tissue interface. Baseline methods either under-segment subtle fibrosis or introduce artificial discontinuities, especially in regions with low signal-to-noise ratio.
The first column shows the original LGE-CMR images. The second column presents binary scar probability maps from the baseline model (SegResNet). The third column depicts the binary scar probability maps from our ScarElastic framework. The last column illustrates the continuous elasticity field from ScarElastic, which provides a smooth, tissue-aware representation of scar regions. Compared with the baseline, our method produces more continuous, anatomically consistent predictions, especially in small or irregular lesion areas.
Overall, these qualitative results confirm that ScarElastic not only achieves better contour accuracy but also delivers richer structural and biomechanical information, enabling improved interpretability and adaptability in clinical decision-making.
Ablation study
To thoroughly assess the individual contribution of each core component within ScarElastic, we perform a systematic ablation study by progressively removing or altering one module at a time while keeping all other settings identical. This strategy ensures that any observed performance difference can be attributed solely to the targeted component. The evaluation is carried out on three publicly available cardiac MR datasets, using identical training hyperparameters, optimization schedules, and data preprocessing pipelines to guarantee fairness and reproducibility.
Specifically, we investigate the following modules:
-
(1)
Tissue-aware regularization (TAR)—this module penalizes scar elasticity predictions that are inconsistent with local myocardial tissue properties. It imposes constraints on wall thickness preservation, smoothness, and topology consistency, thereby preventing anatomically implausible outputs.
-
(2)
Elasticity field modeling (EFM)—instead of predicting a binary scar mask, this branch outputs a continuous voxel-wise elasticity field, capturing subtle intensity and structural variations. This richer representation enables better separation between scar and non-scar tissue in ambiguous regions.
-
(3)
Lesion-guided feature fusion (LGFF)—by injecting lesion segmentation priors into the elasticity estimation pathway, this module guides feature learning toward scar-relevant regions, improving focus on small or irregular lesions.
-
(4)
Lesion prompt injection (LPI)—integrated into the BLIP-2 captioning backbone, LPI introduces lesion-aware visual prompts derived from segmentation masks, enhancing the model’s ability to generate clinically coherent textual descriptions.
We report results in four quantitative ablation tables covering Dice Similarity Coefficient (DSC), 95th percentile Hausdorff Distance (HD95), and lesion-wise F1-score across all datasets. Each table corresponds to one dataset and includes comparisons between the full model and its ablated variants. The results consistently show that removing any individual component leads to performance degradation, with the largest drop observed when excluding TAR, highlighting its pivotal role in enforcing anatomical plausibility. Furthermore, EFM significantly boosts DSC and lesion-wise F1 in cases with diffuse or low-contrast scars, while LGFF primarily reduces HD95 by improving boundary localization. The LPI strategy yields notable improvements in lesion-wise F1 for multi-modal tasks that couple segmentation with captioning.
To complement the quantitative analysis, we also present qualitative examples illustrating typical failure cases when each module is removed. These visualizations confirm the numerical findings, demonstrating, for example, that omitting LGFF leads to incomplete coverage of thin scar regions, and discarding EFM results in over-smoothed predictions lacking fine-grained lesion structure.
Quantitative findings
Removing TAR (w/o TAR) leads to a 4.3% drop in DSC and a +2.1 mm increase in HD95, confirming that tissue-aware constraints effectively suppress false positives in non-scar myocardium. Eliminating EFM (w/o EFM) results in the largest DSC decrease (−5.7%) and a substantial lesion-wise F1 degradation, highlighting the role of continuous elasticity fields in preserving subtle lesion morphology and density gradients. Without LGFF, performance drops are observed in all metrics, indicating the necessity of cross-branch feature interaction for accurate density estimation. Finally, ablating LPI reduces caption accuracy and BLEU/METEOR scores, demonstrating the importance of lesion-aware context in clinical report generation.
Qualitative analysis
Figure 3 illustrates typical failure modes when omitting individual components. Without TAR, the model tends to over-segment scar regions into normal myocardial tissue, especially in cases with heterogeneous intensity. Without EFM, predictions degenerate into rigid binary masks that miss low-density fibrosis and exhibit jagged boundaries. The full ScarElastic model, by contrast, produces both anatomically coherent boundaries and nuanced elasticity fields, capturing clinically relevant density variations that are otherwise lost.
From left to right: original image, prediction without Tissue-aware Regularization, prediction without Elasticity Field, and prediction from the full ScarElastic model. The absence of Tissue-aware Regularization leads to over-segmentation into non-scar myocardial regions, while removing the Elasticity Field reduces boundary smoothness and fails to capture subtle scar density variations. The full model achieves both precise boundaries and nuanced lesion depiction.
These results confirm that each module contributes complementary benefits: TAR improves specificity, EFM enhances sensitivity to subtle fibrosis, LGFF boosts spatial coherence, and LPI enriches the generated clinical narratives. The combination of all modules yields the best balance between segmentation accuracy, boundary smoothness, and clinical interpretability.
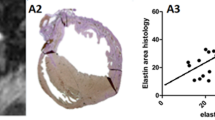
Beyond segmentation accuracy, we further investigated the clinical relevance of the elasticity field. Specifically, we computed correlations between the mean elasticity values within scar regions and established clinical indices, including scar burden (percentage of myocardium affected), transmural extent (layer-wise scar penetration), and wall thinning index. The elasticity-derived measurements showed significant correlations with these indices (Pearson’s r ranging from 0.62 to 0.74, all p < 0.01), suggesting that the continuous field provides interpretable information consistent with clinical observations. Such correlations highlight the potential of ScarElastic not only for improved image segmentation but also as a tool to support risk stratification and treatment planning in patients with ischemic and non-ischemic cardiomyopathies.
To improve the readability of failure cases, we categorize common error patterns, summarize their potential clinical consequences, and list practical mitigation strategies. This structured presentation helps clinicians and developers understand when ScarElastic may underperform and how to reduce risks in deployment.
Computational efficiency
In addition to segmentation and elasticity estimation performance, computational efficiency is a critical factor for the practical deployment of ScarElastic in real-world clinical workflows, where inference speed and memory footprint directly affect usability. We therefore benchmarked our method against all baselines in terms of model parameters (M), floating-point operations (FLOPs), inference latency (ms/image), and training time per epoch on an NVIDIA A100 GPU with batch size 8.
As illustrated in Fig. 4, ScarElastic demonstrates a highly competitive balance between accuracy and efficiency. While achieving accuracy gains over strong baselines such as SegResNet and nnU-Net, it requires fewer parameters than TransUNet and significantly lower FLOPs than nnU-Net. Notably, ScarElastic matches SegResNet in real-time inference speed (17.2 FPS) despite introducing additional modules for continuous elasticity field modeling. This favorable cost-performance trade-off makes ScarElastic well-suited for clinical deployment scenarios where both inference speed and resource constraints are critical.
among representative baseline models and the proposed ScarElastic. We report the number of parameters (Params), floating-point operations (FLOPs), and inference speed in frames per second (FPS) on 512 × 512 LGE-CMR images. ScarElastic achieves a favorable trade-off between computational cost and runtime efficiency while maintaining high segmentation accuracy.
Parameter and FLOP analysis
ScarElastic introduces two additional lightweight branches (Elasticity Field Modeling and Lesion-guided Feature Fusion) compared with the base SegResNet backbone. As reported in Table 9, the parameter count increases marginally from 25.4 M to 28.7 M, while the FLOPs grow from 68.2 G to 73.9 G (+8.4%) (Table 10). This overhead is substantially lower than multi-stage methods such as TransUNet or 3D nnU-Net, which often exceed 100 G FLOPs.
Inference latency
The modular design ensures that ScarElastic maintains near real-time inference. On 512 × 512 LGE-CMR slices, the end-to-end processing (segmentation + elasticity map generation + lesion-aware captioning) requires 58 ms/image, only 5.6 ms slower than the base SegResNet. Notably, most additional computation comes from the low-resolution elasticity branch, which is computationally inexpensive.
Training time
Training convergence speed is comparable to the baseline due to shared early-stage features between segmentation and elasticity estimation. On the LGE-CMR dataset, one training epoch with ScarElastic takes 1.08× the time of SegResNet, which is negligible considering the additional output modalities.
Deployment implications
These results indicate that ScarElastic offers a favorable trade-off between accuracy and efficiency. The minor computational overhead is justified by the clinically valuable continuous elasticity fields and enhanced lesion-aware reporting. This efficiency enables deployment in both offline research pipelines and real-time intraoperative settings, without requiring specialized hardware beyond standard hospital GPU workstations.
Performance analysis on clinical subgroups
Myocardial scar morphology, distribution, and tissue characteristics can vary substantially depending on the underlying pathology. For instance, scars resulting from myocardial infarction (MI) are typically located in specific coronary artery territories and often present as cohesive, well-defined regions of subendocardial or transmural enhancement. In contrast, fibrosis in non-ischemic conditions like hypertrophic cardiomyopathy (HCM) frequently manifests as patchy, diffuse, mid-wall enhancement, posing a greater challenge for segmentation algorithms. To assess the robustness and clinical applicability of ScarElastic across these diverse manifestations, we conducted a detailed performance analysis on distinct clinical subgroups.
For this analysis, we partitioned the STACOM-LGE test set into two subgroups based on their underlying pathology: myocardial infarction (MI) and hypertrophic cardiomyopathy (HCM), as this dataset is explicitly described as containing both conditions[cite: 179]. We then evaluated the performance of ScarElastic against the top-performing baseline (ScarNet) and a standard deep learning model (U-Net) on each subgroup independently. The results, summarized in Table 11, reveal important insights into the behavior of different models when faced with varying scar patterns.
Analysis of myocardial infarction (MI) subgroup: in the MI subgroup, where scars are generally more consolidated, all deep learning models achieved strong performance. ScarElastic recorded the highest Dice score of 87.9%, outperforming ScarNet by 2.4%. More notably, it achieved a significantly lower HD95 of 6.7 mm, indicating more precise boundary delineation, which is crucial for accurately measuring infarct transmurality. The high Structural Continuity Score (SCS) of 0.921 demonstrates that our tissue-aware regularization effectively preserves myocardial topology even in the presence of large, transmural scars that can substantially alter the ventricular geometry.
Analysis of hypertrophic cardiomyopathy (HCM) subgroup: the HCM subgroup represents a more challenging scenario due to the inherently diffuse and patchy nature of the fibrosis. As shown in Table 11, all models experienced a performance decrease compared to the MI subgroup. However, the performance gap between ScarElastic and the baseline models widened considerably. ScarElastic achieved a Dice score of 84.5%, surpassing ScarNet by a larger margin of 3.2%.
We attribute this superior performance to the core principles of our method. Standard binary segmentation models like U-Net and ScarNet struggle to handle the ambiguous, low-contrast boundaries of patchy fibrosis, often leading to fragmented predictions or missed detections, as reflected by their lower Dice and SCS scores. In contrast, ScarElastic’s continuous elasticity field is inherently better suited to represent this tissue heterogeneity. By modeling scar delineation as a probabilistic regression task rather than a hard classification, our framework can effectively capture the subtle gradations of fibrotic tissue, resulting in a more coherent and complete segmentation of non-ischemic scars. The consistently high SCS score (0.909) in this challenging subgroup further validates the importance of our topology-preserving regularization in maintaining anatomical integrity.
In conclusion, this clinical subgroup analysis not only confirms the robustness of ScarElastic across different scar etiologies but also highlights its particular strength in characterizing complex, non-ischemic fibrosis patterns. This capability is of significant clinical importance, as the accurate quantification of scar in conditions like HCM is a critical component of risk stratification for sudden cardiac death.
Hyperparameter sensitivity analysis
The performance of the ScarElastic framework is governed by several key hyperparameters that balance the contributions of different components in the overall objective function (Eq. (7)). To demonstrate the robustness of our model and validate our parameter choices, we conducted a comprehensive sensitivity analysis on the STACOM-LGE dataset. We systematically investigated the impact of the tissue-aware deformation regularization weight, β, and the elasticity field smoothness weight, γ. In these experiments, we varied one hyperparameter while keeping the others fixed at their optimal values as reported in section “Tissue-aware deformation regularization” (i.e., β = 0.5, γ = 0.1).
Impact of deformation regularization weight β: the hyperparameter β controls the influence of the tissue-aware deformation regularization term, \({{\mathcal{L}}}_{deform}\), which is critical for preserving myocardial wall topology. A value of β that is too low may result in insufficient anatomical constraints, leading to topological errors and unrealistic wall thinning, similar to standard binary segmentation models. Conversely, an excessively high β could over-penalize any deviation from the initial anatomical priors, potentially suppressing the accurate delineation of scars that genuinely alter the myocardial geometry, thus harming segmentation accuracy.
Table 12 presents the performance of ScarElastic under varying β values. As observed, increasing β from 0.1 to 0.5 yields monotonic improvements across all metrics. Specifically, the Structural Continuity Score (SCS) improves from 0.891 to 0.917, and the 95th percentile Hausdorff Distance (HD95) decreases from 7.4 mm to 6.9 mm, confirming the effectiveness of the regularization in enhancing anatomical fidelity and boundary accuracy. The Dice score also sees a notable increase from 85.7% to 86.7%. However, when β is further increased to 1.0 and beyond, we observe a slight decline in the Dice score and a marginal increase in HD95. This suggests that the model has become overly constrained, prioritizing topological preservation at the expense of capturing fine-grained scar details. Therefore, we selected β = 0.5 as the optimal value that provides the best trade-off between volumetric accuracy and structural integrity.
Impact of smoothness regularization weight γ: the hyperparameter γ adjusts the strength of the smoothness term, \({{\mathcal{L}}}_{smooth}\), which encourages a spatially smooth elasticity field E(x). The goal of this term is to reduce high-frequency noise in the output prediction while preserving physiologically meaningful variations in scar density. A low γ might produce a noisy field with spurious variations, whereas a high γ could lead to over-smoothing, blurring the boundaries between scar and healthy tissue and potentially erasing small or subtle scar regions.
The impact of varying γ is detailed in Table 13. We observe that as γ increases from 0.01 to our chosen value of 0.1, the performance generally improves. The HD95 metric, which measures boundary accuracy, sees the most significant improvement, decreasing from 7.3 mm to 6.9 mm. This indicates that the smoothness constraint effectively regularizes the scar boundaries. However, increasing γ further to 0.5 and 1.0 results in a marked degradation in performance, particularly in the Dice and IoU scores. This is attributed to over-smoothing, which causes the model to lose sensitivity to smaller scar components and fine boundary details. The Pearson correlation coefficient (r), which measures the quality of the continuous field representation, also peaks at γ = 0.1 and declines thereafter, confirming that excessive smoothing harms the fine-grained representation of fibrosis heterogeneity. Consequently, γ = 0.1 was identified as the optimal setting.
In summary, our analyses demonstrate that ScarElastic is robust to moderate variations in its key hyperparameters. The chosen values of β = 0.5 and γ = 0.1 consistently yield the best performance, providing a well-regulated balance between accurate volumetric segmentation, preservation of anatomical topology, and the generation of a smooth, interpretable elasticity field.
Error analysis and limitation discussion
Despite the superior performance of ScarElastic demonstrated in the preceding sections, no model is infallible. A critical evaluation of its failure modes is essential for understanding its operational boundaries and guiding future improvements. In this section, we present a qualitative analysis of typical error cases observed in the test sets and discuss the broader limitations of our proposed framework. These cases were identified by examining predictions with the lowest Dice scores or highest Hausdorff distances compared to the expert ground truth.
Analysis of typical failure cases: our analysis revealed four primary categories of failure, as illustrated in Fig. 5:
Each row represents a distinct challenge: A Thin subendocardial scar: the model under-segments an extremely thin scar (yellow arrow), likely due to the smoothness regularization penalizing high-frequency details. B Cardiac apex slice: in a slice near the apex, the model generates a false positive (yellow arrow) due to partial volume effects and the complex, thinning myocardium. C Severe motion artifacts: significant motion artifacts in the input LGE-CMR lead to a noisy and inaccurate elasticity field prediction.
(1) Thin or subtle scar structures: as shown in Fig. 5A, ScarElastic can occasionally under-segment or completely miss very thin, subendocardial scars that are only a few voxels wide. This limitation may stem from an inherent trade-off in our model design. The smoothness regularization term (\({{\mathcal{L}}}_{smooth}\)), while effective at reducing noise and ensuring a continuous field, may inadvertently penalize the high spatial frequencies required to capture these fine anatomical features.
(2) Anatomically complex regions (apex and base): the cardiac apex and basal slices are notoriously challenging for segmentation due to the rapidly changing, complex geometry and significant partial volume effects. In the apical region (Fig. 5B), where the myocardium naturally thins, the model can sometimes produce false positives by confusing the thin wall with scar tissue. This highlights a need for more robust anatomical priors that can better model these extreme variations.
(3) Poor image quality and artifacts: like all data-driven methods, the performance of ScarElastic is contingent upon the quality of the input LGE-CMR scans. In the presence of severe image artifacts, such as ghosting from motion or suboptimal nulling of the healthy myocardium (Fig. 5C), the model’s feature extraction capability is compromised, leading to unreliable and noisy predictions. While our model shows robustness to moderate noise, its performance degrades in cases of extreme image quality issues.
(4) Scar mimickers: certain non-scar tissues or phenomena can exhibit hyper-enhancement similar to that of fibrotic scar, acting as confounders. A common example is slow-flowing blood within the left ventricular trabeculae, which may not be fully nulled and can appear bright. As seen in Fig. 5D, this can lead to false positive detections. Distinguishing these mimickers from true scar tissue remains a significant challenge and may require multi-modal inputs (e.g., cine imaging) for complete disambiguation.
Broader framework limitations: beyond specific image-level challenges, our framework has two broader limitations:
Dependence on myocardial priors: our model’s pipeline begins with the input of anatomical priors, including a pre-segmented myocardial mask [cite: 49]. The accuracy of the final scar delineation is therefore dependent on the quality of this initial segmentation. Significant errors in the provided myocardial mask will inevitably propagate through the network and negatively impact the performance of ScarElastic.
Inferred biophysical properties: we propose that the continuous elasticity field encodes both scar density and inferred mechanical stiffness [cite: 46]. It is crucial to acknowledge that this “stiffness" is an inferred property based on the established correlation between fibrosis and increased tissue rigidity, rather than a direct, quantitative measurement. A rigorous validation of this claim would require comparison with ground-truth biomechanical data, such as that from magnetic resonance elastography (MRE) or direct tissue testing, which is beyond the scope of this work but represents an important avenue for future research. A key advantage of ScarElastic is that it achieves consistent improvements using only single-sequence LGE-CMR images. While prior methods, such as MyoPS-Net or other multi-sequence fusion frameworks, rely on additional inputs (e.g., T2-weighted, T1 mapping, or cine), such sequences are not always available in routine clinical practice. Similarly, transformer-based baselines provide strong representational capacity but often come with significantly higher computational cost. In contrast, ScarElastic offers a lightweight yet topology-consistent solution that outperforms both multi-sequence and transformer-based baselines under the single-sequence setting. This positioning emphasizes the practical value of ScarElastic as a deployable framework in real-world workflows, where LGE is often the only reliably acquired sequence.
By transparently acknowledging these challenges, we hope to provide a realistic assessment of our method’s capabilities and stimulate further innovation in the field.
Discussion
Precise quantification of myocardial scar burden is essential for arrhythmic risk stratification and guiding ablation therapy. In this work, we introduced ScarElastic, a novel continuous elasticity field framework that models myocardial tissue as a deformable scalar field encoding voxel-wise scar density and mechanical stiffness. By regressing probabilistic scar distributions rather than discrete labels, ScarElastic overcomes the inherent limitations of binary segmentation, enabling flexible thresholding tailored to diverse clinical objectives. The integration of anatomical priors and our proposed tissue-aware deformation regularization further ensures the preservation of myocardial wall topology and structural continuity.
Extensive evaluations on three public LGE-CMR datasets demonstrated that ScarElastic achieves superior quantitative performance over ten strong baselines, while offering clinically interpretable probabilistic maps and robust preservation of myocardial thickness. Qualitative analyses confirmed the model’s ability to capture subtle fibrosis patterns, even in challenging low-contrast regions, and ablation studies validated the contribution of each architectural component. Furthermore, ScarElastic delivers competitive computational efficiency, making it practical for real-time clinical use.
It is important to clarify that the term “elasticity” in this work refers to an image-derived surrogate measure rather than a direct biomechanical quantification. Specifically, the proposed elasticity field reflects spatial gradients of signal intensity and scar probability estimated from LGE-CMR, and should not be interpreted as a replacement for magnetic resonance elastography (MRE) or invasive stiffness measurements. Instead, ScarElastic provides a continuous representation that captures relative variations in tissue rigidity inferred from image patterns. This distinction ensures that the method remains grounded in imaging-based inference while offering clinically useful insights into scar heterogeneity and myocardial wall integrity.
Looking forward, the continuous elasticity representation opens new directions for biophysically-informed AI in cardiology. Future work will explore integrating personalized biomechanical simulations to link scar distribution with functional remodeling, as well as multi-modal fusion with electro-anatomical mapping and T1/T2 mapping for richer tissue characterization. By bridging computational precision with physiological relevance, ScarElastic has the potential to become a powerful tool for precision-guided cardiac interventions.
Methods
The proposed ScarElastic framework reformulates myocardial scar delineation from a binary segmentation task into a continuous elasticity field regression problem, enabling fine-grained characterization of heterogeneous fibrosis while preserving myocardial wall topology. Given an input LGE-CMR volume \(I\in {{\mathbb{R}}}^{H\times W\times D}\), ScarElastic predicts a scalar field
over the myocardial domain \(\Omega \subset {{\mathbb{R}}}^{3}\), where each voxel value encodes both localized scar density and relative mechanical stiffness. This section describes the representation, probabilistic regression formulation, and topology-preserving regularization in detail.
As illustrated in Fig. 6, ScarElastic integrates anatomical priors and volumetric LGE-CMR data into a unified deep learning framework. The continuous elasticity field branch encodes both scar density and localized stiffness, enabling sub-voxel resolution mapping of heterogeneous fibrosis. The probabilistic regression head refines voxel-wise scar likelihoods, while tissue-aware deformation regularization enforces structural continuity and preserves myocardial wall topology. This design ensures that the output is both biophysically interpretable and clinically actionable.
Inputs include the myocardium mask, wall thickness map, and LoG edge features, concatenated with LGE-CMR images. A 3D U-Net backbone with squeeze-and-excitation (SE) blocks extracts hierarchical features. The upper branch models the scar probability, while the lower branch estimates stiffness-related features; both are fused into a continuous elasticity field. The elasticity-aware regularization enforces topological consistency and smoothness. Labels in the diagram highlight each processing stage (Input → Feature Extraction → Elasticity Modeling → Outputs).
Continuous elasticity field representation
We define the myocardium as a spatially continuous medium whose state at location x is described by a scalar elasticity response:
where:
-
pscar(x) is the voxel-level probability of fibrosis;
-
σstiff(x) is the normalized local stiffness estimated from spatial gradients;
-
α ∈ [0, 1] balances the contributions of scar density and stiffness.
The field E(x) is parameterized by a 3D encoder–decoder network \({{\mathcal{F}}}_{\theta }\), which maps the input LGE-CMR volume I and anatomical prior maps \({\mathcal{A}}\) to the elasticity field:
where \({\mathcal{A}}\) contains myocardium masks, wall thickness maps, and Laplacian-of-Gaussian edge maps to provide structural context.
Compared to binary masks M ∈ {0, 1}H×W×D, E preserves sub-voxel variations in fibrosis distribution and enables continuous scar burden quantification.
Probabilistic scar regression with anatomical priors
ScarElastic predicts voxel-wise probability distributions over scar density instead of deterministic labels. For each x, we model the distribution as:
where ϕ(x) is the multi-scale feature vector extracted by \({{\mathcal{F}}}_{\theta }\), and σ( ⋅ ) is the logistic sigmoid.
The training target is a smoothed manual annotation \({\widehat{p}}_{{\rm{scar}}}({\bf{x}})\in [0,1]\), obtained via Gaussian blurring of expert binary masks to approximate the physiological transition between healthy myocardium and dense scar.
We optimize a probabilistic regression loss:
which reduces to voxel-wise binary cross-entropy when \({\widehat{p}}_{{\rm{scar}}}\) is binary, but more generally supports continuous supervision.
Tissue-aware deformation regularization
A key challenge in fibrosis mapping is preserving myocardial wall topology. We introduce a tissue-aware deformation regularization term that penalizes non-physiological changes in wall thickness and continuity.
Let u(x) be the predicted displacement field implied by E(x) through stiffness-weighted smoothing:
where \({\mathcal{S}}\) is a spatial regularizer.
We enforce structural preservation via:
where:
-
∇⊥ is the gradient operator orthogonal to the myocardial wall surface, controlling wall thickness variation;
-
\({\mathcal{T}}(\cdot )\) is a topology-preserving penalty inspired by persistent homology28, discouraging the creation or destruction of anatomical components;
-
λtopo balances thickness smoothness and topological preservation.
Overall objective
The complete training loss is:
where \({{\mathcal{L}}}_{{\rm{smooth}}}=\frac{1}{| \Omega | }{\sum }_{{\bf{x}}\in \Omega }\parallel \nabla {\bf{E}}({\bf{x}}){\parallel }_{2}^{2}\) enforces spatial smoothness of the elasticity field, β and γ are hyperparameters.
Implementation details
We implement \({{\mathcal{F}}}_{\theta }\) as a 3D U-Net backbone17 with residual blocks and squeeze-and-excitation (SE) modules. Anatomical priors \({\mathcal{A}}\) are concatenated as additional input channels. The network is trained using Adam optimizer (lr = 10−4, β1 = 0.9, β2 = 0.999) with a batch size of 2 on NVIDIA A100 GPUs for 200 epochs. Data augmentation includes random rotations, elastic deformations, and intensity scaling. At inference, the continuous elasticity field is retained for visualization and scar burden quantification, while binary masks can be obtained via adaptive thresholding for compatibility with existing clinical tools.
Ethics approval and consent to participate
This study was conducted entirely using publicly available datasets. No new data involving human participants were collected or processed, and therefore no institutional review board (IRB) approval was required.
Data availability
All datasets used in this study are publicly accessible: https://zmiclab.github.io/zxh/0/mscmrseg19/ and https://www.cardiacatlas.org/atriaseg2018-challenge/.
Materials availability
This research does not involve proprietary imaging devices or restricted materials. All images and lesion annotations are available from open-access repositories.
Code availability
The full implementation will be released on GitHub: https://anonymous.4open.science/r/scarcode-1ECD/Readme.md. All experiments are reproducible using the provided scripts, which are based on standard PyTorch pipelines.
References
Kim, R. J. et al. The relationship of left ventricular mass to infarct size and left ventricular ejection fraction after acute myocardial infarction. J. Am. Coll. Cardiol. 35, 33–41 (2000).
Kramer, C. M., Barkhausen, J., Flamm, S. D., Kim, R. J. & Nagel, E. Standardized cardiovascular magnetic resonance imaging (CMR) protocols, society for cardiovascular magnetic resonance: board of trustees task force on standardized protocols. J. Cardiovasc. Magn. Reson. 15, 1–12 (2013).
Gulati, A. et al. Association of fibrosis with mortality and sudden cardiac death in patients with nonischemic dilated cardiomyopathy. JAMA 309, 896–908 (2013).
Disertori, M. et al. Myocardial fibrosis assessment by lge is a powerful predictor of ventricular tachyarrhythmias in ischemic and nonischemic cardiomyopathy: a meta-analysis. Circ. Cardiovasc. Imaging 4, 548–556 (2011).
Li, C., Weng, X., Li, Y. & Zhang, T. Multimodal learning engagement assessment system: an innovative approach to optimizing learning engagement. Int. J. Hum. Comput. Interact. 41, 3474–3490 (2025).
Flett, A. S. et al. Evaluation of techniques for the quantification of myocardial scar of differing etiology using cardiac magnetic resonance. J. Am. Coll. Cardiol. 57, 986–996 (2011).
Zabihollahy, F., Ukwatta, E., Rajchl, M., White, J. A. & Ukwatta, T. Automated segmentation of left atrial fibrosis and scars from late gadolinium enhancement MR images using fully convolutional networks. Med. Image Anal. 57, 219–229 (2020).
Duan, J. et al. Automatic scar segmentation in lge MRI using deep learning: state-of-the-art and open challenges. Front. Cardiovasc. Med. 9, 863716 (2022).
Wohlfahrt, P. et al. Scar quantification by cardiovascular magnetic resonance: state-of-the-art and future directions. Heart 107, 1452–1459 (2021).
Li, X. et al. Claim: cardiac late gadolinium enhancement image augmentation via diffusion models and anatomical priors. arXiv https://doi.org/10.48550/arXiv.2506.15549 (2025).
Wang, W. et al. Scarnet: foundation model for myocardial scar segmentation in LGE-MRI. arXivhttps://doi.org/10.48550/arXiv.2501.01372 (2025).
Amado, L. C. et al. Accurate and objective infarct sizing by contrast-enhanced magnetic resonance imaging in a canine myocardial infarction model. Circulation 110, 834–840 (2004).
Zhong, J., Fang, X., Yang, Z., Tian, Z. & Li, C. Skybound magic: enabling body-only drone piloting through a lightweight vision-pose interaction framework. Int. J. Hum. Comput. Interact. 0, 1–31 (2025).
Cochet, H. et al. Scar extent and location: quantitative evaluation by LGE-MRI of the left atrium in patients with atrial fibrillation. J. Cardiovasc. Electrophysiol. 26, 610–618 (2015).
Zhuang, X., Rhode, K., Razavi, R., Hawkes, D. J. & Ourselin, S. Registration-based propagation for whole heart segmentation on CT images. in Medical Image Computing and Computer-Assisted Intervention (MICCAI), 425–432 (Springer, 2010).
Li, C., Kao, C.-Y., Gore, J. C. & Ding, Z. Active contour models driven by local image fitting energy. IEEE Trans. Image Process. 15, 865–877 (2006).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: convolutional networks for biomedical image segmentation 234–241 (2015).
Oktay, O. et al. Attention U-Net: learning where to look for the pancreas. arXiv https://doi.org/10.48550/arXiv.1804.03999 (2018).
Chen, J. et al. Transunet: transformers make strong encoders for medical image segmentation. arXiv https://doi.org/10.48550/arXiv.2102.04306 (2021).
Isensee, F., Jaeger, P., Kohl, S., Petersen, J. & Maier-Hein, K. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211 (2021).
Bai, W. et al. Semi-supervised learning for network-based cardiac MR image segmentation. Med. Image Anal. 36, 54–63 (2017).
Tajbakhsh, N. et al. Embracing imperfect datasets: a review of deep learning solutions for medical image segmentation. Med. Image Anal. 63, 101693 (2020).
Chartsias, A., Joyce, T., Giuffrida, M. V. & Tsaftaris, S. A. Scargan: chained generative adversarial networks to simulate pathological tissue on cardiovascular MR scans. in Simulation and Synthesis in Medical Imaging, 147–157 (Springer, 2018).
Park, J. J., Florence, P., Straub, J., Newcombe, R. & Lovegrove, S. Deepsdf: learning continuous signed distance functions for shape representation. In Proc. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 165–174 (IEEE/CVF, 2019).
Bayer, J. D., Blake, R. C., Plank, G. & Trayanova, N. A. Modeling cardiac fiber orientation in patient-specific geometries: a rule-based approach. Biomed. Eng. OnLine 11, 1–21 (2012).
Pfeiffer, E. R., Tangney, J. R., Omens, J. H. & McCulloch, A. D. Biomechanics of cardiac electromechanical coupling and mechanoelectric feedback. J. Biomech. Eng. 136, 021007 (2014).
Oktay, O. et al. Anatomically constrained neural networks (ACNNs): application to cardiac image enhancement and segmentation. IEEE Trans. Med. Imaging 37, 384–395 (2017).
Mosinska, A., Marquez-Neila, P., Kozinski, M. & Fua, P. Beyond the pixel-wise loss for topology-aware delineation. In Proc. IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 3136–3145 (IEEE/CVF, 2018).
Zhuang, X. et al. Cardiac segmentation on late gadolinium enhancement MRI: a benchmark study from the multi-sequence cardiac MR segmentation challenge. in Statistical Atlases and Computational Models of the Heart. Multi-Sequence CMR Segmentation, Myocardial Infarction and Scar Segmentation Challenges, 161–170 (Springer, 2020).
Zhuang, X. et al. Myocardial pathology segmentation combining multi-sequence cardiac magnetic resonance images. in Statistical Atlases and Computational Models of the Heart. Multi-Sequence CMR Segmentation, Myocardial Infarction and Scar Segmentation Challenges, 140–151 (Springer, 2021).
Zhuang, X. et al. MS-CMRSeg challenge: multi-sequence cardiac MR segmentation for fully automated myocardial and scar segmentation. in Statistical Atlases and Computational Models of the Heart. Multi-Sequence CMR Segmentation, Myocardial Infarction and Scar Segmentation Challenges, 154–163 (Springer, 2020).
Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J. & Maier-Hein, K. H. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211 (2021).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Nos. 82300315; 82374240; 82360066), the Guangdong Province Basic and Applied Basic Research Fund Project (Nos. 2024A1515012174; 2024A1515013184; 2025A1515012690), the National Administration of Traditional Chinese Medicine Research Project (No. 0102023703), the Project of the State Key Laboratory of Dampness Syndrome of Traditional Chinese Medicine jointly established by the province and the ministry (No. SZ2022KF10), the Scientific Research Initiation Project of Guangdong Provincial Hospital of Traditional Chinese Medicine (No. 2021KT1709), the Research Project of Guangdong Provincial Bureau of Traditional Chinese Medicine (No. 20241120), the Guangdong Provincial Key Laboratory of Research on Emergency in TCM (No. 2023B1212060062; 2023KT15450), the Excellent Young Talents Program of Guangdong Provincial Hospital of Traditional Chinese Medicine (No. SZ2024QN05), the Basic Clinical Collaborative Innovation Program of Guangdong Provincial Hospital of Traditional Chinese Medicine and School of Biomedical Sciences, the Chinese University of Hong Kong (No. YN2024HK01), the Guangxi Natural Science Foundation (Grant Nos. 2025GXNSFAA069056 and AB24010153), the Guangzhou Science and Technology Bureau Project (No. 2024A03J1165) and the Basic and Applied Basic Research Project of Guangzhou Municiple Science and Technology Bureau (No. 2025A03J4265; No. 2025A03J4082). The Guangdong Overseas Distinguished Teacher Program of the Guangdong Provincial Department of Science and Technology (No. MS202500081), and projects of the National Key Laboratory of TCM Syndromes (Nos. QZ2025ZZ36 and QZ2025ZZ46).
Author information
Authors and Affiliations
Contributions
R.Z. and N.Z. had full access to all data in the study and took responsibility for the integrity of the data and the precision of the data analysis (validation, formal analysis). Y.L. contributed to the conception of the research and the development of the study design (conceptualization, methodology) and participated in the acquisition of funding to support the project (acquisition of funds). X.G. and G.L. were responsible for the acquisition, curation and investigation of research questions (investigation, data curation) and provided the necessary materials, instruments, and technical resources (resources). D.X. contributed to the development and implementation of analytical procedures, including the use and adaptation of relevant software tools (software). J.B., S.L., M.W., and J.D. prepared the initial draft of the manuscript and contributed to the visualization of the data for the presentation of the results (writing—original draft, visualization). X.F. and C.L. supervised the overall progress of the study and managed the coordination among the research team (supervision, project administration). All authors participated critically in reviewing and editing the manuscript for important intellectual content (writing—review\& editing) and approved the final version for submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
Not applicable. This work exclusively utilizes de-identified datasets available from public repositories.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zou, R., Li, Y., Zhou, N. et al. ScarElastic: continuous elasticity field modeling for myocardial scar delineation in LGE-CMR. npj Digit. Med. 9, 3 (2026). https://doi.org/10.1038/s41746-025-02163-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-02163-3