Abstract
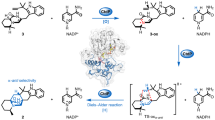
Iminium-catalysed cycloaddition is one of the most prominent examples of organocatalysis, yet a biological counterpart has not been reported, despite the widespread occurrence of iminium adducts in enzymes. Here we present biochemical, structural and computational evidence for iminium catalysis by the natural Diels–Alderase SdnG, which catalyses norbornene formation in sordarin biosynthesis. A Schiff-base adduct between the ε-nitrogen of active site K127 and the aldehyde group of the enal dienophile is revealed by structural analysis and captured under catalytic conditions via borohydride reduction. This Schiff-base adduct positions the substrate into near-attack conformation and decreases the transition-state barrier of Diels–Alder cyclization by 8.3 kcal mol−1 via dienophile activation. A hydrogen-bond network consisting of a catalytic triad is proposed to facilitate the proton transfer required for iminium formation. This work establishes an intriguing mode of catalysis for Diels–Alderases and points the way to the design of iminium-based (bio)catalysts.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Data supporting the findings of this study are available within the Article, its Supplementary Information files, and the Source Data files. All unique biological materials, such as plasmids, generated in the study are available from the authors upon request. Crystal structures of Se–Met-SdnG, SdnG–3NC, SdnG–3C, SdnG–4 and SdnG–8 have been deposited in the Protein Data Bank under IDs 8YIA, 8YHG, 8YJ4, 8YI8 and 8YHM, respectively. Data are available from the corresponding authors upon request. Source data are provided with this paper.
References
MacMillan, D. W. C. The advent and development of organocatalysis. Nature 455, 304–308 (2008).
Ahrendt, K. A., Borths, C. J. & MacMillan, D. W. C. New strategies for organic catalysis: the first highly enantioselective organocatalytic Diels-Alder reaction. J. Am. Chem. Soc. 122, 4243–4244 (2000).
Gefflaut, T., Blonski, C., Perie, J. & Willson, M. Class I aldolases: substrate specificity, mechanism, inhibitors and structural aspects. Prog. Biophys. Mol. Biol. 63, 301–340 (1995).
Fan, P.-H., Sato, S., Yeh, Y.-C. & Liu, H. Biosynthetic origin of the octose core and its mechanism of assembly during apramycin biosynthesis. J. Am. Chem. Soc. 145, 21361–21369 (2023).
Piersen, C. E., McCullough, A. K. & Lloyd, R. S. AP lyases and dRPases: commonality of mechanism. Mutat. Res. Repair 459, 43–53 (2000).
Warren, S., Zerner, B. & Westheimer, F. H. Acetoacetate decarboxylase. Identification of lysine at the active site. Biochemistry 5, 817–823 (1966).
Grell, T. A. J., Young, A. P., Drennan, C. L. & Bandarian, V. Biochemical and structural characterization of a Schiff base in the radical-mediated biosynthesis of 4-demethylwyosine by TYW1. J. Am. Chem. Soc. 140, 6842–6852 (2018).
Erkkilä, A., Majander, I. & Pihko, P. M. Iminium catalysis. Chem. Rev. 107, 5416–5470 (2007).
Mukherjee, S., Yang, J. W., Hoffmann, S. & List, B. Asymmetric enamine catalysis. Chem. Rev. 107, 5471–5569 (2007).
List, B., Lerner, R. A. & Barbas, C. F. Proline-catalyzed direct asymmetric aldol reactions. J. Am. Chem. Soc. 122, 2395–2396 (2000).
Garrabou, X., Beck, T. & Hilvert, D. A promiscuous de novo retro-aldolase catalyzes asymmetric Michael additions via Schiff base intermediates. Angew. Chem. Int. Ed. 54, 5609–5612 (2015).
Xu, G., Crotti, M., Saravanan, T., Kataja, K. M. & Poelarends, G. J. Enantiocomplementary epoxidation reactions catalyzed by an engineered cofactor-independent non-natural peroxygenase. Angew. Chem. Int. Ed. 59, 10374–10378 (2020).
Kunzendorf, A., Xu, G., Saifuddin, M., Saravanan, T. & Poelarends, G. J. Biocatalytic asymmetric cyclopropanations via enzyme-bound iminium ion intermediates. Angew. Chem. Int. Ed. 60, 24059–24063 (2021).
Garrabou, X., Wicky, B. I. M. & Hilvert, D. Fast Knoevenagel condensations catalyzed by an artificial Schiff-base-forming enzyme. J. Am. Chem. Soc. 138, 6972–6974 (2016).
Garrabou, X., Macdonald, D. S. & Hilvert, D. Chemoselective Henry condensations catalyzed by artificial carboligases. Chem. A Eur. J. 23, 6001–6003 (2017).
Leveson-Gower, R. B., Zhou, Z., Drienovská, I. & Roelfes, G. Unlocking iminium catalysis in artificial enzymes to create a Friedel-Crafts alkylase. ACS Catal. 11, 6763–6770 (2021).
Jamieson, C. S., Ohashi, M., Liu, F., Tang, Y. & Houk, K. N. The expanding world of biosynthetic pericyclases: cooperation of experiment and theory for discovery. Nat. Prod. Rep. 36, 698–713 (2019).
Kinsman, A. C. & Kerr, M. A. The total synthesis of (+)-hapalindole Q by an organomediated Diels-Alder reaction. J. Am. Chem. Soc. 125, 14120–14125 (2003).
You, L. et al. Asymmetric total synthesis of propindilactone G. J. Am. Chem. Soc. 137, 10120–10123 (2015).
Wilson, R. M., Jen, W. S. & MacMillan, D. W. C. Enantioselective organocatalytic intramolecular Diels-Alder reactions. The asymmetric synthesis of solanapyrone D. J. Am. Chem. Soc. 127, 11616–11617 (2005).
Bai, Y., Shen, X., Li, Y. & Dai, M. Total synthesis of (−)-spinosyn A via carbonylative macrolactonization. J. Am. Chem. Soc. 138, 10838–10841 (2016).
Horning, B. D. & MacMillan, D. W. C. Nine-step enantioselective total synthesis of (−)-vincorine. J. Am. Chem. Soc. 135, 6442–6445 (2013).
Fage, C. D. et al. The structure of SpnF, a standalone enzyme that catalyzes [4 + 2] cycloaddition. Nat. Chem. Biol. 11, 256–258 (2015).
Zheng, Q. et al. Enzyme-dependent [4 + 2] cycloaddition depends on Lid-like interaction of the N-terminal sequence with the catalytic core in PyrI4. Cell Chem. Biol. 23, 352–360 (2016).
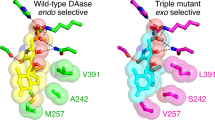
Sato, M. et al. Catalytic mechanism and endo-to-exo selectivity reversion of an octalin-forming natural Diels–Alderase. Nat. Catal. 4, 223–232 (2021).
Cai, Y. et al. Structural basis for stereoselective dehydration and hydrogen-bonding catalysis by the SAM-dependent pericyclase LepI. Nat. Chem. 11, 812–820 (2019).
Sun, Z., Jamieson, C. S., Ohashi, M., Houk, K. N. & Tang, Y. Discovery and characterization of a terpene biosynthetic pathway featuring a norbornene-forming Diels–Alderase. Nat. Commun. 13, 2568 (2022).
Liu, S. H. et al. Biosynthesis of sordarin revealing a Diels-Alderase for the formation of the norbornene skeleton. Angew. Chem. 61, e202205577 (2022).
Eberhardt, R. Y. et al. Filling out the structural map of the NTF2-like superfamily. BMC Bioinformatics 14, 327 (2013).
Zhang, B. et al. Enzyme-catalysed [6 + 4] cycloadditions in the biosynthesis of natural products. Nature 568, 122–126 (2019).
Drulyte, I. et al. Crystal structure of the putative cyclase IdmH from the indanomycin nonribosomal peptide synthase/polyketide synthase. IUCrJ 6, 1120–1133 (2019).
Niwa, K. et al. Biosynthesis of polycyclic natural products from conjugated polyenes via tandem isomerization and pericyclic reactions. J. Am. Chem. Soc. 145, 13520–13525 (2023).
Northrup, A. B. & MacMillan, D. W. C. The first general enantioselective catalytic Diels-Alder reaction with simple α,β-unsaturated ketones. J. Am. Chem. Soc. 124, 2458–2460 (2002).
Ozanne, A., Pouységu, L., Depernet, D., François, B. & Quideau, S. A stabilized formulation of IBX (SIBX) for safe oxidation reactions including a new oxidative demethylation of phenolic methyl aryl ethers. Org. Lett. 5, 2903–2906 (2003).
Bullock, T. L., Clarkson, D. W., Kent, H. M. & Stewart, M. The 1.6 Å resolution crystal structure of Nuclear Transport Factor 2 (NTF2). J. Mol. Biol. 260, 422–431 (1996).
Yu, J., Zhou, Y., Tanaka, I. & Yao, M. Roll: a new algorithm for the detection of protein pockets and cavities with a rolling probe sphere. Bioinformatics 26, 46–52 (2010).
Torosantucci, R., Schöneich, C. & Jiskoot, W. Oxidation of therapeutic proteins and peptides: structural and biological consequences. Pharm. Res. 31, 541–553 (2014).
Lin, J., Cassidy, C. S. & Frey, P. A. Correlations of the basicity of His 57 with transition state analogue binding, substrate reactivity, and the strength of the low-barrier hydrogen bond in chymotrypsin. Biochemistry 37, 11940–11948 (1998).
Hedstrom, L. Serine protease mechanism and specificity. Chem. Rev. 102, 4501–4524 (2002).
Crugeiras, J., Rios, A., Riveiros, E. & Richard, J. P. Substituent effects on the thermodynamic stability of imines formed from glycine and aromatic aldehydes: implications for the catalytic activity of pyridoxal-5′-phosphate. J. Am. Chem. Soc. 131, 15815–15824 (2009).
Thadani, A. N., Stankovic, A. R. & Rawal, V. H. Enantioselective Diels-Alder reactions catalyzed by hydrogen bonding. Proc. Natl Acad. Sci. USA 101, 5846–5850 (2004).
Minami, A. & Oikawa, H. Recent advances of Diels-Alderases involved in natural product biosynthesis. J. Antibiot. 69, 500–506 (2016).
Byrne, M. J. et al. The catalytic mechanism of a natural Diels-Alderase revealed in molecular detail. J. Am. Chem. Soc. 138, 6095–6098 (2016).
Zheng, Q. et al. Structural insights into a flavin-dependent [4 + 2] cyclase that catalyzes trans-decalin formation in pyrroindomycin biosynthesis. Cell Chem. Biol. 25, 718–727.e3 (2018).
van der Helm, M. P., Klemm, B. & Eelkema, R. Organocatalysis in aqueous media. Nat. Rev. Chem. 3, 491–508 (2019).
Kuatsjah, E. et al. Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation. Proc. Natl Acad. Sci. USA 120, e2212246120 (2023).
Ohashi, M. et al. An enzymatic Alder-ene reaction. Nature 586, 64–69 (2020).
Yeh, A. H.-W. et al. De novo design of luciferases using deep learning. Nature 614, 774–780 (2023).
Chen, J. et al. Carbonyl catalysis enables a biomimetic asymmetric Mannich reaction. Science 360, 1438–1442 (2018).
Lubkoll, J. & Wennemers, H. Mimicry of polyketide synthases—enantioselective 1,4-addition reactions of malonic acid half-thioesters to nitroolefins. Angew. Chem. Int. Ed. 46, 6841–6844 (2007).
Zheng, L., Baumann, U. & Reymond, J.-L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32, e115 (2004).
Liu, H. & Naismith, J. H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 (2008).
Gasteiger, E. et al. in The Proteomics Protocols Handbook (ed. Walker, J.) 571–607 (Humana Press, 2005).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution - from diffraction images to an initial model in minutes. Acta Crystallogr. D 62, 859–866 (2006).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
DeLano, W. L. PyMOL: an open-source molecular graphics tool. CCP4 Newslett. Protein Crystallogr. 40, 11 (2002).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Goujon, M. et al. A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res. 38, W695–W699 (2010).
Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Frisch, M. J. et al. Gaussian 16, Revision A.03 (Gaussian, Inc., 2016).
Cossi, M., Rega, N., Scalmani, G. & Barone, V. Energies, structures and electronic properties of molecules in solution with the C-PCM Solvation Model. J. Comput. Chem. 24, 669–681 (2003).
Li, Y.-P., Gomes, J., Mallikarjun Sharada, S., Bell, A. T. & Head-Gordon, M. Improved force-field parameters for QM/MM simulations of the energies of adsorption for molecules in zeolites and a free rotor correction to the rigid rotor harmonic oscillator model for adsorption enthalpies. J. Phys. Chem. C 119, 1840–1850 (2015).
Grimme, S. Supramolecular binding thermodynamics by dispersion-corrected density functional theory. Chem. Eur. J. 18, 9955–9964 (2012).
Luchini, G., Alegre-Requena, J. V., Funes-Ardoiz, I. & Paton, R. S. GoodVibes: automated thermochemistry for heterogeneous computational chemistry data. F1000Res. 9, 291 (2020).
Acknowledgements
This work was supported by NIH (5R01AI141481, to Y.T.), Shenzhen Bay Laboratory Open Program (SZBL2021080601014, to J.Z.) and the NSF (CHE 2153972 and 2409941, to K.N.H.). We thank the staff of beamlines BL17U1, BL18U1 and BL19U1 of Shanghai Synchrotron Radiation Facility for access and help with the X-ray data collection, D. Cascio and M. Sawaya at UCLA-DOE Institute for Genomics and Proteomics for help with discussion of the X-ray data, and Y. Chen at UCLA Molecular Instrumentation Center for help with the peptide MS/MS analysis.
Author information
Authors and Affiliations
Contributions
Z.S., X.Z., Q.Z., M.O., K.N.H., J.Z. and Y.T. developed the hypothesis and designed the study. Z.S. performed compound isolation, chemical synthesis and all biochemical studies. X.Z. performed crystallization and determined all structures. Q.Z. performed all computational studies. Z.S., X.Z., Q.Z., M.O., K.N.H., J.Z. and Y.T. analysed and discussed the results. Z.S. and Y.T. wrote the manuscript. Z.S., X.Z., Q.Z., M.O., K.N.H., J.Z. and Y.T. read, edited and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Catalysis thanks Jan Deska, Hui Ming Ge, Shingo Nagano, Masanobu Uchiyama and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The active sites of SdnG complexed with different ligands are nearly superimposable.
(a) Overlay of active sites of SdnG-3NC (ivory) and SdnG-3C (gray). Noncovalently bound 3 and covalently bound 3 are shown in cyan and green respectively. Water molecules interacting with 3 and Y41 are omitted for clarity. (b) Overlay of active sites of SdnG-3NC (ivory) and SdnG-4NC (gray). 3 and 4 are shown in cyan and green respectively. (c) Overlay of active sites of SdnG-3C (ivory) and SdnG-7 (gray). Covalently bound 3 and adduct 7 are shown in cyan and green respectively.
Extended Data Fig. 2 K127 adducts corresponding to the rDA and Cope rearrangement products of intermediate 7 were observed in the crystal structure of SdnG with 4.
(a) Active site of SdnG-7-5 (the rDA product of 7, chain C of SdnG-4, PDB 8YI8). Adducts 7 and 5 are shown in cyan and green respectively. Residues around 4 Å of the ligands are shown in ivory. The Polder omit map of 7-5 (contoured at 2.0 σ) is shown in blue mesh. Maps contoured at higher levels (2.5 σ and 3.0 σ) are shown in Supplementary Fig. 12. (b) Distance between diene and dienophile (highlighted in purple) of 5 in SdnG-7-5. Active site residues and 5 are shown in ivory and green respectively. Adduct 7 is omitted from the view for clarity. (c) Active site of SdnG-8 (the Cope rearrangement product of 7, PDB 8YHM). Adduct 8 is shown in cyan. The Polder omit map of 8 (blue mash) is contoured at 3.0 σ. Distances between atoms are shown in dashed lines. Bonds corresponding to the diene and the dienophile in 1 are highlighted in purple. Other coloring schemes are the same as in (a). (d) DFT-calculated transition states of rDA and Cope rearrangement of 7. All energies are relative to the ground state energy of 5 (iminium form). Bonds corresponding to the diene and the dienophile in 1 are highlighted in blue in 5 and green in 8. The transition state structure of rDA (TS-5) is the same as that of the forward reaction (TS-4). But the barriers of the forward and reverse reactions differ substantially due to the large energy gap between 5 and 7.
Extended Data Fig. 3 Catalytic activity of SdnG and K127X mutants.
All reactions were carried out for 1 min with 100 µM 1. Values and error bars are obtained from the average and standard deviation of three independent measurements (black circles) respectively (n = 3). (a) Absolute rates of non-enzymatic and enzymatic DA cyclization of 1. Asterisks indicate no measurable substrate consumption during the course of the reaction. (b) Relative activity of SdnG and K127X variants normalized by enzyme concentration. All activities are shown relative to the rate acceleration of the DA reaction exhibited by the wild-type enzyme (WT, 100%). A 0% value (asterisks) indicates no rate acceleration compared to uncatalyzed DA reaction.
Extended Data Fig. 4 Effect of the concentration of 4 on formation of iminium 7 in SdnG and the H72A variant.
SdnG or its mutational variants (5 µM) were mixed with varied amounts of 4 and the mixture was immediately treated with 20 mM NaBH4. The reaction was subsequently analyzed by UHPLC-HRMS and the deconvoluted ESI-MS spectra were shown in the figure. Increased concentration of 4 promotes imine formation of SdnG but not the H72A variant. The result suggests that ligand binding is rate-determining for iminium formation in SdnG but not in the H72A variant. Therefore, diminished iminium formation in H72A is not a result of compromised ligand binding.
Supplementary information
Supplementary Information
Supplementary Tables 1–4, Figs. 1–20, cartesian coordinates of calculated structures and source data.
Supplementary Data 1
Statistical source data for Supplementary Fig. 16.
Supplementary Data 2
Cartesian coordinates of calculated structures.
Source data
Source Data Fig. 3d
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, Z., Zang, X., Zhou, Q. et al. Iminium catalysis in natural Diels–Alderase. Nat Catal 8, 218–228 (2025). https://doi.org/10.1038/s41929-025-01294-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41929-025-01294-w
This article is cited by
-
Uncovering a novel biosynthetic gene cluster for sordarin through genome mining in the fungus Talaromyces adpressus
Bioresources and Bioprocessing (2025)
-
Iminium catalysis meets Diels–Alderase
Nature Catalysis (2025)