Abstract
The Trp metabolite l-kynurenine (KYN) accumulates in numerous solid tumours and mediates potent immunosuppression. Bacterial kynureninases (KYNases), which preferentially degrade KYN, can relieve immunosuppression in multiple cancer models, but immunogenicity concerns preclude their clinical use, while the human enzyme (HsKYNase) has very low activity for KYN and shows no therapeutic effect. Using fitness selections, we evolved a HsKYNase variant with 28-fold higher activity, beyond which exploration of >30 evolutionary trajectories involving the interrogation of >109 variants led to no further improvements. The introduction of two amino acid substitutions conserved in bacterial KYNases reduced enzyme fitness but potentiated rapid evolution of variants with ~500-fold improved activity and reversed substrate specificity, resulting in an enzyme capable of mediating strong anti-tumour effects in mice. Pre-steady-state kinetics revealed a switch in the rate-determining step attributable to changes in both enzyme structure and conformational dynamics. Apart from its clinical significance, our work highlights how rationally designed substitutions can potentiate trajectories that overcome barriers in protein evolution.

Similar content being viewed by others
Main
In numerous cancers, increased tryptophan (Trp) catabolism via indoleamine 2,3-dioxygenase (IDO1) and/or tryptophan 2,3-dioxygenase results in the accumulation of l-kynurenine (KYN) in the tumour microenvironment1,2,3. Elevated concentrations of KYN and its downstream metabolites exert a potent immunosuppressive effect on lymphocytes and myeloid cells and strongly correlate with poor clinical outcomes, as well as increased resistance to immunotherapy4,5,6,7. The pharmacological ablation of KYN accumulation in tumours has attracted substantial interest that in turn has led to multi-billion-dollar efforts aimed at the development of small-molecule drugs that inhibit IDO1 (ref. 8). Unfortunately, clinical results with IDO1 inhibitors have been disappointing, in part because of the sub-optimal pharmacodynamics of the tested inhibitors and additionally because in tumours the action of tryptophan 2,3-dioxygenase, in addition to IDO1, contributes to increased tumoral KYN concentrations9. Therefore, different approaches to achieving a deep and sustained decrease in KYN levels in humans for cancer therapy are urgently needed8,10,11,12,13. We recently showed that the administration of bacterial KYN-hydrolysing enzymes that had been conjugated to polyethylene glycol (PEG) to confer long circulation persistence was capable of complete depletion of circulating and tumoral KYN without affecting serum Trp. Enzyme-mediated depletion of KYN reversed immunosuppression in the tumour microenvironment in preclinical models, resulting in enhanced infiltration by cytotoxic T cell lymphocytes and reduced tumour growth as monotherapy, while enzyme treatment combined with immune checkpoint inhibitor (anti-programmed cell death protein 1 (anti-PD-1) or anti-cytotoxic T lymphocyte-associated protein 4 (anti-CTLA4)) antibodies led to complete tumour ablation and long-term T cell immunity14.
Kynureninases (KYNases) hydrolyse the Cβ–Cγ bond of KYN or 3′-OH-l-kynurenine (OH-KYN) to yield l-alanine (ALA) and either anthranilate or hydroxy-anthranilate (OH-AA) (Fig. 1a). KYNases are pyridoxal 5′-phosphate (PLP)-dependent enzymes, and many prokaryotic and archaeal KYNases are proficient in the hydrolysis of KYN, with the extensively studied enzyme from Pseudomonas fluorescens (PfKYNase) having a catalytic activity kcatKYN/KMKYN in the ∼7 × 104 M−1 s−1 range, where kcat is steady-state turnover rate of the reaction and KM is the substrate concentration for half maximal enzyme activity, and a strong preference for KYN hydrolysis over OH-KYN hydrolysis ((kcatKYN/KMKYN)/(kcatOH-KYN/KMOH-KYN) = 50). In contrast, human, and for that matter all vertebrate KYNases, have a strong preference for the hydrolysis of OH-KYN. Specifically, the human enzyme (Homo sapiens KYNase (HsKYNase)) has reversed substrate specificity ((kcatKYN/KMKYN)/(kcatOH-KYN/KMOH-KYN) = 0.0022) relative to bacterial enzymes and poorly hydrolyses KYN with a kcatKYN/KMKYN of only 110 M−1 s−1 (ref. 15).
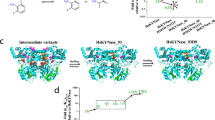
a, Reactions catalysed by KYNases. b, E. coli genetic selection scheme for the isolation of high-catalytic-activity HsKYNase variants from large libraries. c, Schematic showing the directed evolution trajectory leading to the isolation of HsKYNase_66. Blunted arrows denote dead-end evolutionary intermediates, from which higher-catalytic-activity variants could not be selected from numerous libraries. d, Steady-state kinetics parameters of HsKYNase, HsKYNase_46 and their H102W, N333T or H102W/N333T variants with KYN and OH-KYN. The best-fit values ± error for one representative measurement are shown. e, Diagram of mutational strategies to overcome the HsKYNase_46 evolutionary dead end. The grey ribbon (entire protein sequence) denotes sites targeted by whole-gene scanning saturation mutagenesis libraries, error-prone PCR and neutral drift selections and in silico designs. The right half of the image shows HsKYNase_46 mutations as blue sticks and amino acid residues targeted for mutagenesis in two (orange spheres), four to six (dark pink spheres), eight to nine (light pink spheres) or ten additional different libraries (red spheres). f, Steady-state kinetics parameters of HsKYNase_64 and HsKYNase_66, as in d.
In animal models, the administration of high-activity, KYN-preferring bacterial enzymes is capable of mediating a reduction of KYN levels in plasma and tumours, resulting in the reversal of immunosuppression and strong anti-tumour effects in multiple syngeneic murine cancer models14. In contrast, administration of the OH-KYN-preferring HsKYNase enzyme has no such effects. Unfortunately, bacterial enzymes are recognized as foreign by the human immune system, preventing their use for therapeutic applications in humans16,17. This is an especially important consideration for a therapeutic approach whose intended mechanism of action is to stimulate immune responses that, in turn, may promote elicitation of antibodies to foreign (that is, non-human) proteins. Thus, for clinical development, it is critical to deploy a variant of the human KYNase having kinetics comparable to those of the P. fluorescens enzyme to enable therapeutically meaningful depletion of circulating KYN with minimal risk of immunogenicity.
Close examination of the evolutionary and mechanistic differences among KYN- and OH-KYN-selective enzymes underscores the barriers to engineering a HsKYNase variant having high catalytic activity towards its non-preferred substrate. KYN-preferring prokaryotic enzymes and OH-KYN-preferring vertebrate enzymes are close structural homologues, yet they are divergent phylogenetically with an amino acid identity of <50%. PfKYNase and HsKYNase have only 26% amino acid identity (45% amino acid homology), yet their aligned structures show ~1.2 Å Cα root-mean-square deviation (Worldwide Protein Data Bank (PDB) entries 3E9K and 1QZ9). Pre-steady-state kinetics analyses showed that final product release (ALA) is the rate-determining step in the hydrolysis of their respective preferred substrates (that is, KYN for PfKYNase and OH-KYN for HsKYNase). In contrast, a chemical step in the formation of the first product (namely anthranilate for HsKYNase and OH-AA for PfKYNase) is rate limiting in the catalysis of the non-preferred substrate15. Hydrogen–deuterium exchange mass spectrometry (HDX-MS) experiments indicated that OH-KYN hydrolysis by HsKYNase requires stabilization of its active site via hydrogen bonds with the substrate’s hydroxyl group, while the bacterial enzyme does not require such active site stabilization and achieves substrate discrimination primarily by steric effects15.
Here, we show that the adaptive evolution of wild-type HsKYNase for high catalytic activity towards KYN led to the isolation of an enzyme with 28× higher kcatKYN/KMKYN catalytic activity and increased kcatOH-KYN/KMOH-KYN. However, no further increase in KYN catalytic activity could be achieved despite the interrogation of numerous evolutionary trajectories that collectively involved the sampling of >2 × 109 mutants, suggesting that this enzyme represents a dead end or frozen intermediate. We found that the introduction of two amino acid substitutions that are phylogenetically conserved among bacterial KYN-preferring enzymes reduced the fitness of this intermediate, substantially decreasing its kcatKYN/KMKYN without favourably affecting expression, thermodynamic stability or other protein features that have been reported to be associated with productive evolutionary trajectories, but surprisingly served to potentiate the rapid evolution of variants with high KYN activity and selectivity18,19. In particular, the enzyme termed HsKYNase_66, with an overall ~510× higher kcatKYN/KMKYN, fully recapitulates the catalytic and structural properties of prokaryotic KYNases. We further showed that HsKYNase_66 achieves high KYN catalytic activity and selectivity via a switch in the rate-determining step of the catalytic cycle relative to the parental human enzyme and that even though the kinetics properties of HsKYNase_66 closely parallel those of the bacterial KYN-preferring enzymes, the enzyme exhibits protein dynamics that are clearly distinct from those of bacterial KYNase, as well as from the parental human enzyme. The engineering of HsKYNase_66 constitutes a critical step in the clinical development of a KYNase variant that is the subject of Investigational New Drug-enabling studies and may represent an immune checkpoint enzyme for cancer immunotherapy.
Results
Evolving HsKYNase using potentiating mutations to overcome evolutionary dead ends
To enable the isolation of enzymes with increased fitness from very large libraries of HsKYNase variants (Supplementary Table 1), we first developed and validated a sensitive genetic selection. In Escherichia coli, anthranilate synthetase is composed of two polypeptide subunits, TrpE and TrpD, and catalyses the first step in the synthesis of Trp from chorismate, producing anthranilate (Fig. 1b). TrpE mutants are unable to make anthranilate and are auxotrophic for Trp. We found that an E. coli ΔTrpE strain expressing PfKYNase grew well on KYN-supplemented minimal media (lacking Trp), forming colonies after ~1.5 d. In contrast, cells expressing the low-activity HsKYNase formed small colonies only after 4 d of incubation. E. coli expressing variants with threefold lower KYN activity (HsKYNase-H102W/N333T (where H102W represents a mutation transcribing a substitution of the amino acid histidine for tryptophan at residue 102 and N333T represents a mutation transcribing a substitution of the amino acid asparagine for threonine at residue 333) or HsKYNase-H102W; see Fig. 1c,d) could not support growth under these conditions. The inability of the lower-catalytic-activity variant to support growth was not due to impaired protein expression as we observed comparable soluble expression yields for the wild type and the two mutant human enzymes in Trp-containing media (Supplementary Table 2). We sought to assess the enzyme fitness differential that could lead to preferential enrichment of bacteria expressing higher-activity enzymes in competition experiments in liquid media. After six serial passages (approximately 60 generations), cells expressing PfKYNase were enriched >1,000 over cells expressing the Mucilaginibacter paludis KYNase homologue (MpKYNase), which has ~20% lower catalytic activity (kcatKYN /KMKYN = 57,000 M−1 s−1) and similar expression. The ability to enrich clones expressing proteins with relatively small differences in catalytic activity is probably because these competition experiments were performed for six passages, resulting in selection over a high number (~60) of generations. In the lower-activity regimen, cells expressing HsKYNase-F306L (Supplementary Table 2; kcatKYN/KMKYN = ~500 M−1 s−1) were likewise enriched (that is, >1,000 enrichment) from E. coli expressing HsKYNase, which has 4.5-fold lower activity (and again, similar expression) after only two passages.
Ancestral sequence reconstruction has been used successfully as a starting point for the directed evolution of many novel catalytic activities20,21,22. Graphical representation of ancestral sequence predictions (GRASP)23 ancestral reconstruction with inputted phylogenetic trees for KYN- and OH-KYN-preferred KYNases obtained from the W-IQ-TREE webserver24 revealed that the likely ancestral KYN-preferring enzyme has <67% amino acid identity to HsKYNase (Supplementary Fig. 1). In light of the high degree of sequence divergence of the ancestral sequence reconstruction enzyme from the human enzyme, the putative common ancestor was not deemed to be a suitable starting point for a directed evolution campaign, given the goal of developing a high-catalytic-activity enzyme that is as homologous as possible to the human protein. For this reason, we selected the wild-type HsKYNase as the template for directed evolution. We generated libraries of 107–108 transformants over the course of four initial rounds of directed evolution, whereby each library was constructed using a variety of approaches including random mutagenesis, structure- or phylogeny-guided mutagenesis of select residues and DNA shuffling (Supplementary Table 1). Cells expressing each of these libraries were grown for 60 generations (six passages) in selective media, as discussed above, at which point between ~100 and 300 clones were sub-cultured in 96-well plates and the activity of the respective variant enzymes in whole-cell lysate was determined. Clones with a specific activity >20% higher than the parental species were sequenced, the respective enzymes were produced at a preparative scale and the kcatKYN/KMKYN values were determined. Four rounds of enzyme evolution yielded several variants that had around four- to fivefold higher kcat and five- to sixfold lower KM values, with the most active enzyme, HsKYNase_46, having a kcatKYN/KMKYN of 3,100 M−1 s−1 compared with 110 M−1 s−1 for HsKYNase (Fig. 1c,d). Of the nine amino acid substitutions in HsKYNase_46 (Table 1), six were at positions at least 7 Å from the substrate-binding site (Supplementary Fig. 2).
HsKYNase_46 has slightly higher catalytic activity towards OH-KYN relative to the wild-type enzyme (60,000 and 49,000 M−1 s−1, respectively) and is a more promiscuous enzyme ((kcatOH-KYN/KMOH-KYN)/(kcatKYN/KMKYN) = 20 for HsKYNase_46 compared with 430 for HsKYNase; Fig. 1c,d). HsKYNase_46 has slightly better but not statistically different expression compared with the parental HsKYNase and both have high thermal stabilities with melting temperature (Tm) = 75.85 ± 0.28 and 70.40 ± 0.45 °C for HsKYNase and HsKYNase_46, respectively, as determined by differential scanning fluorimetry (Supplementary Table 2). Enzyme variants that are catalytically promiscuous and/or have good stability and expression have been found to be advantageous as a template for the evolution of an improved function in numerous studies18,25,26,27,28,29,30. However, in this case, very extensive efforts involving the screening of libraries encompassing collectively >2 × 109 clones constructed from HsKYNase_46 by various random mutagenesis, structure-guided or Rosetta-assisted computational protein design (in collaboration with Arzeda; https://www.arzeda.com/) approaches failed to yield variants with higher catalytic activity (Supplementary Tables 1 and 2 and Fig. 1e). Notably, no variants with improved catalytic activity could be isolated from a scanning saturation mutagenesis library of every individual amino acid in HsKYNase_46 (a PFunkel library)31. We also generated combinatorial libraries encompassing a large fraction of all of the possible pairwise amino acid substitutions, constructed either by conventional DNA shuffling of the PFunkel library or using the random chimeragenesis on transient templates (RATCHITT) approach, which favours the recombination of near neighbouring positions32. Again, no single or double amino acid substitutions that confer higher activity could be selected. Likewise, the screening of libraries generated by (1) random mutagenesis of HsKYNase_46 via error-prone PCR at different mutation rates, (2) DNA shuffling with variants from previous rounds of evolution, (3) phylogenetic-guided analysis whereby non-strictly conserved residues near the active site (within 7 Å of the H102 and N333 residues) were mutated to amino acids found in high-activity KYN-preferring bacterial variants or (4) extensive focused mutagenesis of the active site residues selected based on structural and phylogenetic considerations again failed to yield any activity improvements (Fig. 1e and Supplementary Tables 1 and 2). We further constructed random mutagenesis libraries of HsKYNase_46 and performed neutral genetic selections in an attempt to generate an altered starting point for further evolution in a manner analogous to what had been reported in the pioneering studies performed by the late Dan Tawfik and his coworkers33. Briefly, we used cycles of low-error-rate, whole-gene random mutagenesis coupled with reduced selective stringency by a reduced number of passages (three passages and ~30 generations instead of six passages and ~60 generations) and periodic supplementation with a 10× higher KYN concentration in the growth media, which we showed independently to decrease the stringency of selection. However, no improved variants were obtained using multiple pools of clones that arose after neutral drift selection, subsequent DNA shuffling and/or additional error mutagenesis followed by purifying selection. In a further effort to explore the effect of altered selective pressure, we developed a selection for heightened KYN hydrolysis activity in a different host (namely complementation of a TRP1-deletion Saccharomyces cerevisiae strain), then screened an HsKYNase_46 random mutagenesis library, but again had no success isolating variants with improved activity. Together, these results support the notion that despite employing a wide variety of directed evolution strategies that had proven successful in numerous earlier enzyme engineering campaigns in the literature, it was not possible to improve the catalytic properties of HsKYNase_46, establishing that this enzyme represents an evolutionary dead end or functionally frozen enzyme with no direct trajectory towards enhanced KYN activity19,34,35.
Earlier elegant biochemical studies by Phillips and coworkers36 have revealed that a H102/N333 motif in the active site of HsKYNase helps to coordinate the OH-KYN substrate through interactions between its aromatic ring and hydroxyl group with H102 (π-stacking interactions) and N333 (hydrogen bond), respectively. All KYN-preferring bacterial enzymes have Trp and Thr, respectively, at these positions. A H102W/N333T substitution in HsKYNase (HsKYNase-H102W/N333T) completely abolished the hydrolysis of OH-KYN (Fig. 1d). With KYN as the substrate, introduction of the H102W/N333T amino acid substitutions resulted in a threefold reduction in kcat, with KM essentially unaffected (Fig. 1d). Stopped-flow, pre-steady-state kinetics analysis showed that the chemical step (that is, formation of the first product, anthranilate) is rate determining in the hydrolysis of KYN by HsKYNase-H102W/N333T, as is the case for the wild-type human enzyme (Supplementary Fig. 3, reaction rate = 0.0007 ± 0.00003 s−1). In light of the phylogenetic conservation of 102W and 333T among KYN-preferring enzymes and their role in substrate selectivity for KYN over its hydroxylated counterpart, we reasoned that despite its effect in reducing catalytic activity and enzyme fitness, HsKYNase-H102W/N333T may constitute a more favourable template for priming the evolution of high KYN activity. However, as mentioned above, HsKYNase-H102W/N333T failed to support the growth of E. coli ΔTrpE on minimal media with KYN, either in liquid media or on agar plates; hence, fitness selections with this template were not feasible. Additionally, 96-well-plate screening of saturation mutagenesis libraries, without previous genetic selection, failed to identify catalytically improved variants relative to HsKYNase-H102W/N333T parental species. Therefore, to enable fitness selections on selective media plates, the H102W/N333T substitutions were introduced into the more active HsKYNase_46 variant. This enzyme (HsKYNase_46-H102W/N333T) has a kcatKYN/KMKYN of 210 M−1 s−1, ~15-fold less than the parental HsKYNase_46, but importantly, around twofold higher relative to the wild-type HsKYNase (Fig. 1c,d). HsKYNase_46-H102W/N333T could support growth both in liquid selective media and on agar plates, thus enabling fitness selections to be carried out.
With HsKYNase_46-H102W/N333T as a starting point, mutagenesis targeting residues near the H102W/N333T dyad and fitness selection led to the isolation of a variant, HsKYNase_64, with ~30-fold higher activity for KYN (Fig. 1c–f, Supplementary Fig. 2 and Table 1). A second and similarly designed round of targeted mutagenesis and screening resulted in the isolation of several variants with further increases in kcatKYN/KMKYN (Fig. 1c,f, Supplementary Tables 1 and 2 and Supplementary Fig. 2). The highest-activity variant, HsKYNase_66, catalyses the hydrolysis of KYN with very similar kinetics to those of the human enzyme for its preferred substrate, OH-KYN (Fig. 1d,f) (HsKYNase_66 kcatKYN/KMKYN = 56,000 M−1 s−1; HsKYNase kcatOH-KYN/KMOH-KYN = 49,000 M−1 s−1). HsKYNase_66 has the reverse substrate selectivity relative to the wild-type human enzyme, strongly preferring KYN over OH-KYN (Fig. 1f). Hence, in terms of catalytic activity and selectivity, this human variant is very similar to PfKYNase and other high-activity, bacterial KYN-preferring enzymes14. Further mutagenesis of active site loops within high-activity HsKYNase variants did not yield variants with improved activity (Supplementary Tables 1 and 2). We note that in a separate experiment we employed the exact mutagenesis and selection scheme employed with HsKYNase_46-H102W/N333T that yielded HsKYNase_64, instead using the dead-end intermediate HsKYNase_46 as a template. This effort failed to produce any improved variants, once more underscoring the critical role of the H102W/N333T potentiating mutations.
The W102/T333 motif that was critical for potentiating the evolution of HsKYNase_66 plays a key role in dictating substrate selectivity. Reversion of Trp102 to His or of Thr333 to Asn reduced its catalytic activity for KYN by 12- and 7-fold, respectively. Similar to HsKYNase_66, both HsKYNase_66-W102H and HsKYNase_66-T333N preferentially hydrolyse KYN over OH-KYN, with (kcatKYN/KMKYN)/(kcatOH-KYN/KMOH-KYN) ratios of 15 and 18, respectively. In contrast, in combination, the W102H/T333N reversion resulted in a dramatic >30-fold increase in kcatOH-KYN/KMOH-KYN. As a result, HsKYNase_66-W102H/T333N acquires inverse substrate selectivity relative to its parental enzyme, preferentially hydrolysing OH-KYN with a kcat/KM that is comparable (less than twofold lower) to that of the native human enzyme (Fig. 1f). This finding is consistent with the proposed critical role of H102 and N333 in coordinating the hydroxyl group of OH-KYN for efficient catalysis. The W102H and T333N mutations exhibit a strong reciprocal sign epistasis effect since they individually impair the OH-KYN activity of HsKYNase_66, whereas in combination they are highly beneficial. Reciprocal sign epistasis is associated with very rugged fitness landscapes37,38,39 and is in line with the absence of a direct evolutionary trajectory from the parental wild-type HsKYNase enzyme to HsKYNase_66. Underscoring the complexity of the fitness landscape, the synergistic effect of these two potentiating substitutions (H102W/N333T) on kcatKYN/KMKYN is highly dependent on the mutational background of the parental species, showing a higher-order positive-sign epistasis40, as described in Supplementary Fig. 4 (ratios of the H102W/N333T mutational effect on intermediate evolved species/effect on HsKYNase were ~0.23 for HsKYNase_46 and 21 for HsKYNase_66).
Pre-steady-state kinetics reveal altered rate-limiting step
Pre-steady-state kinetics analyses were performed by monitoring the fluorescence of the first product, anthranilate or OH-AA, in the msec regimen using a stopped-flow apparatus. In the reaction of HsKYNase with its non-preferred substrate, KYN, anthranilate accumulation was linear over time, indicating that the rate-limiting step is the chemical reaction (or a step that precedes it) leading to anthranilate formation (Figs. 1a and 2a)15. Fitting the pre-steady-state data to a single exponential burst model, we estimated a steady-state rate of 0.04 ± 0.003 s−1, which is in close agreement with the v/[E] steady-state rate measured at the same KYN concentration (v/[E] = ~0.06 s−1 at 800 mM KYN). In contrast with HsKYNase_66 and KYN, we observed a distinct anthranilate burst phase with a decay rate (eigenvalue) of 38.6 ± 1 s−1 during the first 50 msec of the reaction, which was then followed by a linear phase with a steady-state rate of 1.25 ± 0.05 s−1 (Fig. 2b). Since the kcat at steady state is ~1.5 s−1, it follows that a step after the formation of anthranilate, possibly corresponding to the release of the ALA product, is rate limiting. Thus, in the evolution of HsKYNase_66 from the parental human enzyme, the rate-determining step was switched from anthranilate formation in the case of the parental enzyme to product release for the high-KYN-activity-evolved human variant. In HsKYNase_66, the rate of anthranilate formation was accelerated at least 600-fold (38.6/0.06). Of note, product release is the rate-determining step for the KYN-preferring PfKYNase, which shows a very similar burst decay value of 32 ± 0.6 s−1 and a linear, steady-state rate of 5.7 ± 0.02 s−1 (ref. 15).
a–d, Stopped-flow, pre-steady-state fluorescence traces of OH-AA (first product of the reaction) formation with 25 μM HsKYNase with 800 μM KYN (a), 5 μM HsKYNase_66 with 500 μM KYN (b), 5 μM HsKYNase with 500 μM OH-KYN (c) and 12.5 μM HsKYNase_66 with 1000 μM OH-KYN (d). Each trace represents the average of five different experiments. The red traces include all data points. The black line shows the fit to 1 − the exponential burst equation (equation (2) in the Methods).
Similarly, product release is the rate-determining step in the reaction of the parental HsKYNase with its preferred substrate, OH-KYN (Fig. 2c). These findings show that for KYNase enzymes with high activity and selectivity, regardless of whether their preferred substrate is KYN or its hydroxylated form (OH-KYN), the chemical step leading to the formation of the first product is rapid and therefore the steady-state reaction rate is limited by the rates of subsequent intermediate steps that lead to release of the second product, ALA. Conversely, the chemical steps leading to the formation of either anthranilate or OH-AA are rate determining in the catalysis of the non-preferred substrate. This is the case for HsKYNase with KYN, PfKYNase with its non-preferred substrate OH-KYN and HsKYNase_66 with OH-KYN15. As shown in Fig. 2d, for the latter, OH-AA formation is linear over time with a rate constant of 0.13 ± 0.035 s−1. Finally, we note that when the H102W/N333T motif was directly incorporated on HsKYNase, no change in the rate-determining step was detected, while the catalytic rate was greatly diminished (reaction rate = 0.0007 ± 0.00003 s−1; Supplementary Fig. 3).
Structural analysis of HsKYNase_66
The structure of HsKYNase_66 was solved at 3.25 Å resolution (Fig. 3, Supplementary Figs. 5 and 6 and Supplementary Table 3). The enzyme formed crystals in the I4122 space group with two identical polypeptide chains in each asymmetric unit forming a dimer. The overall fold of HsKYNase_66 is almost identical to that of HsKYNase and its close structural homologue of PfKYNase (Supplementary Fig. 7). Compared with PfKYNase, the human enzymes have a loop insertion consisting of residues 50–67. In HsKYNase_66, the N67D mutation forms a salt bridge with R43, thereby stabilizing this long loop relative to the parental human enzyme (Supplementary Fig. 8a).
a, Loop cartoon representation of the crystal structure of HsKYNase_66. Mutations are shown as red spheres on Chain A only, and the PLP cofactor is shown as yellow sticks. Dashed boxes show the homodimer’s two active sites. Omit maps for selected mutations and PLP are shown in Supplementary Fig. 6. b, Magnified snapshot of the HsKYNase_66 active site (PDB: 7S3V; cyan) overlaid against the structure of HsKYNase (PDB: 3E9K; orange) in complex with the 3-hydroxyhippuric acid inhibitor (magenta). Where side chains are shown, C is cyan in HsKYNase_66 and orange in HsKYNase, N is blue and O is red. The phosphate group (PO4) of PLP is shown as burnt orange and red. c, Magnified overlay of the R428–R434 loop of substrate-free HsKYNase_66 (PDB: 7S3V; cyan) and HsKYNase (PDB: 3E9K; orange) in complex with the 3-hydroxyhippuric acid inhibitor (magenta), substrate-free HsKYNase (PDB: 2HZP; pink) and substrate-free PfKYNase (PDB: 1QZ9; green), showing the different conformation of the R434 side chain for the open (2HZP; pink) verses closed (all others) KYNase conformations and the shift of the loop itself in PfKYNase and HsKYNase_66 compared with HsKYNase. d, Magnification of the mutated SSP (A280S–G281S–A282P) active site loop in HsKYNase_66 and its location relative to the I99, W102 and W109 residues, as well as to the internal aldimine moiety and the substrate-binding pocket (shown with the 3-hydroxyhippuric acid inhibitor aligned from 3E9K). e, PLP binding pocket of HsKYNase_66. The asterisks in b, d and e indicate residues that are contributed from the second chain relative to the chain that harbours the W102–T333 motif. Italicized labels show HsKYNase amino acid residues that are mutated in HsKYNase_66.
In HsKYNase_66, most mutations are located at the dimer interface close to the active site pocket, clustering in a hydrophobic region that enhances the recognition of the substrate’s phenyl ring (Fig. 3a,b). Near this pocket, a loop consisting of 16 residues (I97–G112) appears to have undergone a considerable conformational change (Supplementary Figs. 7d and 8b–d). Within this loop, the H102W and E103F amino acid substitutions increase the hydrophobicity of the substrate-binding pocket, and the side chain of F103 in HsKYNase_66 adapts a perpendicular conformation relative to E103 of HsKYNase and, given its location, affects the shape of the substrate-binding pocket (Supplementary Fig. 6a).
The substrate-binding pocket of KYNases can be in open or closed conformations36,41,42. Substrate-free, wild-type HsKYNase is in the open conformation, identifiable by the position of the R434 side chain guanidino nitrogens, which are directed away from the active site and stabilized by a hydrogen bond network with D426, T404 and Y226 (Fig. 3c and Supplementary Fig. 9a)41. In the presence of the 3-hydroxyhippuric acid inhibitor substrate, HsKYNase’s R434 guanidino nitrogens are shifted by ~8 Å, facing the active site36. In this closed conformation, R434 is stabilized by hydrogen bonds with the α carboxyl of 3-hydroxyhippuric acid and D426 and F225 (Fig. 3c and Supplementary Fig. 9b). Notably, substrate-free HsKYNase_66 and PfKYNase42 are also in a closed conformation, with R434 prepositioned in the active site. The backbones of the R428–R434 loop of HsKYNase_66 and PfKYNase are also shifted compared with HsKYNase (Fig. 3c and Supplementary Fig. 9c–e).
A loop very close to the substrate-binding pocket, containing the triad A280–G281–A282 (AGA) in HsKYNase is mutated to S280–S281–P282 (SSP) in the evolved variant HsKYNase_66 (Fig. 3d and Table 1). Whereas an A(S/T)–G–A amino acid triad at positions 280–282 is highly conserved among eukaryotic enzymes (Supplementary Fig. 10), in KYN-preferring KYNases from bacteria, the structurally equivalent positions contain a strictly conserved G/S–G/S–P motif (Supplementary Fig. 11). For example, MpKYNase has an SSP motif (S248–S249–P250 in the M. paludis sequence; UniProt entry H1YAV1) while PfKYNase has a GGP triad (G231–G232–P233). In HsKYNase_66 and other high-KYN-activity enzymes, including PfKYNase, the proline residue that is near the W102 and W109 residues can further increase the local hydrophobicity of this region, which is critical for optimal interactions between the substrate’s aromatic ring and W102, as discussed above. The A282P substitution is located at the turn of this loop and may restrain its flexibility, as is the case with PfKYNase (Supplementary Fig. 12a,b).
Certain mutations found in HsKYNase_66 are located in proximity to the enzyme’s PLP binding site (Supplementary Fig. 6b). More specifically, the introduction of a hydroxyl group at position 136 (A136T) may allow formation of a hydrogen bond (Fig. 3e and Supplementary Fig. 6b), similar to the stabilizing interaction of T96 with the phosphate group of PLP in PfKYNase (Supplementary Fig. 12c,d). On the other side of the PLP, the N333T mutation—a key contributor to substrate discrimination through the disfavouring of OH-KYN binding—could form a hydrogen bond network with Y275 and the phosphate group of the PLP (Fig. 3b and Supplementary Fig. 6)15,36.
Conformational dynamics of HsKYNase_66 during catalysis
The conformational dynamics of HsKYNase_66 were determined in the presence and absence of KYN or OH-KYN and compared with the wild-type enzyme and PfKYNase15. The HDX-MS analyses of HsKYNase_66 were carried out under identical conditions to those described for HsKYNase and PfKYNase, and peptides from regions not involved in binding and/or catalysis showed similar maximum deuteration between HsKYNase_66 and HsKYNase. In HDX-MS experiments, approximately 130 overlapping peptides were analysed after 1, 10 or 100 min exposure to D2O, and for each peptide the difference in the average deuterium uptake in the presence and absence of substrate was calculated (see Supplementary Tables 4 and 5 for deuterium uptake data; only significant differences with P <0.01 and an average |ΔHDX| > 0.3 were considered). Steady-state parameters of HsKYNase_66 determined in deuterated phosphate-buffered saline (D2O-PBS) (kcatKYN = 0.24 ± 0.01 s−1, KMKYN = 0.047 ± 0.0055 mM, kcatOH-KYN = 0.23 ± 0.019 s−1 and KMOH-KYN = 1.37 ± 0.2 mM) suggest that the reaction was under steady-state conditions for the first two time points.
Comparison of deuterium uptake by HsKYNase_66 in the presence and absence of KYN (Fig. 4a,c and Supplementary Fig. 13a) revealed that the region surrounding the critical H102W mutation, which is important for KYN specificity, did not show any deuterium uptake difference upon the addition of substrate (Fig. 4a,c,e; for example, peptide D92–F103). The nearby second-shell active site region A270–I290 that includes the SSP motif, however, showed decreased deuterium uptake (Fig. 4a,c,e; for example, peptide W272–F289). In contrast, the parental HsKYNase showed a statistically significant stabilization of the Y100–I116 region that encompasses the critical H102 residue (only at t = 10 min), with no changes in exchange in the A270–I290 region (Fig. 4b,d)15. Similar to HsKYNase, the KYN-preferring PfKYNase enzyme has also shown stabilization of the region that harbours the cognate W64 residue (only at t = 10 min) and no change in the G231–G232–P233 region (Supplementary Fig. 14a,b). These findings suggest that the hydrophobic region in the HsKYNase_66 active site (I99–W102–F103–W109) that could promote π-stacking interactions with the aromatic ring of KYN is pre-organized for efficient KYN binding and subsequent productive turnover.
a,b, Magnified view of the active sites of HsKYNase_66 (a; PDB: 7S3V) and HsKYNase (b; PDB: 3E9K), coloured by the difference in fractional deuterium uptake (−15 to +15%) between no substrate and with KYN after 1 min exposure to deuterium. Mutations (W102, *T136, *Y225, *S280, *S281, *P282, T333, *L405 and *N408) and important PLP-interacting residues (*F165, *K276 and W305) of HsKYNase_66 located at regions with significant exchange are labelled. The asterisks indicate residues contributed by the second KYNase chain. The respective structurally equivalent residues in the case of HsKYNase are shown in b. Regions with significant exchange were determined based on overlapping peptides and assuming complete back-exchange of the N-terminal residue. Cut-offs for peptide significance are P < 0.01 (as determined by Welch’s test) and average |ΔHDX| ≥ 0.3. Residues without coverage are shown in grey. PLP is shown as sticks and coloured by atom (C is white, N is blue, O is red and P is orange) while the 3-hydroxyhippuric competitive inhibitor is shown as magenta sticks. These data are shown as a volcano plot in Supplementary Fig. 13. c,d, Magnified view of the active sites of HsKYNase_66 (c) and HsKYNase (d), coloured by the difference in fractional deuterium uptake (−15 to +15%) between no substrate and with KYN after 10 min exposure to deuterium. e, Deuterium uptake plots for individual peptides (number ranges in top right of each plot) of HsKYNase_66 reaction with KYN and OH-KYN. A volcano plot for the HsKYNase_66/OH-KYN reaction is shown in Supplementary Fig. 13. The y axis ranges are scaled to 70% of the maximum deuterium uptake assuming that the N-terminal residue undergoes complete back-exchange. The error bars represent ±2σ from three technical replicates. Panels b and d adapted with permission from ref. 15, American Chemical Society.
A reduction in deuterium exchange is also observed for I218–Y226 (Fig. 4a,c,e; for example, peptide F220–T237) and R399–L403 in HsKYNase_66 upon addition of KYN (Fig. 4a,c,e; for example, peptide I390–L403). Both of these regions are involved in hydrogen bonding networks in the closed conformation of the enzyme that is observed following substrate binding by HsKYNase and, alternatively, in the resting state of HsKYNase_66 and PfKYNase (that is, without substrate binding; Supplementary Fig. 9). I218–Y226 contains the V223I and F225Y mutations, and the Y226 hydroxyl group can hydrogen bond with either of R428’s guanidino nitrogens in HsKYNase, depending on the conformation of the R428 side chain, while Q402 and T404 (contained within or adjacent to the R399–L403) form hydrogen bonds with F/Y225 and Y226 backbone carbonyls (Supplementary Fig. 9a–d)36,41,42. The homologous loop in PfKYNase (peptide V333–Y353) showed a decrease in deuterium uptake in the reaction with KYN at t = 10 min, but not with OH-KYN (Supplementary Fig. 14)15. Similar decreases in deuterium uptake were observed when the less preferred OH-KYN was added to HsKYNase_66 (Supplementary Figs. 13b and 15a,c). The conformational dynamics of both of these regions are not affected by substrate turnover (either KYN or OH-KYN) in the HsKYNase enzyme (Fig. 4b,d and Supplementary Fig. 15b,d)15.
We observed a time-dependent increase in deuterium uptake in regions surrounding the PLP cofactor and involved in the dimer interface during the catalysis of either KYN or OH-KYN by HsKYNase_66. These regions include N135–T138 (Fig. 4e; for example, peptide M134–L141), E161–N182 (Fig. 4e; for example, peptides L160–E173 and S174–N182) and H291–L338 (Fig. 4e; for example, peptides 311–329 and 329–337). N135–T138 contains the A136T mutation and is in the vicinity of the phosphate group of the PLP. E161–N182 does not contain any mutations, but F165 and D168 within this region can directly interact with the PLP cofactor. H291–L338 contains the important N333T mutation, as well as an I331C substitution, and it encompasses a long loop at the dimer interface that also interacts with the phosphate groups of PLP. The increased flexibility of N135–T138 that contains the A136T substitution is of interest considering the possible H-bond formation between T136 and the phosphate group of PLP (Fig. 3e). The extent of increased deuterium uptake was similar for the reaction with KYN or with OH-KYN, and in both cases the uptake appeared to increase at 10 min; larger increases were observed for OH-KYN after 100 min when the reaction is no longer predicted to be at steady state (Fig. 4e). We note that the crystal structure of HsKYNase_66 was solved in the absence of substrate, thereby reflecting the resting enzyme state (internal aldimine), whereas HDX-MS samples solution conformational states during catalysis43. Importantly, the destabilization of the pocket surrounding the PLP cofactor was not observed in the wild-type human (Fig. 4b,d and Supplementary Fig. 15b,d) or PfKYNase (Supplementary Fig. 14) enzymes15, representing a unique property of HsKYNase_66. These findings suggest that the evolution of HsKYNase_66 led to substantial shifts in the conformational dynamics during catalysis, as manifested by the increased flexibility of regions in the active site and the PLP binding pocket and further by the selective stabilization of more remote regions upon substrate binding (for direct comparisons of overall deuterium uptake by HsKYNase_66, HsKYNase and PfKYNase in the presence of KYN and OH-KYN, see Supplementary Figs. 16 and 17).
High-activity HsKYNase anti-tumour activity
Next, we determined whether a human KYNase enzyme with high kcatKYN/KMKYN can have similar anti-tumour effects to bacterial enzymes. A variant of HsKYNase_66 named HsKYNase_95 (Table 1 and Supplementary Table 2) having catalytic properties indistinguishable from HsKYNase_66 and whose KYN activity is equally dependent on the H102W/N333T potentiating mutations was expressed at a preparative level, purified to near homogeneity, confirmed to contain low endotoxin and finally conjugated to 5 kDa PEG-NHS ester for prolonged systemic retention in circulation. HsKYNase_95 showed higher stability than HsKYNase_66 during incubation in the presence of 50% serum in vitro—a property that qualitatively correlates with stability in circulation in animal models, making it more suitable for in vivo studies (Supplementary Fig. 18). Compared with HsKYNase_66, HsKYNase_95 has an additional A68T mutation near the predicted salt bridge resulting from the N67D mutation (Supplementary Fig. 8a), two conservative substitutions (I110L and F306W) and an S274N mutation. HsKYNase_95 lacks the A132V and V223I mutations found in HsKYNase_66 (Table 1). These positions all lie within active site loops and, of note, residues 132 and 306 are near the observed area of relative destabilization along the dimer interface and PLP pocket. As we have found that loss of the PLP cofactor is a reason for the deactivation of PLP enzymes in biological fluids, we speculate that the conservative amino acid substitutions in HsKYNase_95 at positions 132 and 306 may play a role in the favourable pharmacokinetics exhibited by this enzyme relative to HsKYNase_66. Syngeneic tumours were established in mice by injection of 5 × 104 CT26 colon carcinoma cells (which are known to express IDO1 and to accumulate KYN in the tumour microenvironment) in the right flank, and treatment was initiated when the tumour size reached 50–100 mm3. While administration of PEGylated wild-type human KYNase (that is, PEG-HsKYNase) at 20 mg kg−1 weight has no effect on tumour growth in syngeneic tumour models14, treatment with PEG-HsKYNase_95 with the identical dosing regimen significantly retarded tumour growth (Fig. 5 and Supplementary Fig. 19). We further compared the effect of PEGylated HsKYNase_95 with that of an antagonistic antibody targeting PD-1—a widely used and successful immune checkpoint inhibition modality. Treatment with the PEGylated HsKYNase_95 variant retarded tumour growth to the same degree achieved by anti-PD-1 antibody at the dose and frequency established to be optimal for this cancer model44 (Fig. 5). No statistical difference in body weight during the course of the experiment was seen across treatment arms (Supplementary Fig. 20).
Time course of CT26 colon carcinoma tumour volumes treated with either vehicle, HsKYNase_95 or anti-PD-1 antibody (n = 8 mice for each experimental arm). The data are presented as means ± s.e.m. Statistical significance was determined by unpaired t-test (two tailed). *P < 0.05 comparing HsKYNase_95 treatment with the vehicle control. The inset shows steady-state kinetics parameters and residual activity after incubation in serum for HsKYNase_66 and HsKYNase_95.
Discussion
Bacterial KYNases have evolved to have high catalytic selectivity for KYN over OH-KYN, whereas their animal orthologous enzymes exhibit the reverse selectivity and catalyse the hydrolysis of OH-KYN. The catalytic properties of prokaryotic and animal enzymes probably reflect their distinct physiological roles. In animals, Trp homoeostasis is maintained by the KYN pathway in the liver and brain. However, under inflammatory conditions the action of IDO1 leads to the accumulation of KYN in tissues where it serves in key immune-suppressive and neuromodulatory roles in trans. Presumably, the high KM of animal KYNases for KYN is important for preventing its consumption in inflammatory settings.
We had shown earlier that administration of PEGylated high-KYN-activity bacterial KYNases can reverse the immune suppression that arises from the tumoral production of KYN14,45. However, the clinical experience with the two approved PEGylated bacterial enzyme therapeutics, pegaspargase46 and pegloticase47, has established that despite PEGylation partially masking bacterial proteins from recognition by the immune system, adverse antibody responses arise in most patients after repeated administration that necessitates discontinuation of treatment. For this reason, an engineered, high-KYN-activity variant of the human KYNase is required for therapeutic purposes.
The considerable degree of sequence divergence (for example, sequence identity of 27% for P. fluorescens and 48% for M. Paludis, the KYN-preferring enzyme with the highest homology to HsKYNase), the different conformational dynamics and the distinct rate-determining steps in the reaction with KYN (that is, ALA release for P. fluorescens versus anthranilate formation for HsKYNase) underscore the difficulty associated with switching the catalytic specificity of the human enzyme. Efforts to engineer a high-KYN-catalytic-activity HsKYNase variant via structure-guided or directed evolution approaches, with the latter involving combinations of a highly sensitive fitness selections and 96-well plate screening, resulted in the isolation of HsKYNase_46. As discussed in the Results section (Fig. 1), very extensive efforts employing numerous approaches and evolutionary trajectories that collectively involved the functional interrogation of >109 variants did not succeed in producing variants of HsKYNase_46 with higher catalytic activity, thus providing compelling evidence that this enzyme represents a dead-end evolutionary intermediate. Having said that, we acknowledge that it is possible that technical limitations may have limited the search of the protein sequence space around HsKYNase_46, such as for example unidentified biological constraints that might have biased the genetic selection.
It was only after the introduction of two potentiating substitutions, H102W and N333T, that a facile path towards high KYN catalytic efficiency could be accessed. Trp at position 102 and Thr at 333 are phylogenetically conserved among all KYN-selective bacterial enzymes and are not found among their non-prokaryotic, OH-selective orthologues. As shown in Fig. 1d, these two substitutions completely abolish OH-KYN activity. Each of these mutations individually or in combination reduces KYN catalytic activity in HsKYNase, and therefore could not be isolated via mutation of the wild-type enzyme by fitness selection alone.
However, the introduction of the deleterious H102W/N333T motif in the context of HsKYNase_46, which has 28× higher KYN activity than HsKYNase, afforded sufficient KYN hydrolysis to support the growth of E. coli ΔtrpE cells in selective media. In the directed evolution literature, intermediates that lead to higher catalytic activity towards a desired substrate frequently show higher expression or thermodynamic stability, which can be instrumental for tolerating the introduction of destabilizing epistatic mutations, and/or they have expanded substrate promiscuity (that is, generalist enzyme intermediates)27,48,49. This was not the case for HsKYNase_46-H102W/N333T, which has comparable stability and expression yield to its parental template, HsKYNase_46, is a bona fide specialist showing no activity towards OH-KYN, and importantly has 15-fold lower KYN activity and substantially lower fitness than its parental evolutionarily frozen variant, HsKYNase_46. Yet, two rounds of mutagenesis of the HsKYNase_46-H102W/N333T variant enabled rapid isolation of HsKYNase_66, with kcatKYN/KMKYN comparable to the most active KYN-preferring bacterial homologues. Of note, employing the identical mutagenesis scheme and screening strategy that led from HsKYNase_46-H102W/N333T to HsKYNase_66 using HsKYNase_46 (lacking potentiating mutations) as the template failed to lead to increased catalytic activity, further underscoring the key role of these two residues. The potentiating effect of H102W/N333T is quite unusual and may be related to complex epistatic effects that prime favourable changes in protein dynamics to favour the catalysis of KYN, as well as structural modifications to mimic its bacterial high-activity homologues.
The crystal structure of HsKYNase_66 suggests that the H102W substitution, along with additional mutations located in its proximity, alters the hydrophobicity and shape of a crucial active site loop to better accommodate KYN (Fig. 3b and Supplementary Fig. 8). In addition, a large shift in HsKYNase_66’s R428–R434 loop occurs as it assumes a closed conformation in which the R434 side chain is prepositioned in the active site, such that it resembles the conformation of substrate-free PfKYNase (Fig. 3c and Supplementary Fig. 9)36,41,42. Structural and HDX-MS data further indicate that the N333T substitution may be important for stabilizing the PLP cofactor during initiation of the KYN catalytic cycle, as well as in playing an important role in the active site’s conformational dynamics during catalysis. The N333T substitution increases the kcat of the evolved HsKYNase_66 by 25-fold (kcat of HsKYNase_66 versus kcat of HsKYNase_66-T333N) while having a minor effect on the kcat of the HsKYNase_46 or the wild-type enzymes (Fig. 1d,f).
It is quite remarkable that HsKYNase_66 has KYN catalytic activity and KYN versus OH-KYN selectivity comparable to the most active bacterial enzymes known. What is more, for HsKYNase_66 the rate-determining step in the reaction with KYN was switched from anthranilate formation (as in the wild-type enzyme) to product release (as is the case with its bacterial orthologues) (Fig. 2). To the best of our knowledge, there are very few examples where a change in the rate-determining step during the course of directed evolution has been inferred50, let alone experimentally demonstrated by pre-steady-state kinetics analyses. Interestingly, the majority of the mutations in HsKYNase_46 are located in second- and third-shell positions (first-shell positions are defined as those residues that directly interact with a substrate or PLP). Similarly, 11 out of 13 mutations that accumulated in the evolution from HsKYNase_46-H102W/N333T to HsKYNase_66 are located in second- and third-shell positions. Interestingly, a correlation between the catalytic efficiency of HsKYNase_66 towards KYN and OH-KYN and the extent of the respective decrease in deuterium uptake was observed in these regions harbouring second- and third-shell mutations, further linking the role of remote mutations to catalytic activity. A large body of literature suggests that flexibility is an important component of catalysis allowing the enzyme to simultaneously sample multiple and catalytically competent conformations51,52,53,54,55. We further established that the introduction of the potentiating mutations alone is not sufficient to switch the rate-determining step in the hydrolysis of KYN (Supplementary Fig. 3). Therefore, the switch of the rate-limiting step and attainment of high KYN activity must be dependent on a broader subset of the amino acid substitutions that accumulated in HsKYNase_66.
Whereas all other KYN-preferring KYNases show <50% sequence identity with eukaryotic enzymes, HsKYNase_66 and _95 are close homologues of the parental human enzyme (95% identity). Consequently, our work shows that high KYN activity and selectivity can be accommodated within the primary sequence of animal enzymes. This finding raises the question of why there is such a large degree of sequence divergence between KYN- and OH-KYN-preferring enzymes in the natural world in the first place. Possibly selective pressure for protein features other than catalysis per se, perhaps relating to the different functions and regulation of KYN in the different kingdoms of life, may have led to distinct evolutionary paths for KYN- and OH-KYN-preferring enzymes. Conceivably, the high KYN catalytic activity and specificity of HsKYNase_66, despite its largely human primary amino acid sequence, might have been co-evolved with the enzyme’s conformational dynamics, predominantly through the incorporation of structural flexibility during catalysis; in a manner not previously observed in HsKYNase or PfKYNase. The role of conformational flexibility in protein evolution has been highlighted in several previous studies53,56,57,58,59 as a key property for the shift of the ensemble of conformational states and the respective functional transition in an evolutionary trajectory. Perhaps most importantly, in addition to the many interesting ramifications entailed in the directed evolution and catalytic properties of KYN-specific human KYNase enzymes, we have shown that such enzymes are capable of mediating a strong anti-tumour effect in a mouse tumour model, with minimal risk of future anti-drug immunogenicity because of an absence of strong T cell epitopes, as determined by computational immunogenicity analysis, and importantly because conjugation to PEG impedes immune recognition60,61. A therapeutic, highly active, PEG-HsKYNase variant is the subject of Investigational New Drug-enabling studies for future clinical evaluation in patients with advanced solid tumours62.
Methods
HsKYNase expression vector construction
The wild-type H. sapiens KYNase (HsKYNase), codon optimized for expression in E. coli and harbouring an amino (N)-terminal 6X histidine tag, was amplified from a previously described plasmid14. This amplicon was inserted into the pMAL-c2x E. coli expression vector in place of the maltose-binding fusion protein (MBP) using Gibson cloning to create an ampicillin-resistant vector (dubbed pMal-noMBP-wtHsKYNase) in which expression of HsKYNase is driven by the Tac promoter63. A modified vector with chloramphenicol resistance (dubbed pMal-CAM) was also constructed such that a HsKYNase gene or library of variants could be inserted between the XmaI and KpnI restriction enzyme sites.
E. coli cloning, enzyme expression strains and strain construction
The E. coli trpE deletion mutant (strain genotype = F-,Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔtrpE772::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514) was obtained from the E. coli Genetic Stock Center at Yale and was thereafter referred to as K12 ΔtrpE. The ΔtrpE772::kan DNA region was transferred into the E. coli T7 Express strain from New England Biolabs (strain genotype = fhuA2 lacZ::T7 gene1 [lon] ompT gal sulA11 R(mcr-73::miniTn10--TetS)2 [dcm] R(zgb-210::Tn10--TetS) endA1 Δ(mcrC-mrr)114::IS10) using P1 phage transduction64.
E. coli strains K12 ΔtrpE, Bl21(DE3), C41(DE3), C43(DE3), T7 Express (New England Biolabs), T7 ΔtrpE and MC1061 were used for routine molecular cloning and plasmid propagation4. E. coli strains were grown at 37 °C with constant shaking in Lenox LB media supplemented with 50 μg ml−1 ampicillin, 50 μg ml−1 kanamycin or 35 μg ml−1 chloramphenicol as necessary.
Initially, library preparation and screening were performed in three strains as follows: (1) transformation of ligated DNA into MC1061 and subsequent extraction of plasmid library DNA by miniprep, followed by (2) transformation of the library into the K12 ΔtrpE strain and selection for improved variants, followed by (3) transformation of individual plasmids into strains Bl21(DE3), C41(DE3), C43(DE3) or T7 Express for HsKYNase expression. Ultimately, the T7 ΔtrpE strain was utilized for all of the steps.
Cloning and transformation procedures
Restriction enzymes were purchased from New England Biolabs and used according to standard protocols. Library preparation routinely required amplification and purification of several DNA sections of the HsKYNase gene, followed by assembly mediated by KOD DNA Polymerase or KOD Hot Start DNA Polymerase. PCR amplicon fragments were routinely incubated at 37 °C with DpnI enzyme to eliminate plasmid template DNA. KOD-mediated assembly reactions were performed without primers and with 1 µg total DNA of the DNA fragments to be assembled and normalized by molar amounts, followed by a second PCR using 5–10 µl of the DNA assembly reaction as a template and the standard addition of primers. The primer sequences used for molecular cloning and library creation can be found in Supplementary Table 6.
Error-prone PCR of an entire HsKYNase gene was performed using Taq DNA Polymerase for 25 cycles in eight 20 µl PCR reactions containing 0.22 mM dATP, 0.20 mM dCTP, 0.27 mM dGTP, 1.88 mM dTTP, 2.75 mM MgCl2, 0.5 mM MnCl2, 0.005 mg ml−1 bovine serum albumin, 20–100 ng HsKYNase plasmid DNA and 0.5 µM of primers JB68/70 (5′-CTCAGGTACCATATGGGCGGTCATCATCACCA-3′ (forward) and 5′-CTGCAGCCGTCGACCTAGTTTTTGGTTTCCGCACTGTCC-3′ (reverse)) or JB270/271 (5′-CACTGTGTGGTACCGAGGTAATACATGGGCGGTCATCATCACCACCATCATGG-3′ (forward) and 5′-CGAGTCAGCCCGGGTAATCCGCGGCTAGTTTTTGGTTTCCGCACTGTCCA-3′ (reverse)).
HsKYNase amplified libraries were gel purified and digested for use in library preparation as described below. Ligation reactions were performed overnight at 16 or 22 °C using T4 DNA Ligase (New England Biolabs). Transformation of E. coli strains was performed using standard electroporator protocols.
Introduction of the bacterial KYNase motifs (H102W and/or N333T) was performed using oligo-mediated site-directed mutagenesis and Gibson assembly of overlapping fragments that contained the mutation(s) into a pMal-CAM plasmid backbone. Reversion of the bacterial KYNase motifs (W102H and/or T333N) were performed similarly.
Large-scale library preparation
High-efficiency library preparation was performed by ligating HsKYNase library DNA amplified by primers JB068/070 and digested with NdeI/SalI-HF/DpnI into the pMal-noMBP-HsKYNase digested with NdeI/SalI-HF or by ligating HsKYNase library DNA amplified by primers JB270/271 and digested with KpnI-HF/XmaI/DpnI into the pMal-CAM vector DNA digested with KpnI-HF/XmaI. Libraries inserted into the pMal-noMBP and pMal-CAM vector backbones were selected for with ampicillin or chloramphenicol, respectively. All ligations contained 100 ng digested HsKYNase library insert and 100 ng digested vector backbone (approximating a 3:1 molar ratio) and were performed overnight before heat deactivation.
Heat-deactivated ligations were dialysed against ultrapure water for 20 min before electroporation into freshly prepared electrocompetent cells. After electroporation, E. coli cells were allowed to recover at 37 °C in 1 ml SOC media with shaking. After recovery, the cells were plated on LB antibiotic plates (Corning) and either grown overnight at 37 °C until visual confirmation of individual colonies or grown overnight at 37 °C in 25 ml LB antibiotic media.
Library size was determined by plating 0.01, 0.1 or 1 µl cells recovered in SOC and counting individual colonies. Library diversity was determined by sequencing single colonies.
Genetic selection for enhanced KYNase variant activity
A defined culture media (dubbed M9-KYN media) was created to allow the selection of higher-activity HsKYNase variants harboured in E. coli K12 or T7 ΔtrpE cells from a pool of cells harbouring less active HsKYNase variants. M9-KYN media contained 1× Difco M9 minimal salts (Becton, Dickinson and Company), 2 mM magnesium sulfate (Thermo Fisher Scientific), 0.1 mM calcium chloride (Sigma–Aldrich), 2% glucose (Sigma Life Science), 0.10 mM isopropyl β-d-1-thiogalactopyranoside (IPTG; Thermo Fisher Scientific), antibiotic (ampicillin or chloramphenicol), between 25 and 100 μM KYN (Sigma–Aldrich) and ultrapure water.
A generalized genetic selection process proceeded as follows. An initial inoculum of 105–1010 E. coli K12 or T7 ΔtrpE cells (tenfold more cells than the calculated library size) harbouring a library of HsKYNase variants was inoculated into 25 ml M9-KYN media and then grown at 37 °C with shaking at 220 r.p.m. until the optical density measured at a wavelength of 600 nm (OD600) was >1.0. Then, 1 ml of the resultant culture was washed three times in sterile DPBS and 20 to 50% of the initial inoculum by cell count was used to inoculate 25 ml fresh M9-KYN media and cultivated at 37 °C with shaking at 220 r.p.m. until OD600 > 0.50. In general, six rounds of selection (inoculating with a reduced number of cells for each round) were performed for each library, before plating on LB antibiotic plates for further screening.
96-well assay of enzyme-specific activity in cell lysate
Individual colonies of E. coli expressing different HsKYNase variants were picked into a 96-well U-bottomed plate containing LB and antibiotic, with controls expressing known HsKYNase variants included in quadruplicate. After overnight growth, 2 μl was used to inoculate a fresh 96-well U-bottomed plate containing 70 μl TB and antibiotic per well. This expression plate was grown at 37 °C for 4 h and then KYNase expression was induced by adding 70 μl TB and antibiotic plus 1 mM IPTG to each well, followed by overnight induction at room temperature.
Induced cells were harvested by centrifugation. The supernatant was discarded, and 60 μl B-Per (Thermo Fisher Scientific) was added to each well. Cell pellets were simultaneously resuspended and lysed by shaking at room temperature for 20 min. Cell lysate was clarified by centrifugation and two aliquots of 25 μl supernatant containing soluble HsKYNase protein were transferred to 96-well plates pre-loaded with 25 μl PBS. Plates were screened by adding 150 μl Kyn substrate in PBS (pH 7.4) (either 500 or 50 μM Kyn concentration) and then monitoring the absorbance at 365 nm for 10–60 min. Specific activity was calculated compared with controls by analysing the slope of Kyn degradation in a plot of time versus absorbance at 365 nm.
Enzyme purification
Enzymes were expressed in BL21(DE3), C41(DE3), C41(DE3), T7 Express or T7 ΔtrpE strains, as described previously14,65. Single colonies of E. coli were used to inoculate 3-ml starter cultures of LB antibiotic and grown overnight at 37 °C. Then, 2.5 ml of the starter cultures was used to inoculate 500 ml Terrific Broth and antibiotic, which was grown to OD600 = 0.80 at 37 °C before induction with 0.5 μM IPTG (Thermo Fisher Scientific) at 25 °C and cultivation overnight. Cells were pelleted and resuspended in 25 ml lysis buffer, then lysed with a FRENCH Press cell disruptor (Thermo Electron Corporation) at 1,500 pounds per square inch.
Buffer formulations were as follows. The lysis buffer contained 100 mM sodium phosphate (pH 8.0), 1 mM PLP (Thermo Fisher Scientific), 300 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (Sigma–Aldrich), 25 mM imidazole (Thermo Fisher Scientific), 0.1% Tween 20 (Sigma–Aldrich) and 25 U ml−1 Universal Nuclease (Pierce). The wash buffer contained 300 mM NaCl, 100 mM sodium phosphate (pH 8.0), 25 mM imidazole and 0.1% Tween 20. The elution buffer contained 300 mM NaCl, 100 mM sodium phosphate (pH 8.0), 5 mM PLP and 300 mM imidazole.
Following lysis, the solid fraction of the lysate was removed by a 1-h centrifugation at 20,000g, then the lysate supernatant was filtered with a 0.22-μm syringe filter and applied via gravity flow to a pre-equilibrated (with wash buffer) Ni-NTA (Qiagen) column. The column was washed with 20 column volumes of wash buffer and then the enzyme was eluted with five column volumes of elution buffer. The eluant was incubated at 37 °C for 3 h, dialysed overnight at 4 °C in 25 mM Tris-HCl (pH 8.5) then buffer exchanged into DPBS.
The purification, PEGylation and endotoxin removal processes for HsKYNase_95 before its use in the in vivo mouse studies were performed as described previously14.
Steady-state kinetics analyses
Michaelis–Menten kinetics parameters for KYNase enzymes against KYN (Sigma–Aldrich) and 3-hydroxy-dl-kynurenine (OH-KYN; Sigma–Aldrich) were determined using substrate concentrations ranging between 0 and 2 mM and enzyme concentrations ranging between 0.10 and 1.00 μM. The enzyme activity was gauged by monitoring the decrease in KYN absorbance at 365 nm or OH-KYN absorbance at 373 nm over time. Values of initial reaction velocity per total enzyme concentration were obtained from a linear regression of reaction progress curves with <10% substrate degraded, then plotted against substrate concentrations in KaleidaGraph (Synergy Software). A nonlinear regression fitting the data to the Michaelis–Menten model (equation (1)) was used to calculate the kcat and KM parameters, where v is the initial reaction velocity and [S] is substrate concentration. For the determination of steady-state kinetics parameters of HsKYNase_66 in D2O-PBS, the purified enzyme was buffer exchanged against D2O-PBS (pD = 7.4). Substrates were prepared in D2O-PBS and the pD was adjusted to 7.0 for subsequent steady-state kinetics analysis.
Pre-steady-state kinetics analyses
Pre-steady-state kinetics traces for the reactions of HsKYNase and HsKYNase_66 with KYN and OH-KYN were followed by stopped-flow fluorescence spectroscopy as described previously15. Briefly, for the reaction of each enzyme with KYN, the rate of formation of the first product anthranilate was measured by exciting at 314 nm and recording the emitted light at 393 nm using single-band bandpass filters (Semrock). Similarly, the produced OH-AA during the reaction with OH-KYN was measured by exciting at 318 nm and detecting the fluorescence at 407 nm. For the reaction with KYN, 5 μM HsKYNase_66 was mixed with 500 μM substrate in the stopped-flow instrument (SF-300X instrument from KinTek Corporation), whereas for the reaction with OH-KYN, 12.5 μM HsKYNase_66 and 1,000 μM OH-KYN were mixed, respectively. For the experiments with HsKYNase, 25 and 5 μM enzyme was mixed with 800 and 500 μM KYN and OH-KYN, respectively. All concentrations were final after mixing the enzyme with the respective substrate in the stopped-flow instrument. All reactions were performed at 37 °C in DPBS and in all plots each trace represents the average of five different reactions. The conversion of fluorescence signals to actual concentrations of anthranilate and OH-AA was done by generating standard curves upon mixing known concentrations of either anthranilate or OH-AA in the stopped-flow apparatus and recording the fluorescence for each concentration. For the calculation of burst and linear rates, the data were analytically fit to a single-burst model described below (equation (2)) by nonlinear regression using the KinTek Explorer program.
where A is the amplitude, kburst is the decay rate of the burst phase (eigenvalue), t is time, Y is rate and kcat is the steady-state turnover rate of the reaction.
Crystallization and structure determination of HsKYNase_66
Following nickel column purification and buffer exchange, HsKYNase_66-Ct6XHis (HsKYNase_66 with a carboxy (C)-terminal 6X histidine tag) was dialysed against a dialysis buffer consisting of 50 mM HEPES (pH 8.0), 50 mM NaCl and 0.2 mM PLP solution for 6 h at 4 °C and further purified through gel filtration using a Sephadex 200 column (GE Healthcare) in dialysis buffer. HsKYNase_66_Ct6XHis was concentrated to a final concentration of 7.4 mg ml−1 using a Vivaspin Turbo 10,000 MWCO ultrafiltration unit before crystallization.
Diffracting crystals were obtained through vapour diffusion in sitting drops by mixing one part 7.4 mg ml−1 HsKYNase_66_Ct6XHis supplemented with 0.005 mg ml−1 trypsin (to remove any flexible terminal ends of the protein) with one part crystallization solution (8% PEG 8000, 100 mM imidazole, 50 mM MgCl2, 0.2 mM PLP and 5% sucrose) followed by incubation at 4 °C. Well-formed crystals were cryo-protected in 30% d-xylitol, 6% PEG 8000, 0.1 M imidazole and 0.2 mM PLP and then flash frozen in liquid nitrogen before data collection.
X-ray diffraction data were collected at the Advanced Photon Source beamline 23-ID-D (Argonne) and processed using the HKL-2000 software suite66. The structure was solved using molecular replacement in Phaser (Phenix) with the previously solved human KYNase structure (PDB: 2HZP) as a search model67,68. Refinement of the initial model was accomplished through iterative model building in COOT69. Models were refined in Phenix with 5% of the diffraction set as a test for Rfree cross-validation. Data collection and refinement statistics are summarized in Supplementary Table 3. Validation of the engineered KYNase structure was determined through visual inspection of individual mutations with reference to the calculated electron density maps in COOT, and molecular figures of the different mutation sites were generated using the PyMOL Molecular Graphics System, Version 2.0 (Schrödinger). The HsKYNase_66 structure was deposited into the PDB with the accession code 7S3V.
HDX-MS
Experiments were performed as per our previous work15. A flash-frozen HsKYNase_66 aliquot (>100 μM) was thawed on ice and incubated with 5 mM PLP at 37 °C for ~3 h. Labelling buffer was prepared by dissolving either KYN, OH-KYN or no substrate in D2O-1× PBS (pD = 7.4), yielding a final concentration of 3.125 mM. Following the PLP incubation, a fraction of HsKYNase_66 was diluted 1:24 (vol/vol) with labelling buffer at a final concentration of 1.6 µM enzyme and 3 mM substrate in 1× PBS at 37 °C. Enzymes were labelled for 0, 1, 10 or 100 min. Each time course was repeated in triplicate with freshly prepared labelling buffer for both substrates. Labelling reactions were quenched 1:1 in 2 M GuHCl and 0.8% (vol/vol) formic acid at 1 °C (pH 2.6). The samples were injected immediately or flash frozen and thawed before online pepsin digestion, trapping and elution.
Liquid chromatography
A Waters M-Class system coupled with a HDX Manager (equipped with a 50 µl loop) was used to perform protein digestion followed by peptide trapping and elution. The chromatography buffer was 0.1% (v/v) formic acid. An Enzymate Pepsin Column (300 Å; 5 µm; 2.1 mm × 30 mm; Waters) was used for online digestion at 15 °C. Digestion and subsequent trapping were performed for 3 min at a flow rate of 100 µl min−1. All peptides were desalted via reverse-phase trap (Waters Protein BEH C4 VanGuard Pre-column; 300 Å; 1.7 µm; 2.1 mm × 5 mm) and separated on a C18 column (Waters BEH C18 Column; 130 Å; 1.7 µm; 1 mm × 100 mm) at 1 °C using an isocratic gradient from 3–40% acetonitrile at 40 µl min−1 for 7 min. Following peptide digestion, a pepsin wash solution consisting of 2 M GuHCl, 4% ACN and 0.8% (vol/vol) formic acid was injected to minimize carryover. Blank injections were performed after each sample injection to ensure that low carryover was maintained.
Mass spectrometry
All data were acquired in resolution mode on a SYNAPT G2-Si Q-TOF mass spectrometer (Waters) using the positive ion mode for either HDMS or HDMSE modes. HDMSE mode was used to collect low- (6 V) and high-energy (ramping 22–44 V) data-independent peptide fragmentation data for downstream identification. HDMS mode was used for low-energy ion data for deuterated samples. The parameters were set as follows: sample sprayer capillary voltage at 2.8 kV, desolvation gas at 650 l h−1 and 175 °C with a source temperature of 80 °C, cone gas flow at 90 l h−1, nebulizer gas at 6.5 bar and sampling cone and source offset at 30 V. The mass correction was performed with [Glu1]-fibrinopeptide B as a reference mass and data were acquired with a 0.4 s scan time and a range of 100–2,000 m/z.
HDX data analysis
Data analysis was employed by exporting the raw files to ProteinLynx Global Server 3.0.2 (Waters) for peptide identification. Low-energy, high-energy and intensity thresholds were set to 250, 50 and 750 counts, respectively. The minimum number of fragment ion matches per peptide was set to three. Peptide and fragment tolerances were both set to automatic, whereas missed cleavages were set to one with a false discovery rate of four. ProteinLynx Global Server peptide lists were exported to DynamX 3.0 (Waters) and used to locate peptides in raw files and assign their weighted relative uptake profiles. DynamX 3.0 thresholds were set to 0.3 minimum products per amino acid and one minimum consecutive product. The file threshold was set to three, utilizing peptide lists from a total of six controls. All other DynamX 3.0 parameters were left unmodified. All DynamX 3.0 results were manually verified and filtered by applying a Welch’s t-test at 99% confidence to identify significant peptides using HD-eXplosion70.
Standards and reagents
Ultrapure-grade water was purchased from EMD Millipore and analytical grade formic acid, methanol, acetonitrile and dimethyl sulfoxide were from Thermo Fisher Scientific. Calibration solutions were from Thermo Fisher Scientific. Deuterated standards were purchased from Cambridge Isotope Laboratories (CIL), Buchem or Acros Organics as follows: 3-hydroxy-dl-kynurenine (5,8,8-D3; 98% purity; Buchem); 3-hydroxyanthranilic acid (4,6-D2; 98% purity; Buchem); anthranilic acid (3,4,5,6-D4; 98% purity; Buchem); dl-glutamic acid (2,4,4-D3; 98% purity; CIL); fumaric acid (2,3-D2; 98% purity; CIL); kynurenic acid (3,4,6,7,8-D5; 98% purity; Buchem); KYN (D4; 98% purity; Buchem); l-tryptophan (indole-D5; 98% purity; CIL); nicotinic acid (D4; 98% purity; CIL); picolinic acid (99% purity; Acros); and quinolinic acid (4,5,6-D3; 99% purity; Buchem).
Differential scanning fluorimetry
A LightCycler 480 instrument (Roche) and SYPRO Orange dye (Sigma–Aldrich) were used to determine the melting temperature following the manufacturer’s protocols, using a simultaneous titration of KYNase enzyme and dye ranging from 0.5–8 μM of enzyme and 5× to 20× of SYPRO Orange. Analysis was performed using the LightCycler 480 Software and confirmed with visual inspection.
Mouse experiments
BALB/cJ mice from The Jackson Laboratories were maintained at the University of Texas in Austin or Ikena Oncology in pathogen-free, ventilated cages with irradiated food and autoclaved water. Institutional animal care and use committees at the University of Texas in Austin or Ikena Oncology approved the experimental procedures. Housing conditions for mice were as follows: 12 h light/12 h dark cycle; 18.3–23.9 °C temperature and 40–60% relative humidity. The mice were monitored daily. Mice were euthanized by CO2 asphyxiation and cervical dislocation at the study endpoints or after showing signs of distress.
A total of eight mice per experimental arm were inoculated with 5 × 104 CT26 cells in the flank and treatments were started when tumour sizes reached 50–100 mm3. PEGylated-HsKynase_95 (20 mg kg−1) or vehicle control was administered by peritumoral injection every 3 d, and 10 mg kg−1 clone RMP1-14 anti-PD-1 antibody (Bio X Cell) was administered by intraperitoneal injection as described previously14. Tumour sizes were measured with callipers three times per week.
Statistical analysis
For statistical significance (P value) analysis, the data were first subjected to a normality test to ensure that parametric statistics could be employed to evaluate of the results. Subsequently, unpaired Student’s t-tests (two tailed) were performed. Statistical significance was denoted as P < 0.05. Statistical analyses were performed with GraphPad Prism.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The datasets generated and/or analysed during the current study are attached in the Supplementary Information and additional data are available from the corresponding authors upon reasonable request. The HsKYNase_66 structure has been deposited in the PDB with the accession code 7S3V. Source data are provided with this paper.
References
Opitz, C. A. et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 478, 197–203 (2011).
Cervenka, I., Agudelo, L. Z. & Ruas, J. L. Kynurenines: tryptophan’s metabolites in exercise, inflammation, and mental health. Science 357, eaaf9794 (2017).
Nguyen, D. J. M. et al. Targeting the kynurenine pathway for the treatment of cisplatin-resistant lung cancer. Mol. Cancer Res. 18, 105–117 (2020).
Frumento, G. et al. Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. J. Exp. Med. 196, 459–468 (2002).
Puccetti, P. et al. Accumulation of an endogenous tryptophan-derived metabolite in colorectal and breast cancers. PLoS ONE 10, e0122046 (2015).
Holmgaard, R. B., Zamarin, D., Munn, D. H., Wolchok, J. D. & Allison, J. P. Indoleamine 2,3-dioxygenase is a critical resistance mechanism in antitumor T cell immunotherapy targeting CTLA-4. J. Exp. Med. 210, 1389–1402 (2013).
Mezrich, J. D. et al. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 185, 3190–3198 (2010).
Prendergast, G. C., Malachowski, W. P., DuHadaway, J. B. & Muller, A. J. Discovery of IDO1 inhibitors: from bench to bedside. Cancer Res. 77, 6795–6811 (2017).
Li, H. et al. The landscape of cancer cell line metabolism. Nat. Med. 25, 850–860 (2019).
Röhrig, U. F., Majjigapu, S. R., Vogel, P., Zoete, V. & Michielin, O. Challenges in the discovery of indoleamine 2,3-dioxygenase 1 (IDO1) inhibitors. J. Med. Chem. 58, 9421–9437 (2015).
Dounay, A. B., Tuttle, J. B. & Verhoest, P. R. Challenges and opportunities in the discovery of new therapeutics targeting the kynurenine pathway. J. Med. Chem. 58, 8762–8782 (2015).
Günther, J., Däbritz, J. & Wirthgen, E. Limitations and off-target effects of tryptophan-related IDO inhibitors in cancer treatment. Front. Immunol. 10, 1801 (2019).
Jennings, M. R., Munn, D. & Blazeck, J. Immunosuppressive metabolites in tumoral immune evasion: redundancies, clinical efforts, and pathways forward. J. Immunother. Cancer 9, e003013 (2021).
Triplett, T. A. et al. Reversal of indoleamine 2,3-dioxygenase-mediated cancer immune suppression by systemic kynurenine depletion with a therapeutic enzyme. Nat. Biotechnol. 36, 758–764 (2018).
Karamitros, C. S. et al. Conformational dynamics contribute to substrate selectivity and catalysis in human kynureninase. ACS Chem. Biol. 15, 3159–3166 (2020).
Pokrovsky, V. S., Kazanov, M. D., Dyakov, I. N., Pokrovskaya, M. V. & Aleksandrova, S. S. Comparative immunogenicity and structural analysis of epitopes of different bacterial l-asparaginases. BMC Cancer 16, 89 (2016).
Mayer, A. et al. Modifying an immunogenic epitope on a therapeutic protein: a step towards an improved system for antibody-directed enzyme prodrug therapy (ADEPT). Br. J. Cancer 90, 2402–2410 (2004).
Trudeau, D. L. & Tawfik, D. S. Protein engineers turned evolutionists—the quest for the optimal starting point. Curr. Opin. Biotechnol. 60, 46–52 (2019).
Goldsmith, M. & Tawfik, D. S. Enzyme engineering: reaching the maximal catalytic efficiency peak. Curr. Opin. Struct. Biol. 47, 140–150 (2017).
Zakas, P. M. et al. Enhancing the pharmaceutical properties of protein drugs by ancestral sequence reconstruction. Nat. Biotechnol. 35, 35–37 (2017).
Thomas, A., Cutlan, R., Finnigan, W., van der Giezen, M. & Harmer, N. Highly thermostable carboxylic acid reductases generated by ancestral sequence reconstruction. Commun. Biol. 2, 429 (2019).
Merkl, R. & Sterner, R. Reconstruction of ancestral enzymes. Perspect. Sci. 9, 17–23 (2016).
Gumulya, Y. et al. Engineering highly functional thermostable proteins using ancestral sequence reconstruction. Nat. Catal. 1, 878–888 (2018).
Trifinopoulos, J., Nguyen, L.-T., von Haeseler, A. & Minh, B. Q. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235 (2016).
Bloom, J. D. & Arnold, F. H. In the light of directed evolution: pathways of adaptive protein evolution. Proc. Natl Acad. Sci. USA 106, 9995–10000 (2009).
Tawfik, D. S. Enzyme promiscuity and evolution in light of cellular metabolism. FEBS J. 287, 1260–1261 (2020).
Khersonsky, O., Roodveldt, C. & Tawfik, D. S. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 10, 498–508 (2006).
Leveson-Gower, R. B., Mayer, C. & Roelfes, G. The importance of catalytic promiscuity for enzyme design and evolution. Nat. Rev. Chem. 3, 687–705 (2019).
Romero, P. A. & Arnold, F. H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 10, 866–876 (2009).
Bloom, J. D., Labthavikul, S. T., Otey, C. R. & Arnold, F. H. Protein stability promotes evolvability. Proc. Natl Acad. Sci. USA 103, 5869 (2006).
Firnberg, E. & Ostermeier, M. PFunkel: efficient, expansive, user-defined mutagenesis. PLoS ONE 7, e52031 (2012).
Coco, W. M. et al. DNA shuffling method for generating highly recombined genes and evolved enzymes. Nat. Biotechnol. 19, 354–359 (2001).
Gupta, R. D. & Tawfik, D. S. Directed enzyme evolution via small and effective neutral drift libraries. Nat. Methods 5, 939–942 (2008).
Weinreich, D. M., Delaney, N. F., Depristo, M. A. & Hartl, D. L. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114 (2006).
Tracewell, C. A. & Arnold, F. H. Directed enzyme evolution: climbing fitness peaks one amino acid at a time. Curr. Opin. Chem. Biol. 13, 3–9 (2009).
Lima, S., Kumar, S., Gawandi, V., Momany, C. & Phillips, R. S. Crystal structure of the Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex: insights into the molecular basis of kynureninase substrate specificity. J. Med. Chem. 52, 389–396 (2009).
Wang, S. & Dai, L. Evolving generalists in switching rugged landscapes. PLoS Comput. Biol. 15, e1007320 (2019).
Kvitek, D. J. & Sherlock, G. Reciprocal sign epistasis between frequently experimentally evolved adaptive mutations causes a rugged fitness landscape. PLoS Genet. 7, e1002056 (2011).
Poelwijk, F. J., Tănase-Nicola, S., Kiviet, D. J. & Tans, S. J. Reciprocal sign epistasis is a necessary condition for multi-peaked fitness landscapes. J. Theor. Biol. 272, 141–144 (2011).
Yang, G. et al. Higher-order epistasis shapes the fitness landscape of a xenobiotic-degrading enzyme. Nat. Chem. Biol. 15, 1120–1128 (2019).
Lima, S., Khristoforov, R., Momany, C. & Phillips, R. S. Crystal structure of Homo sapiens kynureninase. Biochemistry 46, 2735–2744 (2007).
Momany, C., Levdikov, V., Blagova, L., Lima, S. & Phillips, R. S. Three-dimensional structure of kynureninase from Pseudomonas fluorescens. Biochemistry 43, 1193–1203 (2004).
Wales, T. E. & Engen, J. R. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 25, 158–170 (2006).
Toh, J. W. T. et al. The potential value of immunotherapy in colorectal cancers: review of the evidence for programmed death-1 inhibitor therapy. Clin. Colorectal Cancer 15, 285–291 (2016).
Labadie, B. W., Bao, R. & Luke, J. J. Reimagining IDO pathway inhibition in cancer immunotherapy via downstream focus on the tryptophan–kynurenine–aryl hydrocarbon axis. Clin. Cancer Res. 25, 1462–1471 (2019).
Hawkins, D. S. et al. Asparaginase pharmacokinetics after intensive polyethylene glycol-conjugated l-asparaginase therapy for children with relapsed acute lymphoblastic leukemia. Clin. Cancer Res. 10, 5335–5341 (2004).
Guttmann, A., Krasnokutsky, S., Pillinger, M. H. & Berhanu, A. Pegloticase in gout treatment—safety issues, latest evidence and clinical considerations. Ther. Adv. Drug Saf. 8, 379–388 (2017).
Aharoni, A. et al. The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 37, 73–76 (2005).
Soskine, M. & Tawfik, D. S. Mutational effects and the evolution of new protein functions. Nat. Rev. Genet. 11, 572–582 (2010).
Buller, A. R. et al. Directed evolution mimics allosteric activation by stepwise tuning of the conformational ensemble. J. Am. Chem. Soc. 140, 7256–7266 (2018).
Liang, Z.-X., Tsigos, I., Bouriotis, V. & Klinman, J. P. Impact of protein flexibility on hydride-transfer parameters in thermophilic and psychrophilic alcohol dehydrogenases. J. Am. Chem. Soc. 126, 9500–9501 (2004).
Pabis, A., Risso, V. A., Sanchez-Ruiz, J. M. & Kamerlin, S. C. Cooperativity and flexibility in enzyme evolution. Curr. Opin. Struct. Biol. 48, 83–92 (2018).
Klinman, J. P. Dynamically achieved active site precision in enzyme catalysis. Acc. Chem. Res. 48, 449–456 (2015).
Nestl, B. M. & Hauer, B. Engineering of flexible loops in enzymes. ACS Catal. 4, 3201–3211 (2014).
Zhang, J., Balsbaugh, J. L., Gao, S., Ahn, N. G. & Klinman, J. P. Hydrogen deuterium exchange defines catalytically linked regions of protein flexibility in the catechol O-methyltransferase reaction. Proc. Natl Acad. Sci. USA 117, 10797–10805 (2020).
Campbell, E. et al. The role of protein dynamics in the evolution of new enzyme function. Nat. Chem. Biol. 12, 944–950 (2016).
González, M. M., Abriata, L. A., Tomatis, P. E. & Vila, A. J. Optimization of conformational dynamics in an epistatic evolutionary trajectory. Mol. Biol. Evol. 33, 1768–1776 (2016).
Ramanathan, A. & Agarwal, P. K. Evolutionarily conserved linkage between enzyme fold, flexibility, and catalysis. PLoS Biol. 9, e1001193 (2011).
Otten, R. et al. How directed evolution reshapes the energy landscape in an enzyme to boost catalysis. Science 370, 1442–1446 (2020).
Molineux, G. Pegylation: engineering improved pharmaceuticals for enhanced therapy. Cancer Treat. Rev. 28, 13–16 (2002).
Vita, R. et al. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 47, D339–D343 (2019).
Kyn Therapeutics Enters into Global Strategic Collaboration with Celgene to Develop Immuno-oncology Therapies (Ikena Oncology, 2019); https://ir.ikenaoncology.com/news-releases/news-release-details/kyn-therapeutics-enters-global-strategic-collaboration-celgene
Deboer, H. A., Comstock, L. J. & Vasser, M. The Tac promoter—a functional hybrid derived from the Trp and Lac promoters. Proc. Natl Acad. Sci. USA 80, 21–25 (1983).
Thomason, L., Costantino, N. & Court, D. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 79, 1.17.1–1.17.8 (2007).
Miroux, B. & Walker, J. E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 (1996).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Zhang, N., Yu, X., Zhang, X. & D’Arcy, S. HD-eXplosion: visualization of hydrogen–deuterium exchange data as chiclet and volcano plots with statistical filtering. Bioinformatics 37, 1926–1927 (2021).
Acknowledgements
This work was supported by grants from the National Institutes of Health (1 RO1 CA189623), Cancer Prevention and Research Institute of Texas (grant DP150061) and Ikena Oncology (all to G.G. and E.S.), funding from the University of Texas System Proteomics network (to S.D.), grants R01GM104896 and R01GM125882 from the National Institutes of Health (both to Y.J.Z.) and postdoctoral fellowships from the American Cancer Society (grant 128252-PF-15-143-01-CDD to J.B. and grant 123506-PF-13-354-01-CDD to N.M.).
Author information
Authors and Affiliations
Contributions
J.B., C.S.K., K.F., A.Q., C.S., N.M., A.S., B.T., W.-C.L., M.Y.S. and N.A. designed and performed the directed evolution experiments and KYNase enzyme characterizations. C.S.K. and K.A.J. designed and performed the pre-steady-state kinetics experiments. N.T.B. and Y.J.Z. designed and performed the crystallization experiments. C.S.K., K. Murray and S.D. designed and performed the HDX experiments. Y.K., C.L., Y.T., C.S.K., J.B. and C.S. expressed and prepared enzymes for the in vivo and stopped-flow kinetics studies. S.C., M.M., X.M.Z. and K. McGovern designed and performed the in vivo experiments. J.B., C.S.K., G.G., E.S., K.A.J., S.D. and Y.J.Z. interpreted the data. C.S.K., G.G. and J.B. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
J.B., C.S.K., N.M., W.-C.L., G.G. and E.S. are inventors on intellectual property related to this work, including the active patent US9975959B2 and the pending patents US20190350975A1 and US20210207110A1, which are assigned to the Board of Regents of the University of Texas System. G.G., E.S., X.M.Z., K. McGovern, S.C. and M.M. have equity interest in Ikena Oncology, a company pursuing the commercial development of this technology. J.B., C.S.K., C.L. and E.S. have consulted for Ikena Oncology (previously Kyn Therapeutics). The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Catalysis thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–20 and Tables 1–5.
Supplementary Table 6
Sequences of the oligonucleotides used in this study.
Source data
Source Data Fig. 5
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Blazeck, J., Karamitros, C.S., Ford, K. et al. Bypassing evolutionary dead ends and switching the rate-limiting step of a human immunotherapeutic enzyme. Nat Catal 5, 952–967 (2022). https://doi.org/10.1038/s41929-022-00856-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41929-022-00856-6
This article is cited by
-
Physics-based modeling in the new era of enzyme engineering
Nature Computational Science (2025)
-
Mechanistic conformational and substrate selectivity profiles emerging in the evolution of enzymes via parallel trajectories
Nature Communications (2024)
-
Epistasis arises from shifting the rate-limiting step during enzyme evolution of a β-lactamase
Nature Catalysis (2024)