Abstract
Gastric cancer staging is frequently limited by the low sensitivity of routine imaging for occult peritoneal metastasis (OPM), necessitating invasive staging laparoscopy. We developed a Multimodal Model, integrating primary tumor radiomics from CT with clinical factors to non-invasively predict OPM in locally advanced gastric cancer. The model was trained and internally validated in a large cohort (n = 940) and externally validated across two independent multi-center cohorts (n = 309), an incremental cohort (n = 477), and a prospective clinical trial cohort (n = 168). In all cohorts, the model achieved robust performance (AUCs: 0.834-0.857), significantly outperforming single-modality models. Crossover validation showed AI assistance increased the average radiologist AUC from 0.735 to 0.872. Transcriptomic analysis revealed that the model’s low-risk stratification correlated with an enhanced antitumor immune microenvironment (CD8 T cells, TNFα signaling). This validated model provides a practical tool for accurate, non-invasive OPM prediction and individualized treatment planning.
Similar content being viewed by others
Introduction
Gastric cancer (GC) remains one of the leading causes of cancer-related mortality worldwide, particularly in East Asia where the incidence is disproportionately high1,2. A significant proportion of patients present with advanced disease at the time of diagnosis, and peritoneal metastasis is the most common and fatal pattern of dissemination3,4. Occult peritoneal metastasis (OPM) often escapes detection by routine imaging modalities, yet its early identification is essential for optimizing treatment strategy5,6,7,8. Contrast-enhanced computed tomography (CT), the standard first-line imaging for staging, exhibits highly variable sensitivity for detecting peritoneal carcinomatosis, reported as low as 23–76%, depending on lesion size, morphology, and ascites presence9,10,11. Although PET-CT and MRI may marginally improve detection rates, PET-CT often remains limited by low negative predictive value (60%), while MRI is cost-intensive and prone to motion artifacts, limiting practical use12,13. Therefore, many patients with radiologically negative scans still harbor OPM found only during staging laparoscopy.
Staging laparoscopy (SL) remains the gold standard for OPM detection, significantly impacting management decisions14,15. Meta-analyses and cohort studies show SL substantially outperforms CT and PET-CT in sensitivity and specificity, detecting OPM that would otherwise lead to unnecessary laparotomy in 16%–60% of cases16,17,18. However, SL’s invasiveness, procedural cost, need for anesthesia, and limited availability restrict its universal application, especially in healthcare settings with limited resources19. There remains an urgent unmet clinical need for a reliable, noninvasive preoperative tool to predict OPM with high accuracy.
In recent years, radiomics and AI-driven approaches have shown potential to address this gap. Radiomics extracts high-dimensional quantitative features from standard imaging to uncover tumor heterogeneity invisible to the human eye20,21. Several studies have built CT-based radiomics models, with or without clinical features, to predict OPM in advanced gastric cancer. For instance, Shi et al. developed a clinical-nomogram that matches radiomics model performance in predicting OPM22. Similarly, a deep learning-based CT signature was shown to predict OPM and chemotherapy benefit23. A bounding-box-based radiomics model, which simplifies ROI annotation, achieved high discrimination in a multi-center cohort (AUC: 0.87)24. Yet most of these approaches are limited by single-center cohorts, reliance on manual annotation, small sample sizes, and insufficient external or prospective validation. Moreover, they generally lack biological interpretation to support clinical confidence.
To address these gaps, we developed the Multimodal Model, a fully automated “digital biopsy” tool that integrates CT-based primary tumor radiomics with key clinical variables for non-invasive prediction of OPM. The model was trained and internally validated in a large cohort from a high-volume gastric cancer center, and its performance was rigorously tested across two independent multi-center external validation cohorts, an incremental validation cohort assessing cytology positivity, and a prospective clinical trial cohort (NCT06478368). We further performed transcriptomic analysis on propensity score-matched high- and low-risk patients to uncover the underlying immune-biological correlates of the model’s risk stratification. This comprehensive framework not only demonstrates robust predictive accuracy but also provides a rare biological interpretation, thereby offering a clinically practical and mechanistically supported solution for preoperative risk stratification in LAGC patients.
Results
Patient Distribution and Baseline Characteristics
A total of 1726 patients were included, with 940 patients from FHHMU divided into the training (n = 658) and internal validation (n = 282) cohorts (7:3 ratio). Two multicenter cohorts comprised the external validation sets: External cohort I (n = 164) included patients from SJZPH, BDCH, HSPH, and CZPH, while External cohort II (n = 145) included patients from WHPH, AMUFH, NJJLH, NHUFH, and LMUH. Additionally, an incremental validation cohort (n = 477) from FHHMU (Jan 2018–Aug 2020) was utilized to specifically evaluate the model’s ability to predict positive peritoneal cytology, based on patients who underwent both diagnostic laparoscopy and cytology.
Baseline characteristics were well-balanced across all cohorts (Supplementary Table 2). The patient population was predominantly male (65.0%–69.9%) with a median age of 63 years. Regarding tumor characteristics, the lower one-third of the stomach was the most frequent location (56.1%–58.3%), and the diffuse type was the most prevalent Lauren classification (42.7%–50.3%). Crucially, no significant differences were observed among the cohorts for common inflammatory indices.
Development and Performance of the Multimodal Model in the Training Cohort
An ensemble model was first developed based on primary tumor radiomic features to predict OPM in gastric cancer patients. Compared with individual machine learning algorithms, the ensemble model achieved the highest predictive accuracy in the training cohort, with an AUC of 0.811 (95% CI: 0.769–0.852), sensitivity of 0.738, and specificity of 0.757 (Supplementary Figure 4). Multivariate logistic regression analysis identified cT stage (OR = 2.841, P = 0.002), histological type (OR = 3.229, P < 0.001), Borrmann classification (OR = 4.506, P < 0.001), and primary tumor radiomics (OR = 7.493, P < 0.001) as independent predictors of OPM (Supplementary Table 3). These variables were incorporated into the Multimodal Digital Biopsy Model (Multimodal Model), a constructed nomogram used for prediction (Fig. 1a).
a Nomogram integrating independent clinical predictors (cT stage, histology, Borrmann classification) and the radiomic signature for individualized OPM risk estimation. b ROC curves comparing the tumor radiomic model, clinical feature model, and the integrated Multimodal Model. c Precision–recall curves showing superior predictive precision of the Multimodal Model. d Calibration curve showing agreement between predicted and observed probabilities (Brier score = 0.1071). e Confusion matrices for the tumor radiomic, clinical, and Multimodal models in the training cohort, summarizing true positives, false positives, true negatives, and false negatives using the optimal Youden-index cutoff. f Radar plot comparing accuracy, sensitivity, specificity, F1 score, PPV, NPV, PLR, and NLR across models. g Two-tier concentric circle plots showing proportions of OPM+ and OPM− within high- and low-risk groups. h Decision curve analysis demonstrating higher net clinical benefit for the Multimodal Model. i Ten-fold cross-validation performance: bar charts (mean ± SD with per-fold dots) of accuracy, AUC, sensitivity, and specificity for each model, confirming robustness. j-k Clinical impact curves showing the number classified as high risk and those truly with OPM across thresholds. l Kaplan–Meier curves showing worse 5-year OS for high-risk versus low-risk (log-rank P < 0.001). m–o Bootstrap ROC curves (1000 resamples) showing higher mean AUC for the Multimodal Model than single-modality models.
In the training cohort, the Multimodal Model demonstrated an AUC of 0.853 (95% CI: 0.817–0.888), accuracy of 0.793 (95% CI: 0.760–0.824), sensitivity of 0.746, specificity of 0.804, F1 score of 0.572, and Brier score of 0.1071 (Fig. 1b–d, f; Supplementary Table 4). This model correctly identified 46.4% of OPM-positive patients in the high-risk group and 93.3% of OPM-negative patients in the low-risk group (Fig. 1e). Decision curve analysis indicated higher net clinical benefit compared with single-modality models (Fig. 1h). Concentric circle plots showed strong concordance between predicted risk and actual outcomes, with 13.8% OPM-positive patients in the high-risk group and 65.5% OPM-negative patients in the low-risk group (Fig. 1g).
Clinical impact curve analysis demonstrated favorable net benefit across a wide range of threshold probabilities, indicating substantial clinical applicability of the Multimodal Model (Fig. 1j, k). Risk stratification by this model revealed significant survival differences: the 5-year overall survival was 47.4% in the low-risk group versus 27.6% in the high-risk group (Fig. 1l).
Bootstrap analysis with 1000 resamples confirmed that the Multimodal Model (AUC = 0.852, 95% CI: 0.851–0.853; Fig. 1o) outperformed both the clinical model (AUC = 0.747, 95% CI: 0.746–0.749; Fig. 1m) and the tumor radiomic model (AUC = 0.810, 95% CI: 0.808–0.811; Fig. 1n). Ten-fold cross-validation further supported the robustness of the model (Fig. 1i). The Multimodal Model achieved significantly better discrimination and reclassification than single-modality models, with the greatest improvement over the clinical model (IDI = 0.169, 95% CI: 0.123–0.214; NRI = 0.342, 95% CI: 0.077–0.461; both P < 0.05; Supplementary Table 5).
Internal Validation of the Multimodal Model
To evaluate the generalizability of the Multimodal Model, its performance was tested in the internal validation cohort. The model achieved an AUC of 0.849 (95% CI: 0.790–0.909), an accuracy of 0.784 (95% CI: 0.731–0.830), a sensitivity of 0.769, a specificity of 0.787, and an F1 score of 0.567 (Fig. 2a, c, d; Supplementary Table 6). In the high-risk group, the model correctly identified 44.9% of patients with OPM, while in the low-risk group it correctly identified 93.8% of OPM-negative patients (Fig. 2e). The calibration performance was favorable, with a Brier score of 0.1067 (Fig. 2b). Decision curve analysis confirmed that the Multimodal Model provided greater net clinical benefit than any single-modality model (Fig. 2f).
a ROC curves comparing the tumor radiomic model, clinical feature model, and Multimodal Model in the internal validation cohort. b Calibration curve showing good agreement between predicted probabilities and observed OPM outcomes (Brier score = 0.1067). c Precision-recall curves indicating superior predictive precision of the Multimodal Model over single-modality models. d Radar plot comparing accuracy, sensitivity, specificity, F1 score, PPV, NPV, PLR, and NLR among the three models. e Confusion matrices summarizing true positives, false positives, true negatives, and false negatives for each model, using the optimal cutoff derived from the Youden index. f Decision curve analysis demonstrating higher net clinical benefit for the Multimodal Model compared with other models. g Kaplan–Meier survival curves showing significantly poorer 5 year overall survival in the high-risk group compared to the low-risk group (log-rank P < 0.001). h Two-tier concentric circle plots illustrating the proportion of true OPM-positive and OPM-negative patients within each risk group for each model. i Reclassification improvement analysis comparing the clinical feature model and the Multimodal Model in the combined training and internal validation cohorts, showing increased identification of high-risk OPM patients (from 32.8% to 46.0%) and reduced misclassification in the low-risk group (from 9.8% to 6.6%).
Two-tier concentric circle plots showed that the Multimodal Model identified a higher proportion of low-risk OPM-negative patients compared with the clinical model (68.4% vs. 62.1%) (Fig. 2h). Kaplan–Meier analysis demonstrated that the low-risk group had significantly better five-year OS than the high-risk group (48.7% vs. 25.8%, P < 0.001; Fig. 2g). Sensitivity analysis combining results from the training and internal validation cohorts indicated a 13.2% improvement in identifying high-risk patients and a 3.2% reduction in the misclassification rate for low-risk patients (Fig. 2i).
Reclassification statistics demonstrated that the Multimodal Model significantly outperformed both the clinical model and the tumor radiomic model. Compared with the clinical model alone, the NRI was 0.487 (95% CI: 0.236–0.739) and the IDI was 0.200 (95% CI: 0.129–0.273), with all differences being statistically significant (P < 0.001; Supplementary Table 5). DeLong test results further supported the significance of these improvements.
Multicenter External Validation of the Multimodal Model
To further assess the generalizability of the Multimodal Model, two independent external cohorts from nine medical centers in China and Germany were used for validation. In external validation cohort I, the model achieved an AUC of 0.857 (95% CI: 0.792–0.922), sensitivity of 0.750, specificity of 0.795, and accuracy of 0.787 (95% CI: 0.716–0.847) (Fig. 3a, c, e, f; Supplementary Table 7). The high-risk group included 47.1% of correctly identified OPM-positive patients, while the low-risk group included 92.9% of correctly identified OPM-negative patients (Fig. 3k, top). Concentric circle plots indicated strong concordance between predicted risk and OPM status (Fig. 3l). Survival analysis revealed significantly better outcomes in the low-risk group compared with the high-risk group, with five-year OS rates of 47.8% and 25.5%, respectively (Fig. 3m).
a, b ROC curves of the tumor radiomic model, clinical feature model, and Multimodal Model in external validation cohort I (A) and cohort II (B). c, d Calibration curves showing agreement between predicted probabilities and observed OPM outcomes for validation cohort I (C, Brier score = 0.115) and cohort II (D, Brier score = 0.1189). e, j Radar plots comparing accuracy, sensitivity, specificity, F1 score, PPV, NPV, PLR, and NLR across models for validation cohort I (E) and cohort II (J). f, g Precision–recall curves showing superior predictive precision of the Multimodal Model compared with single-modality models in validation cohort I (F) and cohort II (G). h, i Decision curve analyses (DCA) indicating higher net clinical benefit for the Multimodal Model across a range of threshold probabilities in validation cohort I (H) and cohort II (I). k Confusion matrices summarizing true positives, false positives, true negatives, and false negatives for each model in validation cohort I (upper row) and cohort II (lower row), using the optimal Youden-index cutoff. (l) Two-tier concentric circle plots showing the proportions of OPM+ and OPM− cases within high- and low-risk groups for each model in both validation cohorts. m, n Kaplan–Meier survival curves for validation cohort I (M) and cohort II (N), showing significantly poorer 5-year overall survival in the high-risk group versus the low-risk group (log-rank P < 0.05).
External validation cohort II (n = 145), which included patients from four treatment centers in southern China and Ludwig Maximilian University in Germany, also demonstrated robust performance, with an AUC of 0.850 (95% CI: 0.768–0.932), accuracy of 0.786 (95% CI: 0.710–0.850), sensitivity of 0.774, and specificity of 0.789 (Fig. 3b, d, g, j; Supplementary Table 7). The confusion matrix confirmed classification performance, with 50.0% of OPM-positive patients correctly identified in the high-risk group and 92.8% of OPM-negative patients correctly identified in the low-risk group (Fig. 3k, bottom). Risk stratification showed significant survival differences, with five-year OS rates of 47.4% versus 25.0% (Fig. 3n).
Decision curve analysis demonstrated higher net clinical benefit for the Multimodal Model compared with single-modality models in both external cohorts (Fig. 3h, i). In terms of reclassification and discrimination, the Multimodal Model significantly outperformed the clinical model, with IDI values ranging from 0.158 to 0.172 and NRI values ranging from 0.199 to 0.492, all with P < 0.05 (Supplementary Table 5). Sensitivity analysis combining results from both external validation cohorts showed a 15.2% improvement in the identification rate of high-risk patients and a 5.1% reduction in the misclassification rate of low-risk patients (Supplementary Figure 6).
To rigorously assess the stability and generalizability of the Multimodal Model, we integrated patients from the internal and external validation sets into an expanded cohort for sensitivity analysis. The model consistently demonstrated stable and excellent predictive performance, with the AUC uniformly exceeding 0.830 across all examined subgroups (Supplementary Table 8-9). Strong predictive performance was maintained regardless of Lauren classifications (diffuse, intestinal, and mixed types, Supplementary Figure 7) and primary tumor locations (upper, middle, and lower third of the stomach, Supplementary Figure 8). These findings robustly confirm the model’s clinical utility and high generalizability across diverse tumor biological and anatomical characteristics.
Validation in the Incremental Cohort
According to the “seed and soil” theory, free cancer cells within the peritoneal cavity, if not promptly treated, may develop into visible peritoneal metastases and subsequently worsen prognosis25,26. To evaluate whether the Multimodal Model could be extended to predict peritoneal lavage cytology results in newly diagnosed patients, we retrospectively selected 477 gastric cancer patients from FHHMU as an incremental validation cohort. All patients underwent diagnostic laparoscopy and peritoneal biopsy to confirm peritoneal metastasis. For patients without occult peritoneal metastasis (n = 384), peritoneal lavage cytology was performed.
In predicting OPM in this cohort, the Multimodal Model maintained robust performance, achieving an AUC of 0.834 (95% CI: 0.789–0.879), accuracy of 0.799 (95% CI: 0.760–0.834), sensitivity of 0.763, specificity of 0.807, and an F1 score of 0.597 (Fig. 4a, c, d; Supplementary Table 10). The Brier score for calibration was 0.1174 (Fig. 4b). Notably, the Multimodal Model correctly classified 93.4% of OPM-negative patients and 49.0% of OPM-positive patients into the corresponding risk categories (Fig. 4e) and demonstrated higher net clinical benefit across a wide range of threshold probabilities (Fig. 4f). Two-tier concentric circle plots confirmed that, compared with the clinical feature model, the Multimodal Model identified a greater proportion of high-risk patients (14.9% vs. 12.4%) (Fig. 4h).
a ROC curves comparing the tumor radiomic model, clinical feature model, and Multimodal Model for OPM prediction in the incremental cohort. b Calibration curve showing agreement between predicted probabilities and observed OPM outcomes (Brier score = 0.1174). c Precision–recall curves indicating higher predictive precision of the Multimodal Model than single-modality models. d Radar plot comparing accuracy, sensitivity, specificity, F1 score, PPV, NPV, PLR, and NLR for the three models. e Confusion matrices showing true positives, false positives, true negatives, and false negatives for each model, using the optimal Youden-index cutoff. f Decision curve analysis (DCA) showing greater net clinical benefit for the Multimodal Model across a range of threshold probabilities. g Kaplan–Meier survival curves showing significantly worse 5 year OS in the high-risk group versus the low-risk group (log-rank P < 0.001). h Two-tier concentric circle plots displaying proportions of OPM+ and OPM− cases in high- and low-risk groups for each model. i Sankey diagram illustrating reclassification of patients by the Multimodal Model, stratified by OPM status (left) and cytology results (right). j ROC curves comparing models in the subset of OPM-negative patients for predicting positive peritoneal lavage cytology (CY+). k Calibration curve for the Multimodal Model in predicting cytology positivity (Brier score = 0.0856).
Compared with the clinical model, the Multimodal Model achieved the largest incremental improvement in discrimination and reclassification, with an IDI of 0.141 (95% CI: 0.094–0.190) and a NRI of 0.185 (95% CI: 0.148–0.223), both statistically significant (P < 0.05; Supplementary Table 5). Survival analysis based on the Multimodal Model’s stratification showed significantly worse five-year OS in the high-risk group than in the low-risk group (24.8% vs. 48.5%, P < 0.001; Fig. 4g). In a subset of the incremental cohort containing only OPM-negative patients (Fig. 4i), the Multimodal Model also demonstrated strong predictive ability for peritoneal lavage cytology positivity, with an AUC of 0.831 (95% CI: 0.763–0.899) (Fig. 4j, k; Supplementary Table 10).
Prospective Cohort Validation
To further evaluate the generalizability and clinical utility of the Multimodal Model, an external validation was performed using a prospective cohort (Fig. 5a). In the NCT06478368 cohort (n = 168), which included patients with gastric cancer, the model achieved an AUC of 0.839 (95% CI: 0.767–0.910), an accuracy of 0.780 (95% CI: 0.709–0.840), a sensitivity of 0.833, a specificity of 0.768, an F1 score of 0.575, and PPV and NPV of 0.439 and 0.955, respectively, outperforming all baseline models (Fig. 5b, d, f; Supplementary Table 11). Calibration curve analysis demonstrated good model fit, with a Brier score of 0.1166 (Fig. 5c). Decision curve analysis (Fig. 5e) and concentric circle plots (Fig. 5g) confirmed the superior net clinical benefit and predictive consistency of the Multimodal Model compared with other models.
a Patient enrollment flowchart for the prospective validation cohort (NCT06478368), showing inclusion and exclusion criteria, resulting in 168 eligible patients undergoing laparoscopic exploration. b ROC curves comparing the tumor radiomic model, clinical feature model, and Multimodal Model for predicting OPM in the prospective cohort. c Calibration curve demonstrating agreement between predicted probabilities and observed outcomes (Brier score = 0.1166). d Precision–recall curves showing higher predictive precision of the Multimodal Model compared with single-modality models. e Decision curve analysis indicating greater net clinical benefit of the Multimodal Model across a range of threshold probabilities. f Confusion matrices showing true positives, false positives, true negatives, and false negatives for each model using the optimal Youden-index cutoff. g Two-tier concentric circle plots illustrating the proportion of OPM+ and OPM− patients within high- and low-risk groups for each model. h Performance comparison of clinicians with and without AI assistance in the prospective reader crossover study, showing significant improvements in AUC, accuracy, F1 score, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) with model assistance (all P < 0.05). i ROC curves for junior radiologists (<5 years experience) with and without model assistance. j ROC curves for intermediate radiologists (≥5 years experience) with and without model assistance. k ROC curves for senior radiologists (≥10 years experience) with and without model assistance.
Furthermore, a crossover experiment was conducted within this prospective study to assess the model’s performance and its ability to assist clinicians. Ten radiologists with different levels of experience (≥10 years, ≥5 years, and <5 years) independently reviewed the 168 cases in a blinded fashion. As shown in Fig. 5h, the average AUC for all readers significantly increased from 0.735 without model assistance to 0.872 with model assistance, while sensitivity improved from 0.632 to 0.810 and specificity improved from 0.693 to 0.832 (Supplementary Table 12).
Subgroup analysis revealed consistent benefits across all experience levels. For junior radiologists (<5 years of experience), the AUC increased from 0.687 to 0.859 (Fig. 5i); for intermediate radiologists (≥5 years), the AUC improved from 0.730 to 0.883 (Fig. 5j); and for senior radiologists (≥10 years), the AUC rose from 0.806 to 0.893 (Fig. 5k). These findings underscore the model’s potential to enhance diagnostic accuracy and reduce inter-observer variability in real-world clinical practice.
Risk Stratification and Individualized Treatment Strategy
To translate the predictive outcome of the Multimodal Model into clinically actionable risk groups, we applied the quartile method to the predicted probability values of the 237 patients with peritoneal metastasis (the OPM subgroup). This stratified the patients into four quartiles (Q1 to Q4), and their 2 year OS rates were assessed. The results showed that the prognoses of the Q1 group (2 year OS: 39.8%) and the Q2 group (2 year OS: 38.7%) were similar, leading to their combination into the Low-Risk Group. The Q3 group (2 year OS: 13.6%) was designated as the Moderate-Risk Group, and the Q4 group (2 year OS: 5.1%), exhibiting the poorest prognosis, was defined as the High-Risk Group (Supplementary Figure 9a). Kaplan–Meier survival analysis further confirmed a significant divergence among the overall survival curves of these three risk groups (Log-rank P < 0.001), with the high-risk group showing the worst prognosis, thereby validating the accuracy of the risk stratification (Supplementary Figure 9b). The sankey diagram (Supplementary Figure 9c) illustrates a strong correlation between the model-defined risk groups and the conventional peritoneal cancer index (PCI): low-risk patients were primarily clustered at low PCI scores (≤6), while high-risk patients predominantly corresponded to high PCI scores (≥13). These PCI categories (≤6, 7−12, ≥13) were adopted based on recommendations from major consensus guidelines, including those from Asian, NCCN, and other established international consensus groups6,27,28, reflecting thresholds associated with feasibility of complete cytoreductive surgery (CRS). Crucially, the risk stratification derived from our model is highly congruent with the guideline-recommended individualized three-step treatment management strategy (Supplementary Figure 10), which mandates distinct therapeutic pathways, including specific sequences of CRS, hyperthermic intraperitoneal chemotherapy (HIPEC), and chemotherapy, tailored to the patient’s risk level and corresponding PCI range.
Biological Insights into Multimodal Model–Based Risk Stratification
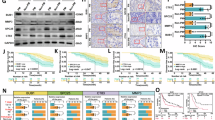
To elucidate the immunobiological basis underlying the Multimodal Model, we performed transcriptome sequencing on 18 paired fresh gastric cancer tumor samples (9 high-risk and 9 low-risk cases) selected through 1:1 propensity score matching. Heatmap analysis of differentially expressed genes revealed marked transcriptional alterations between the two groups (Fig. 6a). Notably, key immune-related genes such as CD244, CD86, TIGIT, and TNFRSF9 were significantly upregulated in the low-risk group (Fig. 6b). Gene set enrichment analysis (GSEA) indicated that low-risk samples were significantly enriched in multiple immune- and tumor-associated pathways, particularly epithelial-mesenchymal transition, inflammatory response, and TNFα signaling via NF-κB (Fig. 6c). These pathways collectively contribute to a more favorable tumor microenvironment that may suppress tumor progression and enhance treatment responsiveness.
a Heatmap of differentially expressed immune-related genes between high- and low-risk groups, including costimulatory and inhibitory checkpoint molecules. b Boxplots showing significantly higher expression of CD244, CD86, TIGIT, and TNFRSF9 in the low-risk group. c Gene set enrichment analysis (GSEA) plots showing pathways enriched in the high-risk group (left) and low-risk group (right); immune-activation pathways such as inflammatory response, TNFα signaling via NF-κB, epithelial–mesenchymal transition, and IL2/STAT5 signaling were enriched in the low-risk group. d Violin plots comparing tumor purity, stromal score, immune score, and ESTIMATE score between the two groups, showing higher stromal and immune scores in the low-risk group. e CIBERSORT analysis of immune cell composition, revealing distinct infiltration patterns between groups. f GSVA heatmap showing pathway enrichment differences, with immune-related and metabolic pathways differentially expressed between high- and low-risk groups. g GSVA bar plot of significantly enriched pathways, highlighting upregulation of IL6/JAK/STAT3, IL2/STAT5, and inflammatory signaling in the low-risk group, and downregulation of hypoxia-related signaling. h ssGSEA heatmap showing enrichment scores of various immune cell subsets in the tumor microenvironment for both groups. i Boxplots comparing specific immune cell infiltration levels between high- and low-risk groups; CD8 T cells, NK cells, and activated dendritic cells were significantly higher in the low-risk group.
The ESTIMATE algorithm showed that immune scores and ESTIMATE scores were significantly higher in the low-risk group compared with the high-risk group (Fig. 6d). CIBERSORT-based immune cell profiling revealed distinct differences in immune infiltration patterns, with multiple immune cell types exhibiting higher infiltration levels in low-risk tumors (Fig. 6e). GSVA enrichment analysis further demonstrated that the low-risk group was significantly enriched for IL6/JAK/STAT3 signaling, IL2/STAT5 signaling, and TNFA/NF-κB pathways (Fig. 6f, g). Finally, single-sample GSEA (ssGSEA) confirmed that the low-risk group had significantly higher activity of CD8 T cells, natural killer cells, and activated dendritic cells (Fig. 6h, i). Collectively, these findings suggest that tumors in the low-risk group possess stronger immune surveillance and antitumor immune responses, which may contribute to reduced tumor progression and improved clinical outcomes.
Discussion
In this multicenter study, we developed and validated a multimodal preoperative prediction model (Multimodal Model) that integrates clinical variables and CT-derived radiomics features of the primary tumor to identify OPM in patients with LAGC. The model demonstrated consistently robust performance across training, internal, two independent external, incremental, and prospective cohorts, with AUCs ranging from approximately 0.83 to 0.86. It also maintained superior net clinical benefit and reclassification improvement compared with unimodal approaches. Importantly, in a prospective reader crossover experiment, the model significantly improved the diagnostic accuracy of radiologists at different experience levels, underscoring its potential as a clinically deployable decision-support tool. Overall, this study provides a scalable “digital biopsy” solution for identifying high-risk OPM patients without additional invasive procedures, thereby reducing non-therapeutic laparotomies and optimizing perioperative decision-making.
Current preoperative pathways are fundamentally limited by the low sensitivity of contrast-enhanced CT for detecting subtle or OPM, especially lesions <5 mm or non-specific signs29,30,31. While SL offers superior accuracy, its routine use is restricted by its inherent invasiveness, necessity for general anesthesia, and potential for procedural morbidity15,32. Our multimodal prediction model is designed to complement SL as a non-invasive risk triage tool, improving patient selection for SL, enhancing diagnostic yield, and alleviating the resource-benefit imbalance in the perioperative workflow. This “digital biopsy” strategy serves as a crucial initial step in a multidisciplinary approach, being complementary to emerging tools like ctDNA. Crucially, the model’s clinical value lies in optimizing therapeutic selection, particularly for intensive locoregional treatments such as CRS/HIPEC, whose success hinges on identifying patients with low-volume disease33,34,35. Our model provides the necessary early, accurate risk stratification for tailored management: low-risk patients proceed directly to curative surgery, while high-risk patients are prioritized for neoadjuvant therapy or specialized trials, thereby ensuring that resource-intensive therapies are optimally reserved for those most likely to benefit.
Recent studies using CT-based radiomics or deep learning to predict OPM, though reporting AUCs around 0.80036,37,38,39,40, are often constrained by single-center design, small samples, manual ROI dependence, and inadequate validation or mechanistic interpretation. Our study substantially overcomes these limitations: (i) we achieved enhanced generalizability through validation on large-scale, multi-regional, multi-institutional datasets, including an international cohort; (ii) we demonstrated sensitivity to minimal disseminated burden via peritoneal cytology-positive prediction; (iii) a prospective reader study confirmed consistent clinical benefit across experience levels; and (iv) we provided transcriptomic evidence linking model-derived risk groups to immune differences, thereby improving interpretability and credibility. The predictive radiomics signature, capturing metrics like large-scale disorder (wavelet-LLL_glszm_ZoneEntropy) and multi-scale texture complexity, provides mechanistic plausibility by reflecting the underlying tumor-stroma/immune interface associated with OPM risk discrimination.
We established a plausible “imaging risk immune microenvironment” hypothesis. Propensity score-matched analysis revealed that the low-risk group was characterized by a more immune-active TME, showing significant enrichment of inflammatory pathways (e.g., TNFα/NF-κB, IL2/STAT5, IL6/JAK/STAT3), higher ESTIMATE scores, and greater abundance of CD8 T cells41,42,43. Conversely, high-risk tumors displayed profiles consistent with immune suppression or exhaustion, correlating with their greater propensity for dissemination and poorer outcomes44,45. These findings suggest that radiomics-derived heterogeneity reflects the underlying immune ecosystem. This stratification hints at differential therapeutic vulnerabilities: the immune-active profile in low-risk tumors suggests better prognosis and potential responsiveness to immunotherapy, optimizing their candidacy for definitive surgery46,47,48. In contrast, the immune-suppressive nature of high-risk tumors suggests they may benefit more from combination therapies or novel immune-enhancing agents to overcome resistance. While providing mechanistic plausibility and guiding future integrated imaging-transcriptomic biomarker development, these interpretations are currently hypothesis-generating and require validation in future prospective and mechanistic studies.
Despite efforts to ensure robustness, several limitations persist. First, the reliance on portal venous CT introduces unavoidable protocol heterogeneity (slice thickness, kernel). While mitigated by ComBat harmonization, achieving regulatory acceptance necessitates establishing a ‘Radiomics Acquisition Standard’ and dedicated standardization studies. Second, the model was built solely on primary tumor features; integrating dual-domain tumor-target features (peritoneal/omental ROIs) may improve sensitivity to minimal dissemination. Third, the prospective external cohort size remains modest, and the assessment of multifactorial downstream clinical endpoints (e.g., decision changes, cost-effectiveness) requires explicit interventional studies. Finally, the transcriptomic immune correlates are currently associative; future studies integrating spatial transcriptomics and single-cell sequencing are essential to establish definitive causal and spatial evidence for the imaging-immune link. Novel modalities like 68Ga-FAPI-04 PET/CT combined with multi-modal data fusion may further enhance OPM visualization and prediction.
In conclusion, we successfully developed and rigorously validated a multimodal OPM prediction model through multicenter, multi-cohort, and prospective evaluations. The model demonstrated stable discrimination, excellent calibration, and a significant boost to diagnostic performance across all radiologist experience levels. This robust performance is mechanistically supported by the correspondence between risk stratification and distinct immune microenvironment profiles, confirming both the interpretability and clinical relevance of the model. Positioned as a complementary, selective tool for staging laparoscopy, this model identifies truly high-risk patients for intensified therapy while reducing unnecessary procedures and delays. Subject to further large-scale, standardized prospective trials, this scalable, interpretable, and translatable “digital biopsy” strategy holds promise as a new cornerstone for individualized OPM management and integrated treatment strategies.
Methods
Study Cohort
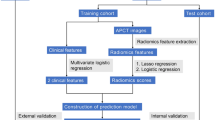
This multicenter study, as outlined in Fig. 7, retrospectively included patients diagnosed with locally advanced gastric cancer who underwent treatment between January 2013 and August 2020 at ten medical centers in China and Germany. A total of 2,565 patients were screened, and 1,726 met the predefined inclusion and exclusion criteria, as shown in Supplementary Figure 1. The participating institutions comprised the Fourth Hospital of Hebei Medical University (FHHMU), Shijiazhuang People’s Hospital (SJZPH), Baoding Central Hospital (BDCH), Hengshui People’s Hospital (HSPH), and Cangzhou People’s Hospital (CZPH) in northern China; Wuhan University People’s Hospital (WHPH), the Fifth Affiliated Hospital of Anhui Medical University (AMUFH), Nanjing Jinling Hospital (NJJLH), and the First Affiliated Hospital of the University of South China (NHUFH) in southern China; and Ludwig Maximilian University Hospital in Munich, Germany (LMUH).
a Data collection: Contrast-enhanced CT scans were acquired within two weeks before treatment initiation. The primary tumor was automatically segmented using a deep learning–based nnU-Net model, followed by three-dimensional region of interest (ROI) reconstruction and extraction of high-dimensional radiomic features. b Feature processing: Radiomic features underwent ComBat harmonization to minimize inter-center variability, followed by Z-score normalization. Features were filtered using the Mann–Whitney U test and Spearman correlation analysis, with the least absolute shrinkage and selection operator (LASSO) logistic regression identifying the most predictive features. An ensemble model combining logistic regression (LR), random forest (RF), extreme gradient boosting (XGB), and support vector machine (SVM) was constructed for radiomic prediction. c Model building: Independent clinical predictors and the radiomic signature were integrated to develop the Multimodal Model, presented as a nomogram for individualized OPM risk estimation. The model outputs were validated against intraoperative findings. d Transcriptomic analysis: Bulk RNA sequencing was performed on propensity score–matched high- and low-risk tumors. Differentially expressed genes were identified, and functional enrichment analyses (including gene set enrichment analysis, GSEA) were conducted to explore immune-biological mechanisms underlying model-based risk stratification.
For model development, patients treated at FHHMU from January 2013 to January 2018 were assigned to the training cohort. The remaining patients were allocated to three independent datasets: an internal validation cohort consisting of other patients from FHHMU within the same period, an external validation cohort I including patients from SJZPH, BDCH, HSPH, and CZPH between January 2017 and December 2019, and an external validation cohort II composed of patients from WHPH, AMUFH, NJJLH, NHUFH, and LMUH between January 2018 and May 2020. All patients underwent diagnostic laparoscopy, and any suspicious peritoneal lesions were confirmed by intraoperative biopsy followed by hematoxylin-eosin staining.
To further assess clinical applicability, an incremental validation cohort was assembled from FHHMU patients treated between January 2018 and August 2020, all of whom received both diagnostic laparoscopy and peritoneal lavage cytology. This dataset, containing 477 patients, was used to examine whether the model could predict positive peritoneal cytology.
A prospective clinical study (NCT06478368) was also conducted at FHHMU from July to December 2024, enrolling 168 patients according to the same inclusion criteria. To evaluate the model in real-world settings, ten radiologists from three national centers were recruited, including three with more than ten years of experience, three with more than five years of experience, and four with one to two years of experience. None of the radiologists participated in data collection or preprocessing, and all were blinded to patient information, CT reports, pathology results, and clinical diagnoses. An independent investigator, blinded to the radiologists’ identities, randomly assigned them in a one-to-one ratio to start either with AI-assisted diagnosis or without AI assistance, followed by crossover after a two-week washout period.
To explore the biological mechanisms underlying the model, fresh tumor samples from eighteen patients matched using 1:1 propensity score matching were collected. The detailed protocols for transcriptomic analysis were performed as previously described in our published literature49,50,51. The study protocol was approved by the Ethics Committee of the Fourth Hospital of Hebei Medical University (approval number: 2025KT151) and received ethics approval from all other participating centers. The study complied with the Declaration of Helsinki. Written informed consent was obtained from all the study participants for the prospective transcriptomic analyses, while retrospective data collection was exempt from the consent requirement. All patients underwent pretreatment abdominal CT and were restaged according to the eighth edition of the AJCC staging system. All authors followed applicable ethical standards to maintain research integrity without duplication, fraud, or plagiarism.
CT Image Acquisition and Segmentation and Radiomic Feature Extraction
All patients underwent contrast-enhanced abdominal CT within two weeks prior to treatment. Portal venous phase images were retrieved for analysis. Automatic segmentation of the primary gastric tumor was performed using a deep learning–based nnU-Net model, which was trained on 200 manually annotated CT scans. The detailed training and preprocessing protocols followed previously established procedures49,50,51. The nnU-Net model achieved a Dice coefficient of 0.890 on the test set. For quality control, only 7.3% (127/1726) of the total cases required partial manual adjustment, confirming the model’s high automation rate, and inter-observer variability analysis yielded a mean Dice coefficient of 0.882 ± 0.034.
Subsequently, 1,130 radiomic features, encompassing first-order, shape, and various texture metrics, were extracted from the segmented 3D-ROIs using PyRadiomics (version 3.1.0) implemented in Python 3.9.1 and integrated into the 3D Slicer platform. Standardized extraction settings included isotropic voxel resampling (1.0 mm³) and a fixed bin width of 25 for gray-level discretization. The effectiveness of ComBat adjustment in reducing clustering effects across imaging centers was visually confirmed (Supplementary Figure 2).
Radiomic Feature Selection and Ensemble Model Development
A three-step feature selection pipeline was implemented to identify radiomic features associated with OPM. First, all features were standardized using Z-scores, and the Mann-Whitney U test was used to identify features with significant differences between OPM-positive and OPM-negative groups. Second, Pearson correlation analysis was performed, and features with an absolute correlation coefficient greater than 0.9 were removed to minimize redundancy and multicollinearity. Finally, the least absolute shrinkage and selection operator (LASSO) logistic regression with ten-fold cross-validation was applied, and the top ten features ranked by regression coefficients were retained for model construction (Supplementary Table 1).
An ensemble learning framework combining logistic regression, extreme gradient boosting (XGBoost), random forest, support vector machine, and a voting regressor was then developed to predict OPM status. The voting regressor served as a meta-estimator, generating the final radiomic prediction score by simple averaging the outputs of individual base models.
In parallel, univariate logistic regression was performed in the training cohort to identify clinical parameters significantly associated with OPM. Significant clinical predictors (P < 0.05) were incorporated into a multivariable logistic regression to establish a clinical prediction model. Independent clinical factors and radiomic features were then integrated into a multimodal nomogram to generate individualized OPM risk scores. The optimal cutoff point for dichotomizing high- and low-risk categories was determined by maximizing the Youden index in the training cohort.
Biological characteristics and immune infiltration
Eighteen tumor tissue samples from the study cohort were collected for exploratory RNA sequencing. Functional enrichment analysis was conducted to investigate the biological relevance of the identified features. Immune cell infiltration within the tumor microenvironment was further analyzed using CIBERSORTx to estimate the abundance of specific immune cell types. The detailed protocols for RNA extraction, sequencing, and bioinformatics analysis have been previously described in our published literature49,50,51.
Statistical analysis
All statistical analyses were conducted using SPSS version 27.0 (IBM Corporation) and R version 4.5.1 (http://www.r-project.org). Continuous variables were compared using the unpaired two-tailed t-test or Mann–Whitney U test, whereas categorical variables were analyzed using the chi-square test or Fisher’s exact test. Inter-observer agreement for manual segmentation correction was quantified using the Dice similarity coefficient. For the predictive model, the optimal cutoff value for risk score was determined in the training cohort by maximizing the Youden index to achieve the best balance between sensitivity and specificity. Model performance metrics included the area under the receiver operating characteristic curve (AUC), accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), positive likelihood ratio (PLR), negative likelihood ratio (NLR), and F1 score.
Net reclassification improvement (NRI) and integrated discrimination improvement (IDI) were calculated to assess the incremental predictive value of different models. The DeLong test was used to compare differences in AUCs between models. Calibration curves were constructed to evaluate the agreement between predicted probabilities and observed outcomes, and prediction error curves along with the Brier score were used for further assessment of model calibration. A two-sided P value less than 0.05 was considered statistically significant.
Data availability
The datasets generated and/or analyzed during the current study are not publicly available due to containing individual patient data and being under license agreement with the providing center but are available from the corresponding author on reasonable request. The requests for access to these data should be made to Qun Zhao, zhaoqun@hebmu.edu.cn.
Code availability
The code used for computation analysis in this study can be found at https://github.com/hebeidpa/DeepComp/tree/main/Gastric-nnUNet. For any additional questions, pleasecon-tact the corresponding author.
References
Siegel, R. L., Kratzer, T. B., Giaquinto, A. N., Sung, H. & Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 75, 10–45 (2025).
Sundar, R. et al. Gastric cancer. Lancet 405, 2087–2102 (2025).
Li, G. Z., Doherty, G. M. & Wang, J. Surgical management of gastric cancer: a review. JAMA Surg. 157, 446–454 (2022).
Bonnot, P. E. et al. Cytoreductive surgery with or without hyperthermic intraperitoneal chemotherapy for gastric cancer with peritoneal metastases (cyto-chip study): a propensity score analysis. J. Clin. Oncol. 37, 2028–2040 (2019).
Gwee, Y. X. et al. Integration of genomic biology into therapeutic strategies of gastric cancer peritoneal metastasis. J. Clin. Oncol. 40, 2830 (2022).
Zhu, Z. et al. Asian consensus on normothermic intraperitoneal and systemic treatment for gastric cancer with peritoneal metastasis. Gastric Cancer 28, 731–748 (2025).
Liu, P. et al. Prediction of occult peritoneal metastases or positive cytology using CT in gastric cancer. Eur. Radiol. 33, 9275–9285 (2023).
Jiang, Y. et al. Noninvasive prediction of occult peritoneal metastasis in gastric cancer using deep learning. JAMA Netw. Open 4, e2032269 (2021).
Veron Sanchez, A. et al. Unravelling peritoneal carcinomatosis using cross-sectional imaging modalities. Diagnostics (Basel) 13, 2253 (2023).
Panagiotopoulou, P. B., Courcoutsakis, N., Tentes, A. & Prassopoulos, P. CT imaging of peritoneal carcinomatosis with surgical correlation: a pictorial review. Insights Imaging 12, 168 (2021).
Campos, N. M. F., Almeida, V. & Curvo Semedo, L. Peritoneal disease: key imaging findings that help in the differential diagnosis. Br. J. Radiol. 95, 20210346 (2022).
Qin, C. et al. 68Ga-DOTA-FAPI-04 PET/MR in the evaluation of gastric carcinomas: comparison with 18F-FDG PET/CT. J. Nucl. Med. 63, 81–88 (2022).
De Vuysere, S. et al. Accuracy of whole-body diffusion-weighted MRI (WB-DWI/MRI) in diagnosis, staging and follow-up of gastric cancer, in comparison to CT: a pilot study. BMC Med. Imaging 21, 18 (2021).
Kubo, N. et al. Risk prediction model of peritoneal seeding in advanced gastric cancer: a decision tool for diagnostic laparoscopy. Eur. J. Surg. Oncol. 49, 853–861 (2023).
Rawicz-Pruszyński, K. et al. Techniques of staging laparoscopy and peritoneal fluid assessment in gastric cancer: a systematic review. Int. J. Surg. 109, 3578–3589 (2023).
Leake, P. A. et al. A systematic review of the accuracy and utility of peritoneal cytology in patients with gastric cancer. Gastric. Cancer 15, S27–S37 (2012).
Schena, C. A. et al. The role of staging laparoscopy for gastric cancer patients: current evidence and future perspectives. Cancers (Basel) 15, 3425 (2023).
Ofluoğlu, C. B. et al. Diagnostic Efficacy of Staging Laparoscopy Compared to CT and PET-CT in Gastric Cancer: A Retrospective Cohort Analysis. Medicina (Kaunas) 60, 2079 (2024).
Chao, T. E., Mandigo, M., Opoku-Anane, J. & Maine, R. Systematic review of laparoscopic surgery in low- and middle-income countries: benefits, challenges, and strategies. Surg. Endosc. 30, 1–10 (2016).
Aerts, H. J. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 5, 4006 (2014).
Sala, E. et al. Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin. Radiol. 72, 3–10 (2017).
Shi, B. et al. Machine learning-driven SERS analysis platform for accurate and rapid diagnosis of peritoneal metastasis from gastric cancer. Ann. Surg. Oncol 32, 7604–7614 (2025).
Dong, D. et al. Development and validation of an individualized nomogram to identify occult peritoneal metastasis in patients with advanced gastric cancer. Ann. Oncol. 30, 431–438 (2019).
Li, Z. Y. et al. Four-point computed tomography scores for evaluation of occult peritoneal metastasis in patients with gastric cancer: a region-to-region comparison with staging laparoscopy. Ann. Surg. Oncol. 27, 1103–1109 (2020).
Mikuła-Pietrasik, J., Uruski, P., Tykarski, A. & Książek, K. The peritoneal “soil” for a cancerous “seed”: a comprehensive review of the pathogenesis of intraperitoneal cancer metastases. Cell Mol. Life Sci. 75, 509–525 (2018).
Fidler, I. J. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer 3, 453–458 (2003).
Japanese Gastric Cancer Association Japanese gastric cancer treatment guidelines 2021 (6th edition). Gastric. Cancer 26, 1–25 (2023).
Ajani, J. A. et al. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 20, 167–192 (2022).
van ‘t Sant, I. et al. Diagnostic performance of imaging for the detection of peritoneal metastases: a meta-analysis. Eur. Radiol. 30, 3101–3112 (2020).
Leeman, M. F., Patel, D., Anderson, J., OʼNeill, J. R. & Paterson-Brown, S. Multidetector computed tomography versus staging laparoscopy for the detection of peritoneal metastases in esophagogastric junctional and gastric cancer. Surg. Laparosc. Endosc. Percutan Tech. 27, 369–374 (2017).
Ho, S. Y. A. & Tay, K. V. Systematic review of diagnostic tools for peritoneal metastasis in gastric cancer-staging laparoscopy and its alternatives. World J. Gastrointest Surg. 15, 2280–2293 (2023).
Ramos, R. F., Scalon, F. M., Scalon, M. M. & Dias, D. I. Staging laparoscopy in gastric cancer to detect peritoneal metastases: a systematic review and meta-analysis. Eur. J. Surg. Oncol. 42, 1315–1321 (2016).
Marano, L. et al. Cytoreductive surgery and hyperthermic intraperitoneal chemotherapy for gastric cancer with synchronous peritoneal metastases: multicenter study of ‘italian peritoneal surface malignancies oncoteam-S.I.C.O. Ann. Surg. Oncol. 28, 9060–9070 (2021).
Brandl, A. & van Sandick, J. W. Treatment of gastric cancer peritoneal metastases: role of cytoreductive surgery and hyperthermic intraperitoneal chemotherapy. Br. J. Surg. 111, znae149 (2024).
Green, B. L. et al. Cytoreduction and HIPEC for gastric carcinomatosis: multi-institutional analysis of two phase ii clinical trials. Ann. Surg. Oncol. 30, 1852–1860 (2023).
Zhu, Z. N., Feng, Q. X., Li, Q., Xu, W. Y. & Liu, X. S. Machine learning-based CT radiomics approach for predicting occult peritoneal metastasis in advanced gastric cancer preoperatively. Clin. Radiol. 80, 106727 (2025).
Wang, L. et al. Novel CT based clinical nomogram comparable to radiomics model for identification of occult peritoneal metastasis in advanced gastric cancer. Eur. J. Surg. Oncol. 48, 2166–2173 (2022).
Huang, W. et al. Radiomics nomogram for prediction of peritoneal metastasis in patients with gastric cancer. Front. Oncol. 10, 1416 (2020).
Zhang, F., Wu, G., Chen, N. & Li, R. The predictive value of radiomics-based machine learning for peritoneal metastasis in gastric cancer patients: a systematic review and meta-analysis. Front. Oncol. 13, 1196053 (2023).
Chen, H. et al. 18F-FDG PET/CT radiomics-based multimodality fusion model for preoperative individualized noninvasive prediction of peritoneal metastasis in advanced gastric cancer. Ann. Surg. Oncol. 31, 6017–6027 (2024).
Chen, Y. et al. Predicting response to immunotherapy in gastric cancer via multi-dimensional analyses of the tumour immune microenvironment. Nat. Commun. 13, 4851 (2022).
Li, F. et al. CD4/CD8 + T cells, DC subsets, Foxp3, and IDO expression are predictive indictors of gastric cancer prognosis. Cancer Med. 8, 7330–7344 (2019).
Li, B., Jiang, Y., Li, G., Fisher, G. A. Jr & Li, R. Natural killer cell and stroma abundance are independently prognostic and predict gastric cancer chemotherapy benefit. JCI Insight 5, e136570 (2020).
Liu, Y., Li, C., Lu, Y., Liu, C. & Yang, W. Tumor microenvironment-mediated immune tolerance in development and treatment of gastric cancer. Front. Immunol. 13, 1016817 (2022).
Ding, J. T. et al. Landscapes and mechanisms of CD8+ T cell exhaustion in gastrointestinal cancer. Front. Immunol. 14, 1149622 (2023).
Tang, Q. et al. Comprehensive analyses of single-cell and bulk RNA sequencing data from M2 macrophages to elucidate the immune prognostic signature in patients with gastric cancer peritoneal metastasis. Immunotargets Ther. 14, 383–402 (2025).
Yu, P. et al. Genomic and immune microenvironment features influencing chemoimmunotherapy response in gastric cancer with peritoneal metastasis: a retrospective cohort study. Int. J. Surg. 110, 3504–3517 (2024).
Sun, Y. et al. Gastric cancer peritoneal metastasis related signature predicts prognosis and sensitivity to immunotherapy in gastric cancer. J. Cell Mol. Med. 27, 3578–3590 (2023).
Ding, P. et al. Interpretable multimodal fusion model enhances postoperative recurrence prediction in gastric cancer. Adv. Sci. (Weinh) 12, e08190 (2025).
Ding, P. et al. Multimodal artificial intelligence-based virtual biopsy for diagnosing abdominal lavage cytology-positive gastric cancer. Adv. Sci. (Weinh) 12, e2411490 (2025).
Ding, P. et al. Radiomics-based ensemble model predicts postoperative recurrence of gastric cancer. BMC Med. 23, 656 (2025.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (No. 82573273, No.82503478), S&T Program of Hebei (23297701Z, 242W7713Z, 25290101D), Hebei Natural Science Foundation (H2025206841), Hebei Province Medical Applicable Technology Tracking Project (GZ20250046), Hengrui-Hebei Innovative Development Medical Cooperation Program Project (HR202501001) and the Hebei Provincial Medical Science Research Project Plan (20260519).
Author information
Authors and Affiliations
Contributions
1. Conception and design: *Q.Z. and L.J.M.*; (II) Administrative support: *Q.Z.*; (III) Provision of study materials or patients: *P.A.D., H.H.G., J.X.Y., S.C., R.J.G., L.L.Z., N.M., X.L.L., Z.J.G., L.J.M., Q.Z.*; (IV) Collection and assembly of data: *P.A.D., S.C., H.H.G., J.X.Y., S.M., Y.H.Y., K.X.G., R.J.G., L.L.Z. Y.L.S., H.L., Z.J.X., N.M., X.L.L., Z.J.G.*; (V) Data analysis and interpretation: *P.A.D., H.H.G., J.X.Y., S.C.*; (VI) Manuscript writing: *P.A.D., H.H.G., J.X.Y., S.C.*; and (VII) final approval of manuscrip: all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, S., Ding, P., Yang, Y. et al. Multimodal digital biopsy for preoperative prediction of occult peritoneal metastasis in gastric cancer. npj Digit. Med. 9, 107 (2026). https://doi.org/10.1038/s41746-025-02268-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-02268-9