Abstract
The remarkable diversity of insect pigmentation offers a captivating avenue for studying evolution and genetics. In tephritids, understanding the molecular basis of mutant traits is also crucial for applied entomology, enabling the creation of genetic sexing strains through genome editing, thus facilitating sex-sorting before sterile insect releases. Here, we present evidence from classical and modern genetics showing that the black pupae (bp) phenotype in the GUA10 strain of Anastrepha ludens is caused by a large deletion at the ebony locus, removing the gene’s entire coding region. Targeted knockout of ebony induced analogous bp phenotypes across six major tephritid agricultural pests, demonstrating that disruption of Ebony alone is sufficient to produce the mutant trait in distantly related species. This functional characterization further allowed a deeper exploration of Ebony’s role in pigmentation and development across life stages in diverse species. Our findings offer key insights for molecular engineering of sexing strains based on the bp marker and for future evolutionary developmental biology studies in tephritids.
Similar content being viewed by others
Introduction
The color palette and patterns displayed by insects are perhaps the most striking evidence of their extraordinary diversity. Insect pigmentation has long fascinated biologists, leading to insights into their evolution, ecology, development, genetics, and physiology1,2,3. A deep understanding of molecular mechanisms underlying pigmentation pathways in insects has not only implications in evolutionary and developmental biology but also provides fertile grounds for the development of genetic-based methods in applied entomology. A textbook example of how mutations affecting pigmentation traits can be implemented in insect pest management comes from genetic studies in tephritid fruit flies.
With nearly 5000 recognized species, the family Tephritidae (true fruit flies) is one of the largest groups within Diptera. In addition to its remarkable diversity and intriguing life-history traits, the family is renowned for accommodating some of the most invasive insects worldwide4,5. Owing to their generalist herbivorous (polyphagous) habits, a few species have become major horticultural and agricultural pests6,7. Females of these species lay their eggs inside a wide range of fruits and vegetables, where hatching larvae will feed to complete their development, resulting in significant consequences on commodity production and trade. These characteristics ensure tephritids a position in food security, sustainable agriculture, and conservation biology debates. The sterile insect technique (SIT), as a component of area-wide integrated pest management (AW-IPM) approaches, is among the most effective and environment-friendly tactics for controlling tephritid pests8. The technique relies on the continuous mass-release of irradiation-sterilized males into target areas to mate with wild females, leading to the production of infertile embryos and suppression of pest populations9. A key determinant to the success of SIT in tephritids has been the linkage of selectable traits to the male sex in genetic sexing strains (GSS). Puparium color based GSS have been developed in several fruit flies using the naturally occurring mutations white pupae (wp)10,11,12 and black pupae (bp)13,14,15. In these strains, females are homozygous for recessive mutations causing atypical white or black puparium pigmentation, respectively, while males display the typical brown puparium color due to a chromosomal translocation of a wildtype rescue allele onto the male Y-chromosome, resulting in a consistent heterozygous state. This sex-linked color dimorphism allows female removal before releases, ensuring cost-effectiveness and efficacy of large-scale SIT programs16.
The white pupae (wp) phenotype in three distantly related tephritids, the Mediterranean fruit fly (medfly) Ceratitis capitata, the oriental fruit fly Bactrocera dorsalis, and the melon fly Zeugodacus cucurbitae, results from parallel mutations in a single, conserved gene encoding a Major Facilitator Superfamily (MSF) transporter protein17. This molecular identity supports early biochemical studies showing that the wp gene is essential to transport hemolymph catecholamines (pigment precursors) to the pupal cuticle18, promoting normal sclerotization and pigmentation. Despite gene conservation, similar wp mutations were never found in Anastrepha—a mega-diverse genus of fruit flies in the American (sub)tropics that includes major fruit-infesting pests, such as the Mexican fruit fly (mexfly) Anastrepha ludens and the South American fruit fly Anastrepha fraterculus. Alternatively, the black pupae (bp) mutant phenotype has been observed in Anastrepha and used to develop GSSs in this group of flies; however, its genetic basis remains largely unknown.
In this study, we present a collection of independent evidence from genetics, transcriptomics, and functional genomics, showing that ebony is the responsible gene for the mutant black pupae trait in A. ludens and likely other tephritids displaying parallel phenotypes. Our findings support the following conclusions: (1) the bp phenotype in the A. ludens GUA10 strain13 is caused by a large deletion at the ebony locus. This deletion removes the entire protein-coding region of the gene, thus disrupting its function; (2) disruption of ebony function is sufficient to recreate analogous bp phenotypes in diverse tephritids, indicating its potential as a candidate gene for other naturally occurring bp mutations within the family; (3) Ebony plays an essential role in inhibiting black melanization in adult fruit flies, which constitutes one of the genetic mechanisms underlying pigmentation differences within and between tephritid species; and (4) the ebony gene may have pleiotropic effects on both embryo viability and adult development in tephritids, ultimately impacting fitness. We discuss these discoveries through the lens of dipteran evolutionary developmental biology19 and their implications for the construction of new GSS using genome editing.
Results
Generation of a mapping population
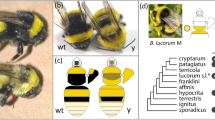
We adopted the A. ludens GUA10 strain as our model to investigate the genetic basis of the black pupae trait in tephritids (Fig. 1a). GUA10 is a GSS with an autosomal recessive bp mutation as a selectable marker, allowing sex separation based on pupal color dimorphism14. Females from GUA10 are homozygous for the bp mutation, exhibiting an atypical dark pupal case. The mutation also induces darkening of larva anal lobes and ectopic melanization of adult cuticle. Conversely, GUA10 males carry an irradiation-derived translocation between the Y-chromosome and autosome 2, which carries the bp-related gene13,14. This Y-2 translocation fragment links a functional bp allele to the male sex. Consequently, males are consistently heterozygous, displaying a wildtype brown pupal case.
a The bp phenotype is the sex-linked trait used for sex-sorting the mexfly GUA10 strain. Females from GUA10 exhibit black larval anal lobes (al) and pupal cases, developing into darker adults. Mutant adults are noticeable due to their darker wing stripes (wg), abdomen (ab), and ovipositor (ov). b To identify the loci responsible for the bp phenotype, we introgressed the GUA10 bp mutation (bp−(GUA10)) into a common wildtype genetic background. Subsequently, we established and whole-genome sequenced a F4 mapping population where siblings develop either wildtype brown (wt) or mutant black (bp) pupal cases. Illustration credit: Alexander Spengler. c Trait mapping analysis (calculated for 100 Kb windows at 20 Kb sliding intervals across the reference genome) located a large interval within the A. ludens chromosome 2 showing significant genetic differentiation (FST) between F4 black and brown pupae siblings (n = 18 from each group). The horizontal line indicates the top 0.25th percentile threshold for significant intervals. d The gene set within this causal region include the candidates yellow-f2 and ebony from the melanin biosynthesis pathway. e RNA-Seq coverage (calculated from merged BAM files of n = 3 biological replicates) revealed the silencing of ebony expression in black pupae females from GUA10. f The ebony gene is differentially expressed between homozygous black pupae female (F) and heterozygous brown pupae male (M) siblings from GUA10 (genotypes in Fig. 1a), while no difference is detectable for yellow-f2. The expression of the white pupae gene, an MFS transporter required to provide pigment precursors to the pupal cuticle, is shown as a reference. Dots represent biologically independent replicates (n = 3), * = FDR < 0.05 and log2FC > 2, and ns = non-significant. The mean ± SD are shown in a gray dot and error bars, respectively. g Semiquantitative RT-PCR assays further validated the RNA-Seq results (n = 3 biological replicates). Amplifications of Rpl18 are internal controls. L = 100 bp DNA ladder (NEB), gDNA = genomic DNA control, and NTC = non-template negative control.
We conducted a series of genetic crosses to introgress the bp mutation into a wildtype genetic background, enabling us to identify the relative genomic location of the bp causative loci in A. ludens using association mapping (Fig. 1b). We selected wildtype (wt) males to provide a reference background, and females from GUA10 as donors for the bp mutation (bp−(GUA10)). Isolated-mating between GUA10 females (bp−/−) and wt males (bp+/+) followed by inbreeding of F1 hybrids (bp+/−) led to the segregation of bp alleles at F2, placing them within a common genetic background. To further increase wildtype representation and recombination between divergent genomes, F2 bp females were individually backcrossed to wt males. Then, the resulting heterozygous F3 (bp+/−) individuals were inbred to generate the F4 mapping population, where siblings share similar genetic makeup with the exception of the bp mutation, and thus develop as black or brown pupae independently of their sexes.
Association mapping implicates pigmentation genes to the black pupae phenotype in A. ludens GUA10
To map the bp−(GUA10) mutation, we generated whole genome sequences (WGS) from black (bp−/−) and brown (bp+/+ or bp+/−) pupae siblings from the F4 mapping population (n = 18 of each phenotype). Resulting short reads were aligned to the A. ludens reference genome (GenBank: GCA_028408465.1) and used to identify DNA variants in each individual. We next calculated measures of genetic diversity along the genome using 100 kb sliding windows with a 20 kb step size. To minimize false identifications, we only considered the top 0.25% of windows from scans. Genome-wide landscape of pairwise genetic differentiation (FST) between black and brown pupae siblings revealed a large island encompassing 8.74 Mb within the A. ludens chromosome 2 exhibiting the most significant genetic difference between phenotypes (Fig. 1c). Therefore, this genomic interval was considered the candidate region of the bp−(GUA10) mutation.
To gain insight into the biological functions within this candidate region, we conducted a gene ontology (GO) enrichment analysis, comparing the gene set in this genomic interval to the complete genome gene set (Supplementary Table 1). Out of the 99 protein-coding genes located at the candidate region, seven exhibited GO terms related to cuticle melanization and sclerotization (Fig. 1d), including five genes within the neuropeptide signaling pathway (GO:0007218; Fisher’s exact test, p = 7.20e-4) and two genes involved in the melanin biosynthetic process from tyrosine (GO:0006583; Fisher’s exact test, p = 3.42e-3). Upon closer examination, we identified yellow-f2 (GenBank: XP_053946313.1) and ebony (GenBank: XP_053946246.1) as candidate genes associated with the black pupae phenotype in mexfly.
In Drosophila melanogaster, mutations in yellow result in a loss of black pigment, underscoring the essential role of the Yellow protein in black melanin production20. The yellow-f2, a member of the Yellow gene family, is implicated in melanization during later pupal and adult development21. In contrast, ebony mutants exhibit increased black pigmentation22. Previous studies have demonstrated that the pattern and intensity of melanization in Drosophila are largely determined by the coordinated expression of Yellow and Ebony23. Therefore, the melanin-promoting gene yellow-f2 and the melanin-inhibiting gene ebony emerged as promising candidates responsible for the black pupae phenotype in A. ludens.
ebony is silenced in black pupae females from A. ludens GUA10
We performed RNA-Seq transcriptome profiling on 1-day-old black (females, bp−/−) and brown (males, bp+/−) pupae siblings from the GUA10 strain currently maintained in Guatemala, under the assumption that transcriptional errors in candidate protein-coding genes result in the mutant trait. Similar to the approach used to identify the wp mutation17, we anticipated that differential read coverage between genotypes would reveal transcript variants or alterations in gene expression. RNA-Seq coverage was similar for yellow-f2; however, only reads from brown pupae males mapped to ebony (Fig. 1e). Differentially expressed gene (DEG) analysis showed significant downregulation of ebony in black pupae individuals (log2 fold-change = 7.4, p = 8.3e-06, and FDR = 0.03; Fig. 1f and Supplementary Table 2). Semiquantitative RT-PCR confirmed the silencing of ebony in black pupae females of GUA10 (Fig. 1g), leading to the hypothesis that ebony is the causative gene behind the mutant phenotype in this strain. Importantly, we located ebony within the polytene chromosomal region 24 of chromosome 2 in wildtype A. ludens using in situ hybridization (Supplementary Fig. 1). This region is part of the Y-2 translocation chromosome of both GUA1014 and its predecessor TAP-713. Therefore, the ebony gene is linked to the male sex in these strains—a requirement for the bp locus candidate.
The black pupae phenotype in A. ludens GUA10 results from a large indel variant at the ebony locus
We next screened the genomic regions surrounding ebony for DNA variants called from WGS short-reads of the F4 mapping population. Apart from single nucleotide and short insertion-deletion polymorphisms (SNPs and indels, respectively) in adjacent sequences, we could not identify variants within ebony (Fig. 2a), indicating that either this locus is identical between brown and black pupae siblings of the mapping population or there is a lack of read coverage in one or both subpopulation phenotypes. To better understand this pattern, we examined the short-read mapping coverage and identified a broad chromosomal interval with no coverage from black pupae individuals (Fig. 2b, Supplementary Fig. 2). This observation suggested that most ebony loci are missing in these flies. To confirm this putative deletion event, we sequenced a single male and female of A. ludens GUA10 strain with PacBio HiFi sequencing. We reasoned the effort could provide evidence of the bp−(GUA10) mutation in single long HiFi reads associated with ebony. Both male and female samples generated approximately 69 Gb of data, representing over 80× coverage of the estimated 820 Mb genome. Mapping of HiFi reads to the reference genome of A. ludens validated our assumption and revealed a complex DNA variant spanning a 20,182 bp region within the ebony locus (Fig. 2c, Supplementary Fig. 2). The variant contains two large deletions of 8,186 bp and 4,310 bp, resulting in the excision of the entire protein-coding sequence and a significant portion of the 5’ upstream region of the gene, respectively. Qualitative end-point PCR encompassing a 360 bp region between ebony exons e1 and e2 confirmed the absence of the gene in bp females from GUA10 (Fig. 2d). Altogether, these data show that the loss of the ebony coding sequence is responsible for the black pupae phenotype in A. ludens GUA10.
a Biallelic DNA variants between brown and black pupae individuals (n = 18 of each phenotype) from the F4 mapping population in the context of the A. ludens genome (NC_071498.1:135,589,966-135,862,362) and GUA10 RNA-Seq coverage (n = 3 males and females). No DNA variants are present within the ebony gene (g15354), coinciding with the absence of RNA-Seq reads from GUA10 females. b WGS read coverage (merged BAM files) from the F4 mapping population shows a large chromosomal interval with no coverage in black pupae individuals, suggesting the entire ebony gene is missing in those flies. c Mapping of HiFi long-reads revealed a 20,182 bp DNA variant (NC_071498.1:g.135,665,293_135,685,474indel; bp−(GUA10)) within the ebony locus of A. ludens GUA10, resulting in the removal of the entire protein-coding region of the gene. The variant is homozygous in black pupae female and heterozygous in brown pupae male, as expected by the genotypes of GUA10 (Fig. 1a). d Qualitative end-point PCR of a 360 bp sequence between ebony exons e1 and e2 confirmed the absence of this region in the GUA10 female genome (arrowheads in Fig. 2c indicate primer locations). Amplifications of Rpl18 are internal controls. L = 100 bp DNA ladder (NEB), M = male, F = female, GUA10 = A. ludens GUA10 genetic sexing strain, WT = wildtype, and NTC = non-template control.
Disruption of ebony creates analogous black pupae phenotypes in diverse tephritid flies
We used CRISPR/Cas9-mediated gene knockout (KO) to functionally validate our findings. Out of 260 A. ludens embryos injected with Cas9-sgRNA ribonucleoprotein (RNP) complexes targeting exon 1 of ebony, 38 (14.6%) survived into the pupal stage (Supplementary Table S3). Among them, 4 (10.5%) displayed the bp phenotype and developed into darker (melanic) adults (Fig. 3a), resembling the original mutant phenotype13 (see Fig. 1a). These mosaic knockout (mKO) flies contained indels in 56-89% of PCR-generated amplicons surround the sgRNA target site (Supplementary Fig. 3), confirming that the disruption of ebony is sufficient to induce the black pupae phenotype in A. ludens. Using a similar approach, we further recreated the mutant phenotype in A. fraterculus (Fig. 3b, Supplementary Fig. 4), demonstrating the functional conservation of ebony in other Anastrepha species.
a We used CRISPR/Cas9 to generate targeted loss-of-function mutations in the ebony gene, aiming to confirm its involvement in the bp phenotype of A. ludens. Surviving flies developed as either wildtype light-brown or mutant black puparia. Adults emerging from the latter also exhibited darker cuticles, mirroring the bp phenotype observed in GUA10. To assess the functional conservation of ebony in other tephritids, we extended our CRISPR experiments to include a close relative, A. fraterculus (b), as well as distantly related species, including C. capitata (c), B. tryoni (d), B. dorsalis (e), and Z. cucurbitae (f). Similar effects were observed in all species, demonstrating that the disruption of ebony is sufficient to induce the bp phenotype in diverse tephritids. wt = wildtype, e = ebony gene, mKO = mosaic knockout, KO = knockout, and Ma = million years. The evolutionary relationships among these species were estimated by Zhang et al.86 using mitochondrial phylogenomics.
Ebony is highly conserved among tephritids (Supplementary Fig. 5); thus, we anticipated that disrupting ebony orthologs would result in the bp phenotype even in distantly related species within the family (Fig. 3c–f). To test this assumption, we conducted additional CRISPR/Cas9 KO experiments on C. capitata, B. dorsalis, and Z. cucurbitae. Across all species, we observed high survival and mutagenesis rates (15-34% and 58-68%, respectively; Supplementary Table 3), and all experiments resulted in G0 individuals exhibiting phenotypes that ranged from mosaic black-brown puparium to complete melanized pupal cases (Fig. 3c–e, f). As expected by the nature of bp mutations in these flies24,25,26, the eclosed adults also displayed an overall darker cuticle. Genotyping of PCR products spanning Cas9 cut sites confirmed the presence of indels at the ebony loci, with modification frequencies ranging from 87-92% (Supplementary Fig. 6). Taken together, these results offered functional evidence supporting ebony as the responsible gene for the bp phenotype in mexfly GUA10 and likely other tephritids displaying parallel phenotypes.
Ebony regulates pigmentation patterning in tephritid flies
Although mutations resulting in the bp phenotype are well-documented in tephritids, their impact on adult morphology has been overlooked. To address this, we established biallelic KO lines to investigate the contribution of ebony to adult pigment patterns. We focused on the conspicuous pictured wings of C. capitata and Z. cucurbitae, and the body pigmentation of B. dorsalis and Z. cucurbitae. These traits are prevalent among tephritids and thus may serve as ideal models for studying the evolution of pigmentation in this group of flies.
The wings of C. capitata exhibit intricate patterns featuring pigmented spots and bands varying in color and shape. Among these, the dominant discal crossband and the costal band are particularly intriguing (Fig. 4a), as their pigmentation is absent in model wings of Drosophila (examples in True et al.27). In ebony mutants, the appearance of those spots changed from brownish-yellow to dark brown; but their pattern remained the same (Fig. 4b). Interestingly, knocking out ebony had no effect on the silver lamina of medfly wings, while mutants of Zeugodacus displayed ectopic pigmentation in the structure (Fig. 4c, d). This ectopic pigmentation closely resembles that observed in D. melanogaster23, with no apparent changes in the apical spot, but a subtle enlargement of the fuscous black dm-cu crossvein blotch. A combination of modifications is observed in the wings of A. fraterculus ebony mutants (Supplementary Fig. 4). Similar to the medfly, the appearance of brownish-yellow bands changes to dark brown, and as in the melon fly, fuscous black edges intensify.
a The wings of C. capitata are one of the most captivating examples of pictured pigmentation in tephritids. Among its intricate patterns, the dominant discal crossband (dc) and the costal band (cb) are particularly intriguing due to their uncommon yellowish-brown color. b In ebony mutants, the appearance of these bands changes to dark-brown, yet their pattern remains the same, with no observable effects on the lamina. c In contrast, the wings of Z. cucurbitae appear nearly clear, except for a few distinct pigmentation patterns, such as the dark apical spot (as) and the dm-cu crossvein blotch. d While a subtle enlargement of the dm-cu blotch appears in ebony mutants of Z. cucurbitae, the most striking phenotype is the ectopic pigmentation of the wing lamina, resulting in a smoky brown appearance. On an interesting note, the appearance of the anal streak (ans) and costal band (cb) in Zeugodacus wings also seems to change from dark-brown to black. Terminology in accordance to White and Elson-Harris7.
The B. dorsalis Punador strain maintained at the USDA-ARS-PBARC facility in Hawaii, exhibits a scutum predominantly black with peripheral reddish-brown areas (Fig. 5a). This pattern closely resembles the predominant morphotype found in the coastal areas of China28, which is the most likely source population that invaded the Hawaiian Islands29. In contrast, its abdomen is predominantly reddish-brown, displaying a distinct black T-shaped marking and narrow fuscous corners on tergites T4 and T5 (Fig. 5b, c). Cas9-mediated loss of Ebony altered the pigment pattern in both body parts, allowing black melanin to spread into areas where it was previously absent or restricted. (Fig. 5d–f). These effects were even more dramatic in ebony mutants of the melon fly. The overall appearance of the thorax changed from orange-brown (Fig. 5g) to black (Fig. 5j), and the narrow abdominal bands (Fig. 5h, i) were significantly enlarged (Fig. 5k, l). These results illustrate the important role of Ebony in controlling the expressivity of black pigments in tephritids by inhibiting melanization, thus ensuring proper pigmentation patterning.
a Wildtype B. dorsalis (Punador strain) displays a predominantly black thorax with a particular reddish-brown area at the posterior edge of the scutum (arrowhead). b, c In contrast, its abdomen is predominantly reddish-brown with a distinct black T-shaped marking accompanied by narrow fuscous corners on tergites T4 and T5 (arrowheads). In ebony mutants, black pigmentation takes over the light areas of the thorax (d) while enlarging abdominal markings (e, f). Wildtype Z. cucurbitae displays a uniform light-brown color in the thorax (g) and abdomen (h, i), except for a few narrow black stripes in the latter. This overall appearance becomes completely distorted in ebony mutants. New pigmentation patterns appear as their thorax turns almost uniformly black (j), and their abdominal stripes become significantly enlarged (k, l). In both species, the absence of Ebony results in new pigmentation patterns. Terminology in accordance to White and Elson-Harris7.
Development of an ebony-null strain for the Queensland fruit fly
The Queensland fruit fly Bactrocera tryoni is a significant agricultural pest in Australia30, where the SIT is applied to manage established populations and eliminate outbreaks in key horticultural regions. Presently, SIT targeting B. tryoni involves releasing both males and females, as no efficient method to remove females before sterilization exists. Therefore, creating a bp strain would represent a significant advancement in managing this important pest. To this end, we expanded our CRISPR/Cas9 experiments to KO ebony in B. tryoni. Out of 661 injected embryos, 12 survived to adulthood, all developing from brown pupal cases. We next backcrossed five G0 adults to the Ourimbah wt strain. All G1 flies were combined and allowed to interbreed. In total, 24 individuals with black puparium were recovered among the G2 offspring, confirming successful germline transmission of Cas9-induced mutations. Fourteen individuals survived to adulthood, all displaying dark cuticles (Fig. 3d). Molecular genotyping of G2 identified four ebony mutant alleles containing indels resulting in frameshift mutations (Supplementary Fig. 7). Initial attempts to develop a stable strain through inter-crossing G2 mutants failed due to non-viable eggs. However, we eventually established an ebony-null mutant strain (named Bt-ebony) after three rounds of backcrossing G2 homozygous mutants to the wt laboratory strain (see crossing scheme in Supplementary Fig. 8). Genotyping of Bt-ebony revealed a homozygous -2 bp deletion at ebony exon 1, resulting in a loss-of-function mutation as evidenced by the complete penetrance of the black pupae phenotype (Supplementary Fig. 7).
Ebony has possible pleiotropic effects on the development of the Queensland fruit fly
We next asked if ebony-null alleles would result in fitness costs over the development of B. tryoni, an important consideration for potential SIT applications. We first conducted phenotype and genotype segregation analyses to test the effects on viability (Supplementary Table 4). Bt-ebony males (e-/-) were mass backcrossed to wildtype females (e+/+), resulting in hybrid offspring (e+/-) with brown pupae phenotypes. Inbreeding of F1 flies produced F2 progeny with wildtype (n = 200) or ebony (n = 60) phenotypes at an approximate 3:1 ratio (X2 = 0.51, p = 0.47, d.f = 1). Genotyping a subset of phenotypically wildtype F2 flies further confirmed the expected 1:2 segregation ratio of wildtype (n = 25) to heterozygous (n = 55) genotypes (X2 = 0.16, p = 0.69, d.f = 1), with no sex ratio bias. These results confirm the complete recessive inheritance of ebony and suggest that the generated mutant alleles do not impose viability costs on B. tryoni.
We further evaluated fecundity and development over three consecutive generations using four genetic crossing combinations: (1) wildtype control crosses; (2) test crossings with Bt-ebony males vs. wildtype females; (3) reciprocal crossings with wildtype males vs. Bt-ebony females; and (4) Bt-ebony crosses. All data were collected from F1 generations (Supplementary Data 3). For fecundity, we collected and counted eggs daily for five days. There were no significant differences in fecundity across all combinations. However, outcrossings (i.e., Bt-ebony vs. wildtype) and ebony crosses generally produced fewer eggs than wildtype control crosses (Fig. 6a). To assess hatchability, we counted eclosed larvae three days after egg collection. The percentage of hatched eggs from ebony crosses (median ± SE; 36.4 ± 5.2%) was significantly lower than in wildtype crosses (80.2 ± 5.5%; p = 0.04, one-way ANOVA Tukey HSD), while outcrossings produced intermediate values (Fig. 6b). Pupation, adult emergence, and partial adult emergence did not drastically differ between crossing combinations (Fig. 6c–e). Yet, ebony parents showed a tendency to produce offspring with more partially emerged adults (Fig. 6d, e). Additionally, the percentage of deformed adults was significantly higher among ebony progeny (3.86 ± 0.71%) compared to wildtype (0.58 ± 0.26%; p = 0.02, one-way ANOVA Tukey HSD; Fig. 6f).
We performed genetic crossings to evaluate whether ebony-null alleles would impose fitness costs over the development of B. tryoni expressed in terms of (a) relative fecundity (no. of eggs / no. of eggs in control experiments), (b) hatchability (no. of larvae / no. of eggs), (c) pupation (no. of pupae / no. of larvae), (d) adult eclosion (no. of fully emerged adults / no. of pupae), (e) partial adult eclosion (no. of partially emerged adults / no. of pupae), and (f) adult deformity (no. of deformed adults / no. pupae). Dots represent data collected from independent replicates (n = 3), colors represent the pupae phenotype of their F1 offspring, and letters indicate significant differences (one-way ANOVA followed by Tukey’s HSD). The mean ± SD are shown in a gray dot and error bars, respectively. ctr = control crossings between wt flies, test = test crossings between Bt-ebony males and wt females, rec = reciprocal crossings between wt males and Bt-ebony females, and e = crossings between Bt-ebony flies.
Discussion
In this study, we combined classical and modern genetics to uncover the molecular basis of the black pupae phenotype (bp) in tephritid fruit flies. We adopted the Mexican fruit fly Anastrepha ludens as our model—since stable bp mutants for the species exist as part of the GUA10 genetic sexing strain14. Trait mapping, gene ontology, and differential gene expression analyses suggest that loss of ebony leads to the bp phenotype in GUA10 females. Since bp is a recessive trait, a functional allele linked to the Y-chromosome rescues the wildtype phenotype in males. Consistent with this model, cytogenetics identified the bp locus in the polytene chromosomal region 24 of A. ludens chromosome 213,14, which is part of the short translocation fragment in GUA10. Using in situ hybridization, we located ebony in this same region. Mapping of Illumina short- and HiFi long-reads revealed a 20,182 bp indel variant at the ebony locus in black pupae individuals from both the F4 mapping population and females from GUA10, confirming the loss of the entire protein-coding region the gene in these flies. This finding aligns with the evolutionary biology principle that mutations causing broad phenotypic effects—such as the bp phenotype in the A. ludens GUA10 strain, which spans all developmental stages and affects all melanic areas in adults (Fig. 1a)—are more likely to occur in the protein-coding regions of developmental genes rather than in their cis-regulatory elements, since loss-of-function mutations would affect all tissues where the protein is expressed31. Finally, functional validation of ebony orthologues via CRISPR/Cas9-mediated KO generates analogous bp phenotypes in A. ludens and diverse tephritid species spanning over 50 Ma of divergent evolution. Collectively, we provide strong evidence that ebony is the responsible gene for the bp phenotype in GUA10 and possibly other tephritids in which parallel phenotypes have been described. In addition to these findings, the creation of ebony-null mutants across distantly related species allowed for fundamental insights into the contribution of Ebony to color patterning and development in tephritids, which have significant importance for dipteran evolutionary developmental biology19, and thus discussed below. We hope that these thoughts will spark renewed interest in the evolution of pigmentation patterns and phenotypic diversity within the family Tephritidae.
Ebony is crucial for insect pigmentation and sclerotization (hardening). It conjugates dopamine (DA) with β-alanine to produce N-β-alanyldopamine (NBAD), the precursor of yellow-tan sclerotin in insect cuticles32,33,34. Null mutants for ebony cannot produce NBAD, leading to an excess of DA, which in turn is diverted into the melanization branch of the pigmentation biosynthesis pathway (as reviewed by Massey and Wittkopp35), resulting in individuals with darker cuticles32,36,37. Consequently, a failure to encode Ebony leads to an increase in melanin (dark pigments) and a decrease in light pigmentation production23. In the classical example of D. melanogaster, ebony loss-of-function causes darkening of larval mouthparts and posterior spiracles, and adults much darker in appearance22. Our KO experiments yielded similar phenotypes, illustrating the functional conservation of Ebony across higher Diptera. Nonetheless, it’s worth mentioning a phenotypic difference between ebony mutants in D. melanogaster and the tephritids we studied. In the former, the loss of Ebony results in an unpigmented or pale pupal case37, while in the latter, it leads to an unusual black puparium. Naturally, the black pupae trait is not unique to tephritids, and similar phenotypes exist in ebony mutants of moths38, mosquitoes39,40, and—to some extent—other Drosophila species41.
The molecular mechanism driving this phenotypic divergence remains unknown (but see Sherald37), though it may involve a preferential utilization of DA during cuticle formation, as discussed by Spana et al.42. In insects, it’s accepted that N-acetyldopamine (NADA) serves as the primary precursor for colorless sclerotin, produced through the conjugation of DA by the speck gene. In contrast, brown cuticles predominantly originate from NBAD33. Thus, the NADA-to-NBAD ratio is expected to be a determinant factor for the final cuticle color intensity. Using the medfly as an example, NBAD is the primary precursor of sclerotization, giving rise to an opaque reddish-brown pupal case. In the absence of NBAD, as seen in ebony mutants, NADA is utilized instead36,43. However, the branch of the pigmentation pathway leading to NADA-sclerotin may not be as dominant as the one leading to NBAD-sclerotin. In ebony mutants, this difference could allow a substantial amount of excess DA to be redirected into the melanin pathway, resulting in the production of black pupal cases in C. capitata. In D. melanogaster, the branch leading to NADA-sclerotin is likely dominant during pupation, as evidenced by their clear, light-brown pupal cases. Therefore, this could prevent free DA from entering the melanization route in ebony mutants. Supporting this idea, the differential expression of speck may be solely responsible for the contrast between the black pupae of Drosophila virilis and the brown pupae of Drosophila americana44. Interestingly, it has been suggested that while DA can be directly converted into black melanin (mediated by Yellow), the conversion of DA into NBAD and then back into DA (mediated by Tan) seems necessary for the production of dark brown melanin23,35. The requirement for this indirect route could potentially explain why tephritid ebony mutants develop black pupae rather than darker brown ones. Finally, it should be noted that similar melanization phenotypes in C. capitata can also arise from the inability to produce β-alanine43, a condition that can be induced by mutations in the black locus45.
Most tephritid flies are adorned with brightly contrasting body colors, and their wings often display elaborate patterns once described as both “lovely and mysterious”46. Wing pigmentation in tephritids can take various forms: from almost completely clear (Bactrocera), across relatively simple spots and blotches (Zeugodacus), and winding stripes shaded in black and brown (Anastrepha) to complex colored pictured wings with many patterns (Ceratitis). After a brief inspection, one could argue that such diversity is perhaps as enigmatic as in Drosophila and yet much less studied.
Here, we showed that Ebony is essential to generate and maintain this diversity. In Zeugodacus, Ebony inhibits melanization of the wing lamina, but it does not directly contribute to the pigmentation of the apical spot or crossvein blotches. Therefore, it is possible that Ebony is uniformly distributed in the species’ wings but less expressed in melanized patterns, and other genes (probably including yellow) are responsible for promoting their black pigmentation. This regulatory model is found in drosophilids exhibiting wing spots, where spatial downregulation of Ebony and upregulation of Yellow are—at least in part—responsible for their melanized patterns23,47,48. By masking melanization, Ebony also contributes to the expressivity of specific phenotypes, including the black crossvein blotch in Z. cucurbitae and the fuscous black edges of the S- and V-bands in the Anastrepha wings. Interestingly, ectopic pigmentation of the wing lamina, resembling that in D. melanogaster ebony mutants23, is also observed in ebony mutants of Zeugodacus and Bactrocera but not Ceratitis and Anastrepha. The mechanism behind this effect is unclear, but it seems to follow a phylogenetic pattern and therefore could be related to a species-specific distribution of pigment precursors through their wing veins.
The role of Ebony in the pigmentation of C. capitata wings was investigated by Pérez et al.49. Using biochemical assays, the authors showed that Ebony is spatially pre-patterned in the yellowish-brown bands of the medfly wings during early adult development, maintaining an extracellular activity long after epithelial cells disappear from the lamina. As observed in Drosophila27, pigment precursors present in hemolymph circulation gradually diffuse out from wing veins and are conjugated into NBAD by the pre-patterned Ebony to promote the coloration of these bands. Ebony activity is detectable in the yellowish-brown spots of C. capitata wing but not in the silver-paint lamina49, suggesting that the enzyme is only responsible for the pigmentation of the former. Our results provide functional evidence to their findings, showing that Ebony is necessary to produce the normal coloration of C. capitata wing bands and, in its absence, their appearance changes from yellowish-brown to dark brown—but their pattern remains the same, and no effects appear in the lamina. Notably, Anastrepha ebony mutants exhibit similar phenotypic changes in their wings, including the transformation of their once brownish-yellow S- and V-bands into a deep brown shade. The appearance of a dark-brown color in the wings of these ebony mutants can be explained by DA being shunted to the melanization branch of the pigmentation pathway and used to produce brown melanin through a mechanism open to debate. Some authors propose that the production of black and brown melanins occurs depending on the activity of different members of the Yellow gene family50. Others suggest that DA can be converted into brown melanin through a process involving phenol oxidases (POs; responsible for the oxidation of pigment precursors in the cuticle) but not the yellow gene23,35, and thus the indirect route must be taken (as discussed before). Our results seem to confront the latter idea, suggesting that brown pigmentation can be achieved through different molecular mechanisms in pupae and adults. These observations illustrate that the regulation of wing coloration in tephritid flies can be as mysterious as their pattern, and thus deserves more attention.
Natural populations of B. dorsalis exhibit extreme variation in adult pigment patterns28,51. This intraspecific diversity is particularly noticeable in their thorax (scutum), which ranges from pale reddish-brown to black, often with lanceolate-patterned intermediates. Similarly, their abdomen can vary from predominantly pale to predominantly black due to the expressivity of the typical ‘T’ pattern and the dark markings on tergites 4 and 5. The difference between morphological variants (morphs) is so dramatic that invasive populations of B. dorsalis in Africa were classified as a new species (Bactrocera invadens) and later synonymized52. Although the ecological relevance—if any—of these melanin patterns remains unknown, the geographic distribution of morphs seems to form irregular pigmentation clines28. The ectopic melanization in ebony mutants fairly overlap with some of these natural patterns, suggesting that this diversity in B. dorsalis may, in part, result from variations in the spatial expression of Ebony. The genetic background for ectopic expression also seems to contribute to the depth of these new patterns. For example, ebony mutants of the B. dorsalis Punador exhibit enlarged but distinct abdominal markings, which are nearly unrecognizable in predominantly black natural morphs51. In contrast, both thorax and abdominal segments of ebony mutants of the B. tryoni Ourimbah turn entirely black.
Within its genus, Z. cucurbitae is one-of-a-kind regarding adult morphology. Adults display a pale reddish-brown scutum and narrow abdominal bands, showing little intraspecific variability in natural populations53. These traits become completely distorted in ebony mutants; the scutum turns almost entirely black, and abdominal markings extend to a point where it resembles other related species (e.g., Zeugodacus tau). These shifts further illustrate how changes in Ebony expression may also confer phenotypic divergence between fruit fly species.
How do these phenotypic variations arise and how are they maintained? Based on our observations, and other more comprehensive studies23, we speculate these phenotypes are largely determined by the coordinated expression of Ebony and other pigmentation genes (such as yellow and tan), and changes in their expression patterns result in pigmentation diversity. Expression differences within and between species often arise from changes in either cis- or trans-acting elements important to these genes. In Drosophila, changes in cis-regulatory regions of ebony are responsible for the naturally occurring variation in abdominal coloration54,55 and, at some degree, to the intensity of the thoracic “trident” phenotype35. Similarly, differences in expression levels of ebony and tan contribute to pigmentation divergence between the dark-bodied Drosophila americana and its light-colored sister species Drosophila novamexicana, which seems to be controlled in part by noncoding changes in these loci56. While divergence of pigment patterns within and among species might arise due to ecological and sexual selection pressures3, it remains uncertain whether it serves as raw material for speciation in species complexes with varying pigmentation, such as B. dorsalis.
The initial difficulties in establishing an ebony mutant line for B. tryoni could have been caused by off-target mutagenesis or genetic linkage. To mitigate and potentially eliminate these effects, we backcrossed Ebony mutants to wildtype flies for three consecutive generations before evaluating their relative fitness in terms of fecundity and development. Despite the effort, we found that Bt-ebony mutants exhibited a significant reduction in egg-hatching rates across three generations. Presumably, one disruption causes the other, suggesting that loss-of-function mutations in ebony impact traits other than pigmentation in the species. In insects, pleiotropy (a single gene affecting multiple traits) is often associated with pigmentation genes, and effects on behavior, physiology, and life-history traits have been extensively documented (reviewed by True1, Wittkopp and Beldade2, and Takahashi57). Although pleiotropic effects are difficult to prove, our results indicate that disruption of Ebony might compromise embryo viability in B. tryoni. Lower egg-hatching rates have also been reported in ebony mutants of the diamondback moth58 and silkworm59. In the latter, larvae seem to develop normally—but have trouble breaking out from their eggshell. It’s unclear whether ebony mutants of B. tryoni have impairments regarding larvae development or eclosion capabilities. Nevertheless, similar effects were observed in early adulthood, where Bt-ebony mutants appear to have difficulty emerging from their pupa, and a significant number of flies emerge with wing deformities. The absence of NBAD in ebony mutants is known to cause decreased cuticle stiffness due to inefficient sclerotization (see Andersen60), which could potentially contribute to abnormal wing development in B. tryoni mutants at eclosion. Interestingly, heterozygous individuals displayed intermediate egg-hatching and deformity rates, indicating semi-dominant phenotypes contrasting the apparent complete dominance of Ebony in pigmentation.
We also cannot discard the possibility of a higher frequency of copulation failure by ebony mutants, thus leading to an increased number of unfertilized eggs. Mutations in ebony have been correlated to abnormal mating behavior in Drosophila, likely due to defects in circadian rhythm and visual system (two well-known phenotypes of ebony mutants) and perhaps courtship behavior (for which ambiguous phenotypes exist)57. Furthermore, their relative abundance of cuticular hydrocarbons—that can function as conspecific short-range pheromones61—is also influenced by Ebony41,62. Additional studies, including more replicates and monitoring of mating trials, are needed to elucidate the influence of Ebony on the mating behavior of B. tryoni.
Despite these non-specific phenotypes, the Bt-ebony strain has been maintained for over 30 generations in small colonies under typical laboratory conditions, demonstrating that these mutants are stable and suitable for future experimentation. However, our results suggest that in a potential GSS with ebony as a marker, homozygous recessive females would experience higher mortality than heterozygous males—although, at this point, the impacts on (semi-)mass rearing conditions are uncertain. Further experiments are needed to investigate whether additional backcrossings to wildtype could alleviate these deleterious effects and if the linkage of ebony to the male Y-chromosome could rescue them. In the same context, it’s reasonable to think that genome modifications at regulatory sequences could lead to the development of lines with fewer fitness costs—as regulatory mutations might minimize pleiotropic effects relative to coding mutations1,31. Therefore, there is a need to investigate the regulatory networks governing the spatiotemporal expression of pigmentation genes in tephritids. Technologies for profiling the transcriptome and epigenome at single-cell resolution (single-cell ATAC-Seq + RNA-Seq) could facilitate the identification of regulatory factors associated with their spatiotemporal expression. In theory, such investigation could help us manipulate the temporal expression patterns of these genes, thereby bypassing the downsides associated with their disruption. Fitness costs are also likely to arise from random radiation-induced mutagenesis. Potentially, this could be overcome by CRISPR/Cas9-mediated knock-in or chromosomal translocation of the rescue allele into the Y-chromosome—as previously proposed17,63—for which standardized protocols remain to be established.
Methods
Flies
Anastrepha ludens flies (wildtype) used for microinjections were obtained from the USDA-APHIS Moore Air Base Facility in Edinburg, TX, USA. Flies are reared at 23 °C and 60% RH under a 14/10 h light/dark cycle. Adults are fed with a standard dry diet (1 yeast: 3 sugar) and water. Larvae are reared on a meridic diet consisting of 15.6% pelletized corncob, 8.4% granulated white sugar, 2.8% torula yeast, 7% wheat germ, 4.6% toasted soy flour, 2.8% corn flour, 0.2% vitamin mix, 1.9% citric acid, 0.1% sodium benzoate, 0.2% methylparaben, and 0.2% bravo WS (1%), all dissolved in water. Samples of the A. ludens GUA10 strain and F4 mapping populations, used for WGS, RNA-Seq, and HiFi sequencing, were obtained from the USDA-APHIS MOSCAMED San Miguel Petapa Fruit Fly Rearing and Quarantine facility in San Miguel Petapa, Guatemala.
Anastrepha fraterculus sp. 1 flies (Vacaria strain) were obtained from the Insect Pest Control Laboratory in Seibersdorf, Austria. Flies are maintained at 25 °C and 48% RH under a 12/12 h light/dark cycle. Adults are fed with a standard dry diet (1 yeast: 3 sugar) and water. Larvae are reared on a carrot diet consisting of 8.06% brewer’s yeast, 0.76% sodium benzoate, 0.8% (v/w) HCl, 23.9% carrot powder, and 67.17% fresh carrots, all dissolved in water.
Ceratitis capitata (rearing strain), Bactrocera dorsalis (Punador strain), and Zeugodacus cucurbitae (rearing strain) were obtained from the USDA-ARS-PBARC in Hilo, HI, USA. All species are maintained at 25 °C and 60% RH under a 12/12 h light/dark cycle. Adults are fed with a standard dry diet (1 yeast: 3 sugar) and water. Larvae are reared on a species-specific diet. The C. capitata larval diet consists of 59% wheat mill feed, 27.3% granulated white sugar, 7.6% torula yeast, 5.2% citric acid, and 0.5% each of the preservatives nipagen and sodium benzoate. The B. dorsalis larval diet consists of 63% wheat mill feed, 28.5% granulated white sugar, 8% torula yeast, 0.3% nipagen, and 0.2% sodium benzoate. The Z. cucurbitae larval diet consists of 73.7% wheat mill feed, 17.4% granulated white sugar, 8.4% torula yeast, and 0.3% each of the preservatives nipagen and sodium benzoate. All diets are dried and mixed with water before being provided to larvae.
Bactrocera tryoni flies (Ourimbah strain) were obtained from the New South Wales Department of Primary Industries in Ourimbah, Australia. Flies are reared at 25 °C and 65% RH under a 14/10 h light/dark cycle. Adults are fed with a standard dry diet (1 yeast: 3 sugar) and water. Larvae are reared on a gel diet containing 20.4% brewer’s yeast, 12.1% sugar, 0.2% methyl p-hydroxy benzoate, 2.3% citric acid, 0.2% wheat germ oil, 0.2% sodium benzoate, and 1% agar.
Mapping population
The crossing scheme shown in Fig. 1b was used to introgress the mexfly black pupae mutation into a wildtype reference genetic background and generate a mapping population for genome-wide genetic differentiation analysis. Briefly, mexfly black pupae females were isolated from the GUA10 strain14 during pupal stage and individually outcrossed to wildtype males to produce hybrid offspring with non-sex-linked variation in pupal color. The resulting F1 population displayed the wildtype brown pupae phenotype and was allowed to intercross freely to recover the black pupae phenotype in the next generation. Resulting F2 black pupae females were individually backcrossed to wildtype males. Heterozygous flies at F3 were let to inbreed, and black and brown pupae siblings were selected at F4. Individuals were separated by pupae color, snap frozen in liquid nitrogen upon adult emergence, fixed in absolute ethanol and kept at -80 °C until further analysis.
Association mapping
Total DNA was extracted from brown (n = 18) and black (n = 18) pupae individuals of the F4 mapping population using the NucleoMag Tissue kit (Macherey Nagel) on a Kingfisher Flex system. Extractions were quantified by Qubit dsDNA BR assay (Invitrogen), normalized to 50 ng on a 96-well plate, and subjected to library preparation using the RipTide High Throughput Rapid DNA Library Preparation kit (iGenomX). Libraries were pooled and sequenced on a single lane of HiSeq 4000 (Illumina) in a 150 bp paired-end run. Raw data was demultiplexed and processed in-line to remove barcodes using the fgbio toolkit (https://github.com/fulcrumgenomics/fgbio).
Demultiplexed whole-genome sequencing (WGS) reads were then filtered using fastp v0.23.264 and mapped against the mexfly reference genome using BWA-MEM v.2.2.165. SAMtools v1.1766 fixmate was used to update mate information and duplicated pairs were marked using SAMBLASTER v0.1.2667. Alignments were filtered using SAMtools view with the following parameters: skip alignments with mapping quality ≤ 30, only keep reads mapped in a proper pair (-f 0×0002), and discard unmapped reads (-F 0×0004), reads with unmapped mates (-F 0×0008), and reads marked as duplicates (-F 0×0400). Genome Analysis Toolkit v4.4 (GATK, https://gatk.broadinstitute.org/) RealignerTargetCreator and IndelRealigner were used to locally realign reads around insertions and deletions (InDels) in filtered alignments. Variant calling was performed using GATK HaplotypeCaller, CombineGVCFs, and GenotypeGVCFs with default parameters. Variants were hard filtered using GATK SelectVariants and VariantFiltration following the recommended parameters for SNPs and InDels to pass. Remaining variants were further filtered using VCFtools v0.1.16-968 with the following parameters: maf 0.05, mac 2, min-alleles and max-alleles 2, min-meanDP 0.2 and max-meanDP 1.8 (based on 5th and 95th percentiles of mean depth per site), hwe 1e-5, max-missing 0.5, and minQ 30. Genome-wide genetic differentiation index (FST) between black and brown pupae siblings from the mapping population were calculated in 100 Kb windows at 20 Kb sliding intervals using VCFtools and visualized with the qqman package in R v4.3.2. The final, filtered VCF (Variant Call Format) file was examined in the Integrative Genomics Viewer (IGV) tool v2.16.2-069.
Genome annotation
RepeatModeler v2.0.470 was used to identify repetitive sequences and construct a de novo repeat library for the A. ludens reference genome. This library was combined with Drosophila repetitive sequences extracted from RepBase (RepeatMasker Edition v.20181026, https://www.girinst.org/server/RepBase/index.php) and used with RepeatMasker v4.1.5 (https://www.repeatmasker.org/) to mask repetitive sequences in the genome. Gene prediction was carried out using RNA-Seq alignment data as extrinsic evidence for gene models. Publicly available RNA-Seq raw reads, encompassing most of A. ludens development stages (Supplementary Table 5), were filtered using fastp v0.23.264 and mapped to the masked genome with STAR v2.7.10b71 in two-pass spliced alignment mode. The resulting alignments were used as input for BRAKER2 pipeline72. The longest isoform of each predicted gene model was retrieved using AGAT v0.8.0 (https://github.com/NBISweden/AGAT) function agat_sp_keep_longest_isoform.pl resulting in an initial set of 18,230 protein coding genes. BUSCO v5.4.5 analysis73 found 96.9% of the 3,285 single-copy orthologues (in the diptera_odb10 database) to be complete (96.3% single-copied and 0.6% duplicated genes), 0.8% fragmented, and 2.3% missing. Functional annotations were performed by mapping this gene set against the UniProtKB database (https://www.uniprot.org/) using BLASTp (e-value ≤ 1e-6), and the Pfam, PANTHER, Gene3D, SUPERFAMILY, SMART, and CDD databases using InterProScan v5.64-96.074. Orthology assignments were performed with the DIAMOND (e-value ≤ 1e-5) mapping mode implemented in eggNOG-mapper v2.1.12-075 in the context of Diptera taxonomic scope. The final gene set for A. ludens genome contained 13,114 full-length protein-coding genes with annotated gene ontology (GO) terms.
Enrichment analysis
Gene ontology (GO) enrichment analysis of biological processes terms was performed using the Bioconductor package topGO v2.54.0-0 (https://bioconductor.org/packages/topGO) on the gene set within the black pupae causal region on mexfly chromosome 2, using all genes with annotated GO terms in the genome as the background set. The elim method was used to reduce redundancy and searches were limited to categories containing at least 10 annotated genes. Overrepresentation significance of GO terms was calculated using the classical Fisher test adopting a cutoff threshold of p-value ≤ 0.05.
RNA isolation
Specimens were euthanized at -20 °C, fixed in RNAlater (Invitrogen) and stored at 4 °C until further processing. Before extractions, samples were mixed with one volume of 1x PBS (pH 7.4) and centrifuged at top speed for 10 min at 4 °C, allowing removal of solution excess by pipetting. Pre-processed samples were homogenized in TRISure (Bioline Meridian Bioscience), and total RNA was isolated using Direct-zol-96 MagBead RNA kit (Zymo Research). Extractions were DNase treated in a 50 µL reaction containing 2 U of TURBO DNase (Invitrogen), 50 U of RiboGuard RNase Inhibitor (Lucigen), 1x TURBO reaction buffer and 5-10 µg of total RNA. Reactions were performed at 37 °C for 30 min, stopped by the addition of 15 mM of EDTA (pH 8.0) followed by an incubation at 75 °C for 10 min, and purification using the RNA Clean & Concentrator-5 kit (Zymo Research).
RNA-Seq
RNA sequencing (RNA-Seq) libraries were generated from 250 ng of DNase-treated RNA with the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB) following the Poly(A) mRNA Magnetic Isolation Module protocol. Sequencing was performed on a AVITI platform (Element Biosciences) in a 150 bp paired-end run. Raw reads were filtered using fastp v0.23.264 and mapped against the A. ludens reference genome with STAR v2.7.10b71 using the two-pass spliced alignment mode along with gene models generated by BRAKER2. Mapped reads were quantified by featureCounts76 implemented in subread v2.0.4 (https://subread.sourceforge.net/) and resulting count matrix used as input for differential expression analysis with the Bioconductor package edgeR v4.0.1277. Briefly, counts were first filtered by the filterByExpr function and normalized using the TMM-method (Trimmed mean of M-values). Differences between brown (n = 3) and black (n = 3) pupae samples were estimated with the quasi-likelihood (QL) F-test against the threshold of log2 fold-change (logFC) > 2 using the glmTreat function. Genes were considerate differentially expressed when false discovery rate (FDR) < 0.05. Read counts were converted to RPKM (reads per kilobase of transcript per million reads mapped) with the rpkm function and used as a descriptive measure of gene expression for visualization.
HiFi long-read mapping
High molecular weight (HMW) DNA was extracted from single adult flies using the MagAttract HMW DNA Kit (QIAGEN). Samples were quantified using the Qubit dsDNA BR assay (Invitrogen) and size checked in the Femto Pulse System (Agilent Technologies). Extractions were further subjected to a 2x bead cleanup, and their purity determined on the basis of OD 260/230 and 260/280 ratios estimated in a DS-11 spectrophotometer (DeNovix). Clean HMW DNA samples were sheared to a mean size of 20 kb using the Megaruptor 2 (Diagenode), and size checked on the Fragment analyzer (Agilent Technologies) with the HS Large Fragment kit. SMRTBell libraries were prepared using approximately 1 µg of sheared DNA with the SMRTBell Prep Kit 3.0 (Pacific Biosciences). The prepared libraries were bound and sequenced on a 24 M SMRT Cell on a Revio system (Pacific Biosciences) using a 24 h movie collection time. Circular consensus sequences were obtained using SMRTLink v13.0 on the Revio instrument. Highly accurate long-reads (HiFi reads) were filtered for adapter contamination with HiFiAdapterFilt v.3.0.178 and mapped to the A. ludens reference genome with minimap2 v.2.2479 (-ax map-hifi). Mapping files were examined in the IGV tool v2.16.2-069.
Manual gene annotation
Gene models for ebony orthologues (Supplementary Data 1) were manually curated by mapping the D. melanogaster Ebony peptide sequence (FlyBase: FBgn0000527) against each reference genome using tBLASTn (BLOSUM45 matrix, e-value ≤ 1e-5). Highly similar genomic regions were retrieved with BEDTools v2.31.1-080 and intron-exon boundaries modelled using Exonerate v2.4.0-781 protein2genome mode (--refine full). Intron-exon boundaries were manually annotated, coding sequences were translated using the ExPASy translate tool (https://web.expasy.org/translate/), and signature motifs identified using DELTA-BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) against the NCBI’s non-redundant protein database.
Semiquantitative RT-PCR and qualitative end-point PCR
For RT-PCRs, first-strand cDNAs were synthesized from 1 µg of DNase-treated RNA using the MMLV Reverse Transcriptase Synthesis Kit (Lucigen) protocol with the Oligo(dT)21 primer. Reverse transcriptions were performed at 37 °C for 1 h, terminated at 85 °C for 5 min and stored at -20 °C. Reverse Transcription PCRs (RT-PCR) were carried out for a final volume of 12.5 µL containing 1x OneTaq HS Quick-Load Master Mix with Standard Buffer (NEB), 0.2 µM of each forward and reverse Intron-flanking primers (Supplementary Table 6), and an equivalent amount of cDNA synthetized from 50 ng of total RNA. Amplifications were carried out in the following conditions: 94 °C for 3 min, 40 cycles of [94 °C for 30 s, 55 °C for 30 s and 68 °C for 1 min], and a final extension at 68 °C for 5 min. All experiments included independent biological (n = 3) and technical (n = 3) replicates, no template controls (cDNA omitted from the reaction and volume adjusted with nuclease-free water), and a genomic DNA control (total DNA extraction from a single A. ludens wildtype individual). Amplifications were resolved in 1% agarose gels stained with Midori Green Advance DNA Stain (at 1:15000 dilution factor; Nippon Genetics) in 0.5x TBE buffer. Images were acquired in a Gel Doc XR+ Gel Documentation System (Bio-Rad) using predefined setups in the software Image Lab v.6.0.1. Contrast and light corrections were made in the same software. For end-point PCRs, genomic DNA was extracted from whole adult flies using the NucleoMag Tissue Kit (Macherey-Nagel), and a total of 50 ng was used as a template for PCR amplifications encompassing ebony exons e1 and e2 with OneTaq HS Quick-Load Master Mix with Standard Buffer (NEB) as described before.
Cytogenetics
Third-instar larvae salivary glands of wildtype A. ludens were dissected in 45% acetic acid and fixed in glacial acetic acid: water: lactic acid (3:2:1) solution for about 5 min. Preparations were stored overnight at -20 °C and dipped into liquid nitrogen in the next day. Slides were dehydrated in absolute ethanol, air dried, and stored at 4 °C. DNA probes for fluorescent in situ hybridization (FISH) of ebony were prepared by PCR for a final volume of 25 µL containing 1x Platinum II Hot-Start Green PCR Master Mix (Invitrogen), 0.2 µM of each forward and reverse primer (Supplementary Table 6), and 100 ng of total DNA. Amplifications were carried out as follows: 94 °C for 5 min, 35 cycles of [94 °C for 45 s, 56 °C for 30 s and 72 °C for 1.5 min], and a final extension at 72 °C for 1 min. Probe labeling was performed according to the DIG DNA Labelling Kit (Roche) protocol, and slides were prepared for fluorescence detection as previously described17. Hybridizations in isolated chromosomes were photographed in a Leica DM2000 LED microscope using a Leica DMC5400 digital camera and analyzed with the LAS X software v3.7.0 in the context of the mexfly salivary gland chromosome maps82.
CRISPR/Cas9
Knockout experiments in A. ludens, C. capitata, B. dorsalis, and Z. cucurbitae were performed according to a proposed standard protocol for Cas9-mediated gene disruption in non-model tephritids63. Briefly, single guide RNAs (sgRNAs) were designed against the ebony gene using CRISPOR v.5.0183 and their reference genomes (Supplementary Table 6). Templates for sgRNAs were generated by PCR with Phusion High-Fidelity DNA Polymerase (NEB), purified using the QIAquick PCR purification kit (QIAGEN), and used for T7 in-vitro transcription with MEGAshortscript Kit (Invitrogen). Purified Cas9 protein conjugated with a nuclear localization signal (NLS) was obtained commercially (PNA Bio). Microinjection mixes contained end-concentrations of 360 ng/µl Cas9, 200 ng/µl sgRNA, 1x injection buffer (0.1 mM Sodium phosphate buffer, 5 mM KCl), and 300 mM KCl in a final volume of 10 μL in nuclease-free water. Ribonucleoprotein (RNP) complexes were pre-assembled at 37 °C for 15 min and delivered through the chorion into the posterior end of preblastoderm embryos within the first 60 min after egg-laying (AEL). Mosaic black-brown puparia at G0 were inbred to establish biallelic mutant lines.
Microinjections in A. fraterculus followed the same protocol with minor modifications. The sgRNA was designed using CHOPCHOP v.384 in combination with Geneious v.2023.0.2 (https://www.geneious.com/) in the context of A. fraterculus draft genome. Both sgRNA (Merck) and purified Cas9 protein (PNA Bio) were obtained commercially. RNPs were pre-assembled at 37 °C for 15 min, followed by 5 min at room temperature. Embryos were collected every 30 min, dechorionated with a 30% hypochlorite solution for 80 s, and microinjected within 120 min AEL. Injected G0 flies were individually backcrossed to wildtype flies. The resulting G1 offspring were inbred, and all G2 displaying the bp phenotype were intercrossed. Adult individuals at G3 were genotyped using non-lethal methods, and flies harboring the -7 bp deletion allele were inbred to establish a homozygous mutant strain at G4.
For knockout experiments in B. tryoni, purified Cas9 protein (IDT Alt-R S.p. Cas9 Nuclease 3NLS) and guide RNAs (IDT customized Alt-R crRNAs and universal Alt-R tracrRNA) were obtained commercially. Two customized crRNAs were designed using CRISPOR v.5.0183 in the context of the species reference genome and a multiple sequence alignment of target regions to avoid polymorphisms in the injection population. Two dual-guide RNA (dgRNA) duplexes were annealed separately by mixing 40 μM of each specific crRNA with 40 μM of universal tracrRNA in Nuclease-Free Duplex Buffer and heating at 95 °C for 5 min before cooling to room temperature. The final injection mix contained 300 ng/µL Cas9 protein, 10 μM of each dgRNA, and 1x injection buffer (0.1 mM sodium phosphate buffer pH 6.8, 5 mM KCI) in a final volume of 10 µL in Nuclease-Free Duplex Buffer. The injection mix was incubated at room temperature for 5 minutes to allow RNP formation and delivered into the posterior end of embryos within 60 min AEL. Surviving G0 adult flies were individually backcrossed to wildtype flies, and their G1 progeny were combined and allowed to interbreed. Biallelic ebony mutants were recovered at G2 and used to stablish the Bt-ebony homozygous mutant strain (Supplementary Fig. 7, 8).
Genotyping
Mosaic knockouts (mKO) of A. ludens, C. capitata, B. dorsalis, and Z. cucurbitae were genotyped by deep-sequencing of indexed amplicons surrounding the Cas9 cut sites, as previously described63. Briefly, total DNA was extracted from whole adult flies using the NucleoMag Tissue Kit (Macherey-Nagel) and used as a template for a two-step PCR with Phusion High-Fidelity DNA polymerase (NEB) and primers in Supplementary Table 6. Indexed amplicons were purified with the QIAquick PCR purification kit (QIAGEN), pooled in equimolar ratios, and sequenced on Illumina iSeq 100 system (150 bp paired-end reads). Targeted genome modifications were inspected using CRISPResso2 v2.2.14-085. Biallelic KO mutants examined in phenotypic analysis were genotyped in the same way.
Total DNA from whole G2 adults of A. fraterculus was extracted using the ExtractMe Genomic DNA kit (QIAGEN) and used as a template for PCR amplifications with Platinum Green Hot Start PCR Master Mix (Invitrogen) and primers in Supplementary Table 6. Reactions were cycled as follows: 94 °C for 2 min, 34 cycles of [94° C for 15 s, 59 °C for 15 s, and 68 °C for 15 s], and a final extension at 68 °C for 5 min. Amplification products were purified using the ZR-96 DNA Clean-up kit (Zymo Research) and Sanger sequenced on an ABI 3730XL DNA Analyzer system (Applied Biosystems). Site-specific modifications were inspected in a multiple sequence alignment produced by Geneious v.2023.0.2. Non-lethal genotyping of G3 flies was carried out using single adult legs with Platinum Direct PCR Universal Master Mix (Invitrogen).
Amplification of DNA surrounding the Cas9 cut sites in B. tryoni was performed directly from single adult legs using the Phire Animal Tissue Direct PCR Kit (Thermo Scientific) and primers in Supplementary Table 6. Amplification conditions were as follows: 98 °C for 5 min, 35 cycles of [98 °C for 5 s, 60 °C for 5 s, and 72 °C for 20 s], and a final extension at 72 °C for 1 min. For the identification of heterozygous G2 brown pupal flies, amplifications were submitted to cleavage assays using the T7 Endonuclease 1 (T7E1) enzyme protocol (NEB). PCR products of confirmed G2 heterozygous (n = 48) and final ebony-null mutants (n = 30) were Sanger sequenced on an ABI 3730XL DNA Analyzer system (Applied Biosystems). Targeted genome modifications were inspected in a multiple sequence alignment produced by Geneious v.2023.0.2.
Performance assays
Segregation analysis was performed to measure the viability of B. tryoni individuals carrying ebony-null alleles. To account for environmental variations, data were collected after the 1st and 2nd rounds of mass backcrossing between ebony males and wildtype females during the establishment of the Bt-ebony strain (Supplementary Fig. 8). All F1 individuals were interbred, and phenotypic segregation at F2 was tested against the 3:1 Mendelian inheritance ratio of phenotypes. A subset of phenotypically wildtype F2 flies were further genotyped by T7E1 assays, and the segregation of wildtype to heterozygous was tested against a 1:2 inheritance ratio of genotypes. Pearson’s chi-square goodness-of-fit tests were used to determine significant deviation from expected ratios as implemented in the stats package in R v4.3.2.
Fitness analysis was performed to measure the relative fecundity and development of B. tryoni ebony mutants. Virgin females and naive males from the Bt-ebony and wildtype Ourimbah strains were sorted within three days after adult emergence and kept separated until experimentation. The following crosses were performed using 8-to-11-days-old flies (10 males and 10 females): (1) wildtype males vs. wildtype females; (2) ebony males vs. wildtype females; (3) wildtype males vs. ebony females; and (4) ebony males vs. ebony females. Assays were performed over three consecutive generations to account for environmental variations (Supplementary Fig. 8). The first batch of eggs was collected 48 h after experiment setups. Eggs were collected for 24 h, and the number of eggs laid was counted daily for 5 days to assess fecundity ratios. Collected eggs were placed on larval diet and resulting larvae were counted 3 days after incubation to estimate embryonic hatching rates. The number of larvae that reached the pupal stage was used to estimate pupariation rates. Emerging adults were recorded daily until no new eclosions were observed and categorized as fully emerged, partially emerged (remaining in the puparium), and deformed (emerged flies with twisted or deformed wings). Significant differences between groups at p ≤ 0.05 were determined by one-way ANOVA followed by a Tukey’s Honest Significant Difference (HSD) tests using the stats package in R v.4.3.2.
Statistics and reproducibility
The sample size for association mapping analysis was defined based on the number of F4 offspring displaying the mutant and wildtype phenotype and normalized to the first. Similarly, the sample size for fitness analysis was defined based on the number of offspring obtained from each crossing scheme. The significant threshold for FST was set to the 0.25th percentile to mitigate false-positive windows due to linkage disequilibrium associated with limited recombination. RNA-Seq was performed with three biological replicates, and DEGs were considered at logFC > 2 and FDR < 0.05. Semiquantitative and qualitative analyses were performed with three biological and technical replicates, and amplifications of targets and controls were resolved in the same gel, while in situ hybridizations were analyzed using replicates prepared in the same microscope slide. Mean differences between samples or groups were considered statistically significant at p < 0.05 (after corrections). All relevant information for the reproducibility of this study is reported in the figure legends and detailed in the corresponding methods section.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The following reference genome assemblies were used in this study: A. ludens (GenBank: GCA_028408465.1), C. capitata (GenBank: GCA_000347755.4), B. dorsalis (GenBank: GCA_023373825.1), B. tryoni (GenBank: GCA_016617805.2), and Z. cucurbitae (GenBank: GCA_028554725.2). All raw sequencing data generated in this study are deposited in the NCBI Sequence Read Archive (SRA) database under the BioProject PRJNA1139181. SRA accession numbers for each sample are detailed in Supplementary Table 5. Final de novo genome annotation files for A. ludens are archived on figshare repository at https://doi.org/10.6084/m9.figshare.26376841. All primers used in this study are listed in Supplementary Table 6. Manually curated annotations of ebony orthologues can be found in Supplementary Data 1. TMM-normalized RPKM expression data can be found in Supplementary Data 2. The source data for fitness analysis of B. tryoni ebony strain is available as Supplementary Data 3. Full-length gels are shown in Supplementary Fig. 9.
References
True, J. R. Insect melanism: the molecules matter. Trends Ecol. Evol. 18, 640–647 (2003).
Wittkopp, P. J. & Beldade, P. Development and evolution of insect pigmentation: genetic mechanisms and the potential consequences of pleiotropy. Semin. cell Dev Biol. 20, 65–71 (2009).
Wittkopp, P. J., Carroll, S. B. & Kopp, A. Evolution in black and white: genetic control of pigment patterns in Drosophila. TRENDS Genet. 19, 495–504 (2003).
Duyck, P., Jourdan, H. & Mille, C. Sequential invasions by fruit flies (Diptera: Tephritidae) in Pacific and Indian Ocean islands: A systematic review. Ecol. Evol. 12, e8880 (2022).
Malacrida, A. R. et al. Globalization and fruitfly invasion and expansion: the medfly paradigm. Genetica 131, 1–9 (2007).
Clarke, A. R. Why so many polyphagous fruit flies (Diptera: Tephritidae)? A further contribution to the ‘generalism’debate. Biol. J. Linn. Soc. 120, 245–257 (2017).
White, I. M. & Elson-Harris, M. M. Fruit Flies of Economic Significance: Their Identification and Bionomics. (CAB international, 1992).
Suckling, D. M. et al. Eradication of tephritid fruit fly pest populations: outcomes and prospects. Pest Manag. Sci. 72, 456–465 (2016).
Knipling, E. F. Possibilities of insect control or eradication through the use of sexually sterile males. J. Econ. Entomol. 48, 459–462 (1955).
Robinson, A. S. & Van Heemert, C. Ceratitis capitata—a suitable case for genetic sexing. Genetica 58, 229–237 (1982).
McInnis, D. O. et al. Development of a pupal color-based genetic sexing strain of the melon fly, Bactrocera cucurbitae (Coquillett)(Diptera: Tephritidae). Ann. Entomol. Soc. Am. 97, 1026–1033 (2004).
McCombs, S. D. & Saul, S. H. Translocation-Based Genetic Sexing System for the Oriental Fruit Fly (Diptera: Tephritidae) Based on Pupal Color Dimorphism. Ann. Entomol. Soc. Am. 88, 695–698 (1995).
Zepeda-Cisneros, C. S. et al. Development, genetic and cytogenetic analyses of genetic sexing strains of the Mexican fruit fly, Anastrepha ludens Loew (Diptera: Tephritidae). BMC Genom. Data 15, 1–11 (2014).
Ramírez-Santos, E. et al. A novel genetic sexing strain of Anastrepha ludens for cost-effective sterile insect technique applications: improved genetic stability and rearing efficiency. Insects 12, 499 (2021).
Meza, J. S., Bourtzis, K., Zacharopoulou, A., Gariou-Papalexiou, A. & Cáceres, C. Development and characterization of a pupal-colour based genetic sexing strain of Anastrepha fraterculus sp. 1 (Diptera: Tephritidae). BMC Genet. 21, 1–9 (2020).
Rendón, P., Lance, D. & Stewart, J. Medfly (Diptera: Tephritidae) genetic sexing: large-scale field comparison of males-only and bisexual sterile fly releases in Guatemala. J. Econ. Entomol. 97, 1547–1553 (2004).
Ward, C. M. et al. White pupae phenotype of tephritids is caused by parallel mutations of a MFS transporter. Nat. Commun. 12, 491 (2021).
Wappner, P. et al. White pupa: a Ceratitis capitata mutant lacking catecholamines for tanning the puparium. Insect Biochem. Mol. Biol. 25, 365–373 (1995).
Dion, W. A., Steenwinkel, T. E. & Werner, T. From Aedes to Zeugodacus: A review of dipteran body coloration studies regarding evolutionary developmental biology, pest control, and species discovery. Curr. Opin. Genet. Dev. 69, 35–41 (2021).
Morgan, T. H. & Bridges, C. B. Sex-Linked Inheritance in Drosophila. (Carnegie institution of Washington, 1916).
Han, Q. et al. Identification of Drosophila melanogaster yellow-f and yellow-f2 proteins as dopachrome-conversion enzymes. Biochem. J. 368, 333–340 (2002).
Bridges, C. B. & Morgan, T. H. The Third-Chromosome Group of Mutant Characters of Drosophila Melanogaster. (Carnegie Institution of Washington, 1923).
Wittkopp, P. J., True, J. R. & Carroll, S. B. Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns. (2002).
Rössler, Y. & Koltin, Y. The Genetics of the Mediterranean Fruitfly, Cerotitis capitata: Three Morphological Mutations. Ann. Entomol. Soc. Am. 69, 604–608 (1976).
McCombs, S. D. & Saul, S. H. Linkage analysis of three new alleles affecting puparium morphology in the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 85, 799–804 (1992).
McCombs, S. D., McInnis, D. O. & Saul, S. H. Genetic studies of the melon fly, Bactrocera cucurbitae. in Fruit Fly Pests 237–241 (CRC Press, 1996).
True, J. R., Edwards, K. A., Yamamoto, D. & Carroll, S. B. Drosophila wing melanin patterns form by vein-dependent elaboration of enzymatic prepatterns. Curr. Biol. 9, 1382–1391 (1999).
Schutze, M. K. et al. One and the same: integrative taxonomic evidence that B actrocera invadens (D iptera: T ephritidae) is the same species as the O riental fruit fly B actrocera dorsalis. Syst. Entomol. 40, 472–486 (2015).
Zhang, Y. et al. Genomes of the cosmopolitan fruit pest Bactrocera dorsalis (Diptera: Tephritidae) reveal its global invasion history and thermal adaptation. J. Adv. Res. 53, 61–74 (2023).
Sultana, S., Baumgartner, J. B., Dominiak, B. C., Royer, J. E. & Beaumont, L. J. Potential impacts of climate change on habitat suitability for the Queensland fruit fly. Sci. Rep. 7, 13025 (2017).
Stern, D. L. Perspective: Evolutionary Developmental Biology and the Problem of Variation. Evolution 54, 1079–1091 (2000).
Wright, T. R. The genetics of biogenic amine metabolism, sclerotization, and melanization in Drosophila melanogaster. Adv. Genet. 24, 127–222 (1987).
Hopkins, T. L. & Kramer, K. J. Insect cuticle sclerotization. Annu. Rev. Entomol. 37, 273–302 (1992).
Walter, M. F. et al. Catecholamine metabolism and in vitro induction of premature cuticle melanization in wild type and pigmentation mutants of Drosophila melanogaster. Archives of Insect Biochemistry and Physiology: Published in Collaboration with the Entomological Society of America 31, 219–233 (1996).
Massey, J. H. & Wittkopp, P. J. The genetic basis of pigmentation differences within and between Drosophila species. Curr. Top. Dev Biol. 119, 27–61 (2016).
Wappner, P., Kramer, K. J., Manso, F., Hopkins, T. L. & Quesada-Allue, L. A. N-β-alanyldopamine metabolism for puparial tanning in wild-type and mutant Niger strains of the Mediterranean fruit fly, Ceratitis capitata. Insect Biochem. Mol. Biol. 26, 585–592 (1996).
Sherald, A. F. Sclerotization and coloration of the insect cuticle. Experientia 36, 143–146 (1980).
Futahashi, R. et al. yellow and ebony are the responsible genes for the larval color mutants of the silkworm Bombyx mori. Genetics 180, 1995–2005 (2008).
Li, M. et al. Germline Cas9 expression yields highly efficient genome engineering in a major worldwide disease vector, Aedes aegypti. Proc. Natl Acad. Sci. 114, E10540–E10549 (2017).
Feng, X. et al. Evaluation of gene knockouts by CRISPR as potential targets for the genetic engineering of the mosquito Culex quinquefasciatus. CRISPR J. 4, 595–608 (2021).
Lamb, A. M., Wang, Z., Simmer, P., Chung, H. & Wittkopp, P. J. ebony affects pigmentation divergence and cuticular hydrocarbons in Drosophila americana and D. novamexicana. Front. Ecol. Evol. 8, 184 (2020).
Spana, E. P. et al. speck, first identified in Drosophila melanogaster in 1910, is encoded by the arylalkalamine N-acetyltransferase (AANAT1) gene. G3: Genes, Genomes. Genetics 10, 3387–3398 (2020).
Wappner, P. et al. Role of catecholamines and β-alanine in puparial color of wild-type and melanic mutants of the Mediterranean fruit fly (Ceratitis capitata). J. insect Physiol. 42, 455–461 (1996).
Ahmed-Braimah, Y. H. & Sweigart, A. L. A Single Gene Causes an Interspecific Difference in Pigmentation in Drosophila. Genetics 200, 331–342 (2015).
Phillips, A. M., Smart, R., Strauss, R., Brembs, B. & Kelly, L. E. The Drosophila black enigma: the molecular and behavioural characterization of the black1 mutant allele. Gene 351, 131–142 (2005).
Sivinski, J. & Pereira, R. DO WING MARKINGS IN FRUIT FLIES (DIPTERA: TEPHRITIDAE) HAVE SEXUAL SIGNIFICANCE? flen 88, 321–324 (2005).
Gompel, N., Prud’homme, B., Wittkopp, P. J., Kassner, V. A. & Carroll, S. B. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433, 481–487 (2005).
Arnoult, L. et al. Emergence and Diversification of Fly Pigmentation Through Evolution of a Gene Regulatory Module. Science 339, 1423–1426 (2013).
Pérez, M. M., Bochicchio, P. A., Rabossi, A. & Quesada-Allué, L. A. Extracellular activity of NBAD-synthase is responsible for colouration of brown spots in Ceratitis capitata wings. J. Insect Physiol. 107, 224–232 (2018).
Futahashi, R. & Osanai-Futahashi, M. Pigments in Insects. in Pigments, Pigment Cells and Pigment Patterns (eds. Hashimoto, H., Goda, M., Futahashi, R., Kelsh, R. & Akiyama, T.) 3–43 (Springer, Singapore, 2021). https://doi.org/10.1007/978-981-16-1490-3_1.
Leblanc, L., Hossain, M. A., Khan, S. A., San Jose, M. & Rubinoff, D. A Preliminary Survey of the Fruit Flies (Diptera: Tephritidae: Dacinae) of Bangladesh. (2013).
Schutze, M. K. et al. Synonymization of key pest species within the Bactrocera dorsalis species complex (Diptera: Tephritidae): taxonomic changes based on a review of 20 years of integrative morphological, molecular, cytogenetic, behavioural and chemoecological data. Syst. Entomol. 40, 456–471 (2015).
De Meyer, M. et al. A review of the current knowledge on Zeugodacus cucurbitae (Coquillett) (Diptera, Tephritidae) in Africa, with a list of species included in Zeugodacus. Zookeys 539–557 https://doi.org/10.3897/zookeys.540.9672 (2015)
Rebeiz, M., Pool, J. E., Kassner, V. A., Aquadro, C. F. & Carroll, S. B. Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 326, 1663–1667 (2009).
Pool, J. E. & Aquadro, C. F. The genetic basis of adaptive pigmentation variation in Drosophila melanogaster. Mol. Ecol. 16, 2844–2851 (2007).
Cooley, A. M., Shefner, L., McLaughlin, W. N., Stewart, E. E. & Wittkopp, P. J. The ontogeny of color: developmental origins of divergent pigmentation in Drosophila americana and D. novamexicana. Evol. Dev. 14, 317–325 (2012).
Takahashi, A. Pigmentation and behavior: potential association through pleiotropic genes in Drosophila. Genes Genet. Syst. 88, 165–174 (2013).
Xu, X., Harvey-Samuel, T., Yang, J., You, M. & Alphey, L. CRISPR/Cas9-based functional characterization of the pigmentation gene ebony in Plutella xylostella. Insect Mol. Biol. 30, 615–623 (2021).
Sun, J., Zheng, X., Ouyang, G., Qian, H. & Chen, A. Ebony plays an important role in egg hatching and 30k protein expression of silkworm (Bombyx mori). Arch. Insect Biochem. Physiol. 113, e22014 (2023).
Andersen, S. O. Cuticular sclerotization and tanning. in Insect molecular biology and biochemistry 167–192 (Elsevier, 2012).
Billeter, J.-C. & Levine, J. The role of cVA and the Odorant binding protein Lush in social and sexual behavior in Drosophila melanogaster. Front. Ecol. Evol. 3 (2015).
Massey, J. H. et al. Pleiotropic Effects of ebony and tan on Pigmentation and Cuticular Hydrocarbon Composition in Drosophila melanogaster. Front. Physiol. 10 (2019).
Paulo, D. F. et al. A Unified Protocol for CRISPR/Cas9-Mediated Gene Knockout in Tephritid Fruit Flies Led to the Recreation of White Eye and White Puparium Phenotypes in the Melon Fly. J. Econ. Entomol. 115, 2110–2115 (2022).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Vasimuddin, M., Misra, S., Li, H. & Aluru, S. Efficient architecture-aware acceleration of BWA-MEM for multicore systems. in 2019 IEEE international parallel and distributed processing symposium (IPDPS) 314–324 (IEEE, 2019).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Faust, G. G. & Hall, I. M. SAMBLASTER: fast duplicate marking and structural variant read extraction. Bioinformatics 30, 2503–2505 (2014).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinforma. 14, 178–192 (2013).
Flynn, J. M. et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl Acad. Sci. 117, 9451–9457 (2020).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Brůna, T., Hoff, K. J., Lomsadze, A., Stanke, M. & Borodovsky, M. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom. Bioinforma. 3, lqaa108 (2021).
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A. & Zdobnov, E. M. BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38, 4647–4654 (2021).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P. & Huerta-Cepas, J. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829 (2021).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. bioinformatics 26, 139–140 (2010).
Sim, S. B., Corpuz, R. L., Simmonds, T. J. & Geib, S. M. HiFiAdapterFilt, a memory efficient read processing pipeline, prevents occurrence of adapter sequence in PacBio HiFi reads and their negative impacts on genome assembly. BMC Genom. 23, 1–7 (2022).
Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Slater, G. S. C. & Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinforma. 6, 1–11 (2005).
Garcia-Martinez, V. et al. Mitotic and polytene chromosome analysis in the Mexican fruit fly, Anastrepha ludens (Loew) (Diptera: Tephritidae). Genome 52, 20–30 (2009).
Concordet, J.-P. & Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018).
Labun, K. et al. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic acids Res. 47, W171–W174 (2019).
Clement, K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019).
Zhang, Y. et al. Mitochondrial phylogenomics reveals the evolutionary and biogeographical history of fruit flies (Diptera: Tephritidae). Entomologia Generalis (2022).
Acknowledgements
The authors would like to thank N. Barr, E. Braswell, T. Todd, H. Conway, K. Pina, and A. Arellano for their support with microinjections and rearing of A. ludens at the USDA-APHIS Moore Air Base Facility (Texas, USA). We thank D. Rubinoff and C. Doorenweerd for their insights into the adult color patterns of B. dorsalis. We would also like to acknowledge C. Sylva for her assistance in fly rearing at the USDA-ARS-PBARC (Hawaii, USA), M. Okamoto for confirming Bt-ebony genotypes, and A. Spengler for the scientific illustrations displayed in Fig. 1b (created in Adobe Illustrator using photos in Fig. 1a as references). This research was supported by the in-house appropriated USDA-ARS project Advancing Molecular Pest Management, Diagnostics, and Eradication of Fruit Flies and Invasive Species (no. 2040-22430-028-000-D), the USDA Plant Protection Act Section 7721 project Develop Techniques to Rapidly Generate Non-Transgenic Female Lethal Genetic Sexing Strains in Tephritid Pests (no. 6.0179), the HSF 18-6 Hermon Slade Foundation grant, and the Insect Pest Control Subprogramme of the Joint FAO/IAEA Centre of Nuclear Techniques in Food and Agriculture. This study also benefitted from discussions at meetings for the Coordinated Research Project Generic Approach for the Development of Genetic Sexing Strains for SIT Applications (no. D44003), funded by the International Atomic Energy Agency (IAEA), and used resources provided by the SCINet project of the USDA-ARS projects no. 0201-88888-003-000D and 0201-88888-002-000D. USDA is an equal opportunity provider and employer. Mention of trade names does not imply an endorsement from USDA or the Federal Government.
Author information
Authors and Affiliations
Contributions
The initial evidence linking ebony to the black pupae phenotype in the A. ludens GUA10 strain was independently conceived by DFP and SMG at the USDA-ARS-PBARC (Hawaii, USA) and by CMW, TNMN and SWB at the University of Melbourne (Melbourne, AUS). This work combines and expands the data of these two preliminary studies. DFP, TNMN, CMW, AC, PC, KB, SWB, SBS and SMG conceptualized this study. SBS and SMG designed and supervised the establishment of the A. ludens F4 mapping population in Guatemala. PR and REYR established the mexfly mapping population, collected and pre-processed samples for WGS and RNA-Seq. RLC prepared samples and libraries for WGS, RNA-Seq, and HiFi sequencing with the assistance of ANK. DFP performed the bioinformatics analyses with insights from SBS and SMG. GG performed the in situ hybridization of ebony in A. ludens chromosomes. DFP performed microinjections in A. ludens with support from ANK. CRISPR experiments in A. fraterculus were made by AASC under the supervision of KB and CS, and strains were maintained by AASC. DFP performed CRISPR experiments in C. capitata, B. dorsalis, and Z. cucurbitae. DFP established and maintained KO strains with the assistance of ANK. TNMN performed CRISPR experiments in B. tryoni. The Bt-ebony mutant strain was established and maintained by AO, AC, EF and TNMN. TNMN, AO, AC, and EF performed segregation and fitness analysis of Bt-ebony mutants. PC, KB, CS, SBS, SWB, and SMG supervised, administered, and/or secured funding for this work. The same authors assisted with data interpretation and manuscript revision. DFP wrote the manuscript with substantial insights from the initial draft written by TNMN. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Virginie Courtier and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Mengtan Xing.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Paulo, D.F., Nguyen, T.N.M., Ward, C.M. et al. Functional genomics implicates ebony in the black pupae phenotype of tephritid fruit flies. Commun Biol 8, 60 (2025). https://doi.org/10.1038/s42003-025-07489-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-07489-y