Abstract
The reprogrammed osteogenic phenotype of vascular smooth muscle cells (VSMCs) is considered a critical mechanism of vascular calcification (VC) in chronic kidney disease (CKD). Currently, the RNA N6-methyladenosine (m6A) modification is deciphered to be dynamically and reversibly participated in functional regulation of VSMCs. Here, we discover that serum m6A levels in RNA are dramatically reduced as VC progressed in patients with CKD, and this m6A demethylation is mainly due to the downregulation of methyltransferaselike-3 (METTL3). Functionally, METTL3 depletion exacerbates, whereas its overexpression attenuates calcification progression and osteogenic reprogramming. Mechanistically, Runx2, a crucial osteogenic gene, is identified as a key downstream target of METTL3-mediated m6A methylation. METTL3 negatively regulates Runx2 expression through the m6A modification. Overexpression of METTL3 exacerbates Runx2 mRNA degradation, which is orchestrated by the m6A reader YT521-B homology domain family 2 (YTHDF2) through specifically recognizing its m6A sites in the 3′UTR region. Finally, in vivo METTLs inhibitor SAH treatment aggravates VC and osteogenic conversion in aortas of CKD rats, accompanied by Runx2 expression upregulation. These above data reveal an underlying mechanism by which the m6A writer METTL3 regulates Runx2 expression through YTHDF2-mediated mRNA degradation and suggest a potential therapeutic strategy to reverse the osteogenic reprogramming of VSMCs.
Similar content being viewed by others
Introduction
Vascular smooth muscle cells (VSMCs), a vital cellular component of arteries, play an essential role in the maintenance of vascular integrity and function due to their contractile properties1. Extensive studies have shown that diverse pathological stimuli or environmental cues (such as hyperphosphatemia) promote cthe alcification process, driving VSMCs to display prominent phenotypic plasticity and undergo osteogenic reprogramming2. Different from previous perspectives on the spontaneous deposition of high concentration calcium phosphate, this phenomenon in VSMCs is now widely recognized as an active, highly organized process3. This osteogenic trans-differentiation is typified by the loss of specific contractile markers such as α-smooth muscle actin (α-SMA) and smooth muscle 22 actin (SM22α)4,5, as well as the increased expression of osteogenic markers, including osteopontin (OPN) and collagen type I α1 (Col1α1)6,7. The resulting cells develop a mineralization matrix, thus, the osteogenic reprogramming of VSMCs is the essential event for vascular calcification (VC) involved in multiple pathological conditions, such as chronic kidney disease (CKD)8, aging, diabetes mellitus, and hypertension9. Moreover, VC increases the risk of cardiovascular disease morbidity and mortality in advanced CKD patients, but anti-VC interventions based on causal therapy are not effective due to a lack of a comprehensive understanding of the molecular mechanisms10. Therefore, it is pivotal to uncover the mechanisms by which VSMCs undergo osteogenic reprogramming, which remain unascertained.
Runt-related transcription factor 2 (Runx2) is an indispensable transcription factor for osteogenesis and chondrogenesis, the expression of which in cardiovascular cells commands the intracellular signaling transduction cascades of osteochondrogenic reprogramming and vascular calcification11,12. By responding the activation of osteoblast differentiation marker genes, Runx2 facilitates calcification and matrix protein production and dominates the cell plasticity in several biological processes and diseases, including bone regeneration13, osteoarthritis14, and arteriosclerotic disease15. Previously, Runx2 had been reported to be markedly increased in calcified animal models and human aortic tissues and induced phenotype switch of VSMCs in patients with atherosclerotic plaques16. Moreover, a recent study proposed modulating Runx2 expression as a prospective therapeutic target to treat cardiovascular diseases via supplement of rate-limiting enzyme for tryptophan metabolism17. Exactly, in order to develop biological strategies which can ameliorate VC by targeting Runx2, it is necessary to perform multifaceted studies on the regulation of Runx2 during osteogenic reprogramming.
Dynamic RNA modifications that allow the fine-tuning of protein-coding genes and cellular processes have been discovered as a notable regulatory mechanism for gene expression18. N6-methyladenosine (m6A) methylation is the most abundant and prevalent internal chemical modifications of mammalian mRNA and was identified to control almost all aspects of RNA metabolism through involvement in precursor mRNA processing, RNA stability, splicing, export, and translation19,20. Emerging evidence suggests that m6A methylation modification is involved in the development of CKD by modulating the abnormal expression of genes involved in epithelial-mesenchymal transition, fibrosis and inflammation21. Accumulating studies have revealed m6A methylation as a new research hotspot, and its regulatory processes in cardiovascular diseases have received wide attention22. Published study hinted that the m6A modification might also associate with Runx2 expression23,24. Furthermore, a recent study highlighted the vital role of m6A modifications in the development of intimal hyperplasia and arterial restenosis via regulating the proliferation and migration of VSMCs25. Nonetheless, whether m6A modification is involved in the regulation of Runx2 during VSMCs osteogenic reprogramming remains to be further illuminated.
The current study aimed to dissect the potential function and mechanism of m6A in the osteogenic reprogramming of VSMCs and its value for the prediction and clinical management of VC in patients with CKD. Here, we verified that the m6A methyltransferaselike-3 (METTL3) restrained Runx2 expression through mRNA destabilization mediated by YT521-B homology domain family 2 (YTHDF2). The current findings may help to decipher a novel mechanism of m6A modification in the Runx2-induced osteogenic reprogramming of VSMCs, and may also provide potential targets for therapeutic medication towards VC in CKD.
Results
Aberrant m6A methylation levels mediated by METTL3 were observed in vascular calcification of chronic kidney disease
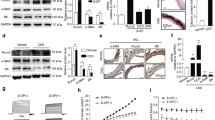
Initially, to expose modification alterations during VC, serum m6A levels were assessed in CKD individuals (accepted maintenance hemodialysis) with or without artery calcification. Patients with VC presented lower m6A levels than those without VC, and the m6A levels were progressively decreased as the severity of VC increased (Fig. 1A, B). Furthermore, the association between m6A levels and calcification scores were analyzed, and the findings suggested a significant negative correlation between m6A levels and calcification degree, as evidenced by a coefficient of determination (R2) of 0.7867 (P < 0.0001) (Fig. 1C).
A Serum RNA m6A levels in CKD patients with VC (n = 6) or without VC (n = 12). B Serum RNA m6A level in subgroups defined by calcification Agatston score (n = 6 per group). C Correlation between serum RNA m6A level and calcification Agatston score from CKD patients with coronary arteries calcification. D Relative mRNA expression of m6A enzymes in serum from patients with CKD with VC or without VC (n = 6 per group). E RT-qPCR analysis of METTL3 mRNA expression level in subgroups defined by calcification Agatston score (n = 20 for without VC; n = 20 for mild; n = 22 for severe). F Correlation matrix of METTL3 and calcification factors. G Receiver operating characteristic (ROC) curve for detection of calcification based on METTL3. H Representative von Kossa staining and immunofluorescence for METTL3 in radial arteries slides from CKD patients with VC or without VC (n = 6 per group). **p < 0.01, ***p < 0.001. Statistical significance between the groups was determined by unpaired two-tailed student’s t tests and one-way ANOVA. VC vascular calcification, NS not significant, Pi phosphate, Ca calcium, ALP alkaline phosphatase, iPTH intact parathyroid hormone, CACs coronary artery calcification score.
To verify the regulators of reduced m6A in VC, we then detected the expression levels of certain known enzymes relevant to m6A methylation such as METTL3, METTL14 and demethylases ALKBH5, FTO. The result suggested that METTL3 was markedly decreased in VC, while the transient and inconsistent changes of other enzymes could not fully explain the abnormal and sustained decrease in m6A levels in the serum of VC patients (Fig. 1D). Moreover, to explore the relationship between METTL3 and VC, METTL3 mRNA levels in the serum of CKD patients with or without VC were investigated. The general characteristics of the study individuals were listed in Supplementary Table 1. Compared with without VC control, METTL3 displayed a substantial decrease, and gradually downregulated as the severity of VC increased (Fig. 1E). A negative correlation was analyzed between METTL3 levels and calcification score (Fig. 1F). Of note, the ROC curves proposed that METTL3 harbored a promising performance to predict VC (AUC = 0.83; Fig. 1G). Von Kossa assays were conducted to confirm VC, while immunofluorescence (IF) staining for METTL3. In the tunica media of radial artery from hemodialysis patients, METTL3 expression exhibited significantly lower expression in calcified artery specimens (Fig. 1H).
Consistent with clinical observations, m6A levels and METTL3 expression were decreased in calcified aortas in adenine diet-induced CKD rats, respectively (Supplementary Fig. 1A, B). These data provide strong evidence that the reduction of m6A modification and its key enzyme, METTL3, were conserved in different species between human and rat under CKD conditions. Taken together, these results verified that METTL3 reduction could be a central molecular hallmark that might explain the reduced m6A levels which was correlated with VC progression in the clinical setting.
METTL3 suppressed osteogenic reprogramming and calcification formation of VSMCs
Considering the osteogenic conversion of VSMCs plays a crucial role in VC, we subsequently investigated the potential function of METTL3 in this process. To explore whether METTL3 knockdown could reprogram VSMCs toward osteogenic phenotype and induce calcification, we knocked down METTL3 under both normal (N) and pro-calcific (10 mM high phosphorus, HP) conditions via transfecting with a specific short hairpin RNA targeting METTL3 (sh-METTL3). The knockdown efficiency of METTL3 was confirmed at mRNA levels. As shown by RT-qPCR results, METTL3 was significantly downregulated in both normal and HP cultured VSMCs compared with the corresponding control group (Fig. 2A). Depletion of METTL3 strikingly promoted the intracellular calcium nodule formation and mineralization of the VSMCs compared with the negative control shRNA (sh-NC) group (Fig. 2B, C). Moreover, the downregulation of METTL3 reduced the expression of contractile-specific markers α-SMA and SM22α, and increased the expression of osteogenic markers Col1α1 and OPN (Fig. 2D).
A RT-qPCR showed transfection efficiency of sh-METTL3 in normal and HP mediated VSMCs. B Alizarin red S staining and (C) calcium content quantification of VSMCs were performed in different experimental groups for detecting mineralization. D Western blot and quantitative densitometry analysis of osteogenic reprogramming markers in N + sh-NC or N + sh-METTL3 and HP + sh-NC or HP + sh-METTL3 groups. E RT-qPCR showed transfection efficiency of OE-METTL3 in normal and HP mediated VSMCs. F Alizarin red S staining and (G) calcium content quantification of VSMCs were performed in different experimental groups for detecting mineralization. H Western blot and quantitative densitometry analysis of osteogenic reprogramming markers in N + OE-NC or N + OE-METTL3 and HP + OE-NC or HP + OE-METTL3 groups. (n = 3 per group). *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance between the groups was determined by unpaired two-tailed student’s t-tests and one-way ANOVA. NS not significant, N normal, HP high phosphorus, sh-NC control shRNA, sh-METTL3 shRNA for METTL3, OE-NC control plasmid, OE-METTL3 plasmid overexpressing METTL3.
As METTL3 can be downregulated during VC, it is intriguing to address whether METTL3 overexpression might alleviate the VC progression. As expected, a plasmid encoding METTL3 (OE-METTL3) could upregulate the METTL3 mRNA levels in both normal and HP conditions (Fig. 2E). Ectopic METTL3 expression cohorts indicated dramatically decreased calcium deposition as evidenced by Alizarin red S staining and quantification of calcium content (Fig. 2F, G). Simultaneously, forced expression of METTL3 negated the reduction of α-SMA and SM22α caused by high-phosphorus medium and downregulated Col1α and OPN in vitro (Fig. 2H). Collectively, both gain- and loss-of-function assays exhibited that METTL3 deficiency bestowed a powerful pro-calcification function, whereas exogenous overexpression of METTL3 attenuated calcification formation by impeding osteogenic reprogramming of VSMCs in vivo.
METTL3 inversely regulated Runx2 expression in VSMCs
We next aimed to uncover the underlying mechanism by which METTL3 mitigated calcification deposition. Because the osteogenic reprogramming of VSMCs was highly administrated by Runx2 through activating the transcription of osteoblast differentiation marker genes26,27, we then investigated whether METTL3 mediated the process of calcification through Runx2. The expression level of Runx2 mRNA in VSMCs cultured with high-phosphate were preliminarily evaluate by RT-qPCR (Fig. 3A). Consistently, the Runx2 protein expression displayed a substantial increase in VSMCs incubated with pro-calcific medium at various intervals compared with that treated without HP, and gradually elevated with the duration extended, which presented an opposite pattern to METTL3 (Fig. 3B). Meanwhile, further relationship between Runx2 and METTL3 was analyzed, and the results indicated a forceful inverse correlation between Runx2 and METTL3 (Fig. 3C).
A Relative mRNA expression of Runx2 in HP mediated VSMCs at different time points. B Representative blot images of METTL3 and Runx2 in HP stimulated VSMCs at different time points. C Correlation analysis between the protein expression of METTL3 and Runx2. Relative Runx2 mRNA (D) and protein (E) expression in N + sh-NC or N + sh-METTL3 and HP + sh-NC or HP + sh-METTL3 groups. Relative Runx2 mRNA (F) and protein (G) expression in N + OE-NC or N + OE-METTL3 and HP + OE-NC or HP + OE-METTL3 groups. H Representative immunofluorescence pictures and quantification of METTL3 and Runx2 in sh-METTL3 or OE-METTL3 treated VSMCs. (n = 3 per group). *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance between the groups was determined by unpaired two-tailed student’s t-tests and one-way ANOVA. NS not significant, D day, N normal, HP high phosphorus, sh-NC control shRNA, sh-METTL3 shRNA for METTL3, OE-NC control plasmid, OE-METTL3 plasmid overexpressing METTL3.
To determine whether METTL3 declined Runx2 expression, its regulation levels under METTL3 knockdown or overexpression were detected. After silencing METTL3, the RT-qPCR results exposed that Runx2 expression in normal cultured VSMCs was obviously increased. Additionally, when VSMCs were stimulated with HP after knocking down METTL3, the expression of Runx2 also presented an increase than corresponding control group (sh-NC) (Fig. 3D). Similarly, western blot also indicated ablation METTL3 increased Runx2 protein expression both in normal and HP culture conditions (Fig. 3E). Conversely, overexpression of METTL3 decreased Runx2 both at mRNA and protein levels in the VSMCs with or without high phosphorus stimulation (Fig. 3F, G). Furthermore, IF staining showed the fluorescence intensity of METTL3 (green fluorescence) exhibited significantly decreased in METTL3 knockdown group, along with reciprocal changes in Runx2 (red fluorescence) compared with the control group, while overexpression of METTL3 effectively reversed the increase in Runx2 fluorescence intensity (Fig. 3H). Altogether, our data suggested that METTL3 could negatively regulate Runx2 expression in VSMCs.
METTL3 regulated mRNA m6A methylation and instability of Runx2
Since METTL3 was the critical m6A methyltransferase in VC and negatively regulated the osteogenic facilitator Runx2, we predicted that METTL3 could mediate m6A modification of Runx2 and further impact its mRNA stability. Firstly, the m6A modification level of Runx2 mRNA in high phosphorus-induced calcified VSMCs was detected by m6A RIP-qPCR. The results showed that the m6A methylated Runx2 mRNA was enriched at a high level in VSMCs under normal conditions by reaction with m6A-specific antibody, and the m6A methylation level of Runx2 gradually decreased as the duration increased of high phosphorus stimulation (Fig. 4A, B). Of note, the data of correlation analysis further confirmed that Runx2 m6A methylation was positively correlated with METTL3 protein and negatively associated with Runx2 mRNA, suggesting that METTL3 might affect Runx2 mRNA levels via regulating its m6A methylation (Fig. 4C, D). To further determine whether METTL3 could regulate m6A modification on Runx2 mRNA in VSMCs, METTL3 knockdown and overexpression experiments were performed and the alterations of Runx2 m6A methylation were examined. As demonstrated by m6A RIP-qPCR, the m6A methylation level of Runx2 was lower in the METTL3 knockdown group than the sh-NC group (Fig. 4E, F). Moreover,the overexpression of METTL3 led to an increase in m6A methylation of Runx2 mRNA, indicating that METTL3 was responsible for the induction of m6A modification of Runx2 (Fig. 4G, H). To investigate whether METTL3 regulated the mRNA degradation of Runx2, RNA stability assay was conducted by using actinomycin D (5 μM) to inhibit transcription. Compared to the relevant control VSMCs, the half-life of Runx2 mRNA was prominently longer in METTL3-knockdown VSMCs, while significantly shortened in METTL3-overexpressing VSMCs (Fig. 4I, J). Overall, these results revealed that m6A-mediated Runx2 down expression was associated with the regulation of mRNA decay.
A, B m6A modification level of Runx2 mRNA in HP mediated VSMCs at different time points. C Correlation analysis between the expression of METTL3 protein and the m6A methylation level of Runx2. D Correlation between the expression of Runx2 mRNA and its m6A methylation level. E, F m6A modification level of Runx2 in sh-NC or sh-METTL3 treated VSMCs. G, H m6A modification level of Runx2 in OE-NC or OE-METTL3 treated VSMCs. I Degradation curves and the t1/2 analysis of Runx2 mRNA in sh-NC or sh-METTL3 treated VSMCs. J Degradation curves and the t1/2 analysis of Runx2 mRNA in OE-NC or OE-METTL3 treated VSMCs. (n = 3 per group). *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance between the groups was determined by unpaired two-tailed student’s t-tests and one-way ANOVA. D day, sh-NC control shRNA, sh-METTL3 shRNA for METTL3, OE-NC control plasmid, OE-METTL3 plasmid overexpressing METTL3.
METTL3 reduced Runx2 mRNA stability through m6A-YTHDF2-dependent way
To explore how m6A activities dominated Runx2 mRNA degradation, we further altered the expression of YTHDF2, which had been deciphered to be a central RNA m6A reader responsible for accelerating the mRNA destabilization of its target gene by mediating recognition of m6A28,29. The knockdown efficiency of sh-YTHDF2 and the overexpression efficiency of OE-YTHDF2 were validated at both the mRNA and protein levels. The results showed that Runx2 mRNA and protein levels were strikingly higher by silencing YTHDF2 compared with control VSMCs, whereas overexpression of YTHDF2 effectively reversed the increase in Runx2 expression caused by HP (Fig. 5A–D). Furthermore, the m6A modification levels of Runx2 had no marked difference between the YTHDF2 knocking down and control VSMCs, eliminating the possibility that YTHDF2 might alter the m6A status on Runx2 (Fig. 5E, F). Meanwhile, the mRNA half-life results of Runx2 suggested that the mRNA stability of Runx2 was also enhanced after the knockdown of YTHDF2, while reduced after overexpression of YTHDF2, compared with that in the control VSMCs (Fig. 5G, H). Subsequently, RIP-qPCR was performed to investigate the direct interaction between YTHDF2 and the m6A methylation on Runx2. The result identified that Runx2 mRNA could be obviously immunoprecipitated by a YTHDF2-specific antibody compared to the IgG pull-down group, and Runx2 mRNA enrichment in the METTL3 knocking down VSMCs was prominently lower than that of the control group (Fig. 5I, J). As YTHDF2 destabilized Runx2 mRNA by recognizing its m6A modification, we hypothesized that deleting the m6A methylation of Runx2 could inhibit the degradation effect of YTHDF2 on the mRNA of the target gene. Expectedly, the rescue experiment also confirmed that the knockdown of METTL3 partially rescued the reduced expression of Runx2 induced by YTHDF2 at both mRNA and protein levels (Fig. 5K, L), as well as the attenuation of osteogenic differentiation and calcification (Fig. 5M). The above data indicated that METTL3 accelerated Runx2 mRNA degradation in an m6A-YTHDF2-dependent manner.
A Relative mRNA and (B) protein expression of YTHDF2 and Runx2 in sh-NC or sh-YTHDF2 treated VSMCs. C Relative mRNA and (D) protein expression of YTHDF2 and Runx2 in OE-NC or OE-YTHDF2 treated VSMCs. E, F m6A modification level of Runx2 mRNA in sh-NC or sh-YTHDF2 treated VSMCs. G Degradation curves and the t1/2 analysis of Runx2 mRNA in sh-NC or sh-YTHDF2 treated VSMCs. H Degradation curves and the t1/2 analysis of Runx2 mRNA in OE-NC or OE-YTHDF2 treated VSMCs. I, J YTHDF2 RIP-qPCR analysis of Runx2 mRNA in sh-NC or sh-METTL3 treated VSMCs. K Relative mRNA expression of Runx2 in HP + OE-YTHDF2 or HP + OE-YTHDF2 + sh-METTL3 treated VSMCs. L Representative blot images of Runx2 and osteogenic reprogramming markers in HP + OE-YTHDF2 or HP + OE-YTHDF2 + sh-METTL3 treated VSMCs. M Alizarin red S staining was performed to detect mineralization in HP + OE-YTHDF2 or HP + OE-YTHDF2 + sh-METTL3 groups. (n = 3 per group). *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance between the groups was determined by unpaired two-tailed student’s t-tests and one-way ANOVA. NS not significant, sh-NC control shRNA, sh-YTHDF2 shRNA for YTHDF2, sh-METTL3 shRNA for METTL3, OE-NC control plasmid, OE- YTHDF2 plasmid overexpressing YTHDF2.
YTHDF2 recognized specific m6A methylation sites in the Runx2 3′ UTR region
As distinct m6A readers, YTHDF2 can recognize and bind to mRNA transcript targets through their specific m6A modification sites and play a pivotal function in mRNA stabilization30. Preliminarily, specific m6A modification records on Runx2 mRNA were retrieved from the m6AVar database to predict the potential m6A sites that might regulate Runx2 expression31. The result exhibited that the m6A sites with high confidence were located in chr 6:45547738 and chr 6:45547767, which were both in the Runx2 3′ UTR region (Fig. 6A). Based on the predicted results, the mutation constructs at m6A sites of chr 6:45547738 (Runx2-Mut1) or chr 6:45547767 (Runx2-Mut2) were established, and the corresponding wild-type (Runx2-WT1-2) expression plasmids were accompanied (Fig. 6B). To confirm the m6A sites of Runx2, Runx2 wild-type or mutant plasmids were transfected in YTHDF2-overexpressed 293 T cells. The results indicated that the mRNA and protein levels of both Runx2-WT1-2 and Runx2-Mut1 were decreased in YTHDF2-overexpressed cells, while those for Runx2-Mut2 groups were resistant to the effect of YTHDF2 augment (Fig. 6C–F). In addition, to further ascertain the specific m6A motifs bound by YTHDF2, RIP-qPCR was conducted by using an anti-YTHDF2 antibody. The results uncovered that YTHDF2-enriched Runx2 was remarkably reduced in the Runx2-Mut2 transfected 293 T cells but not in the Runx2-WT1-2 and Runx-Mut1 groups (Fig. 6G–J). Taken together, these results revealed that YTHDF2 was responsible for reading specific m6A modified sites in the Runx2 3′ UTR region.
A Schematic representation of m6A sites of Runx2 mRNA in the 3′ UTR region. B Schematic representation of mutated m6A sites of Runx2 mRNA. Adenines in chr 6:45547738 (Runx2-Mut1) and chr 6:45547767 (Runx2-Mut2) were mutated to guanine. C Relative Runx2 mRNA expression in Runx2-WT1 or Runx2-Mut1 transfected 293 T cells treated with OE-NC or OE-YTHDF2. D Relative Runx2 mRNA expression in Runx2-WT2 or Runx2-Mut2 transfected 293 T cells treated with OE-NC or OE-YTHDF2. E Representative blot images of Runx2 in Runx2-WT1 or Runx2-Mut1 transfected 293 T cells treated with OE-NC or OE-YTHDF2. F Representative blot images of Runx2 in Runx2-WT2 or Runx2-Mut2 transfected 293 T cells treated with OE-NC or OE-YTHDF2. G, H YTHDF2 RIP-qPCR analysis of Runx2 mRNA in Runx2-WT1 or Runx2-Mut1 transfected 293 T cells. I, J YTHDF2 RIP-qPCR analysis of Runx2 mRNA in Runx2-WT2 or Runx2-Mut2 transfected 293 T cells. (n = 3 per group). *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance between the groups was determined by unpaired two-tailed student’s t-tests. NS not significant, OE-NC control plasmid, OE- YTHDF2 plasmid overexpressing YTHDF2, WT wild type, Mut mutation type.
Inhibition of METTL3 expression facilitated osteogenic reprogramming and calcification in arteries of CKD rats
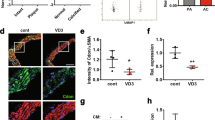
Finally, we further explored the in vivo effect of METTL3 inhibition on VSMCs osteogenic phenotypic transdifferentiation and calcium deposit in the aortas of CKD rats. The METTL3 inhibitor SAH was administered in an adenine-induced CKD model, and the specific scheme of animal model construction was shown in Fig. 7A. After fed with adenine diet, the rats successfully developed severe renal failure, which was verified by loss of body weight, increased creatinine, urea and urine protein levels, and pathological staining of the kidneys (Supplementary Fig. 2). It could be seen in Fig. 7B that serum phosphate, calcium and parathyroid hormone (PTH) levels were raised in the CKD group compared with the control group, and showed tendencies of alteration levels after METTL3 inhibition. Moreover, METTL3 repression could further aggravated CKD-induced vascular calcification in the aortas, according to the results of calcium staining and quantification (Fig. 7C–E). Again, METTL3 was effectively decreased in the VSMCs of SAH (S-Adenosylhomocysteine) treated CKD rats, accompanied with the elevation of Runx2 at the same localization (Fig. 7F). These effects paralleled a similar regulation of osteogenic differentiation in aortic tissues. Inactivation of METTL3 increased osteogenic markers Col1α1 and OPN, and decreased the expression of contractile makers α-SMA and SM22α (Fig. 7G, H). Therefore, these findings suggested that METTL3 could be the potential target to trigger osteogenic reprogramming, and aggravate the progression of VC in CKD.
A Scheme of the construction of the CKD-associated VC rat model. B Serum levels of P, Ca, and PTH in control or CKD or CKD + SAH rats (n = 6). Data are shown as mean ± standard deviation. a, means the difference between the control group; b, means the difference between the CKD group. Representative images of Alizarin red S staining (C) and von Kossa staining (D) to reveal the calcium deposition in the thoracic and abdominal aortas of control or CKD or CKD + SAH rats. E Calcium content quantification of aortas to detect mineralization of control or CKD or CKD + SAH rats (n = 3). F Representative immunofluorescence pictures and quantification of METTL3 and Runx2 in aortas of control or CKD or CKD + SAH rats (n = 3). G Representative blot images of METTL3, Runx2, and osteogenic reprogramming markers in aortas of control or CKD or CKD + SAH rats (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001. Statistical significance between the groups was determined by one-way ANOVA. SAH S-Adenosylhomocysteine, P phosphate, Ca calcium, PTH parathyroid hormone.
Discussion
The heterogeneity of VSMCs and their close response with the bloodstream make it challenging to illuminate the mechanism of VC. To the best of our knowledge, the current study was the first to show that the RNA m6A levels in the serum of CKD calcification patients were strikingly decreased along with VC progression, and downregulation of METTL3 mediated this observed m6A reduction. Moreover, METTL3 deficiency exacerbated osteogenic reprogramming and calcification formation in HP-stimulated VSMCs, whereas exogenous overexpression of METTL3 bestowed an obvious anti-calcification effect. Further investigation revealed a novel mechanism that METTL3 methylated the osteogenic facilitator Runx2 and negatively regulated its expression through coordinating the m6A-YTHDF2-dependent mRNA degradation manner. In addition, inhibition of METTL3 promoted the osteogenic transdifferentiation capacity and calcification of VSMCs in vivo.
m6A is the most abundant internal post-transcriptional modification in eukaryotic mRNA32,33, and participates in various biological and cellular processes such as cell pluripotency, differentiation, reprogramming, and metabolism34. Recently, the role of m6A methylation in regulating the development and progression of vascular dysfunction, arteriosclerosis, and angiogenesis has been explored. As reported by Meng et al. and Shao et al., the reduction of m6A modification mediated by ALKBH5 upregulation35 and METTL14 downregulation36 had been exhibited to act as an important role in diabetic cardiomyopathy. Similarly, we also found that the epigenetic m6A modification levels were significantly decreased in the serums of calcified CKD patients and negatively correlated with the severity of VC. Moreover, m6A levels in the arterial tissues of CKD rats model exposed the concordant decreasing pattern with humans. Namely, serum m6A modification, at least in part, might reflect the corresponding systematic change of that in VSMCs. Alternatively, the opposite trend of m6A levels had been reported in radial arteries of ESRD patients and indoxyl sulfate-stimulated human artery smooth muscle cells37, indicating that the alteration of m6A was caused by environmental signal modulation, experimental set-up, and cell type. The m6A modification is a dynamic and reversible process mediated by methyltransferases and demethylases, among which METTL3 has been reported as the core enzyme exerting methyltransferase activity38. Notably, our study observed that METTL3 was the most significantly downregulated enzyme compared with other m6A-related proteins (METTL14, FTO, and ALKBH5), which was consistent with the finding that decreased m6A level was due to the downregulation of endothelial METTL3 in atherosclerosis39. Previous studies had demonstrated the certain accuracy of METTL3 in predicting infertile stage IV endometriosis40. We also analyzed the ability of METTL3 expression to predict VC, and the ROC curve highlighted the diagnostic value of METTL3. These explanations were limited due to the lack of control data in healthy populations and CKD patients with different etiologies. Moreover, the findings in CKD patients with VC were observational and required further validation. Despite these limitations, our preclinical data provided at least an initial indication that METTL3 levels might serve as a potential biomarker for the early diagnosis of VC.
Accumulating evidence has revealed that RNA m6A modification plays a critical role in the regulation of the pathophysiological function of VSMCs through effecting cell proliferation, migration, and phenotypic transdifferentiation41. METTL3, as the most important m6A methyltransferase subunit, has been extensively investigated for its effect on cellular osteogenic phenotype transdifferentiation, especially in bone marrow mesenchymal stem cells (BMSCs), and indicates that METTL3 promotes the cellular osteogenic differentiation of metabolic bone diseases, such as osteoporosis, ossification, and maxillofacial bone defects42,43. Although studies have suggested that METTL3 is also involved in the regulation of osteogenic differentiation in cardiovascular diseases, its role in cardiovascular cells remains controversial. In human aortic valve interstitial cells, METTL3 has been reported to promote osteogenic differentiation44, whereas in primary human aortic smooth muscle cells, METTL3 inhibits this progression45. In our present study, gain- and loss-of-function experiments clearly manifested the evidence for causal METTL3 involvement in the osteogenic differentiation of VSMCs. Specifically, METTL3 deficiency significantly exacerbated the osteogenic differentiation of VSMCs, which was confirmed by the elevation in osteoblast markers expression (including Col1α1 and OPN), osteogenic phenotype, and calcium deposition. In comparison, METTL3 overexpression hindered this pathological process. Thus, the protective role of METTL3 in VC amelioration was potentially mediated by suppressing the osteogenic reprogramming of VSMCs.
It is known that Runx2 localized both in the nucleus and cytosol is upregulated in VC, and its core function in VSMCs osteogenic transdifferentiation has been fully documented46. Nonetheless, it remains unclear whether VSMCs can adopt m6A modification in the regulation of Runx2 during osteogenic reprogramming. In the current study, for the first time to our knowledge, we ascertained that METTL3 is a critical effector for VSMCs to regulate Runx2 expression and subsequent osteogenic differentiation. We discovered that mRNA and protein expression of Runx2 were significantly increased in a METTL3 ablation VC model in vivo and in vitro and vice versa. More importantly, our results indicated that METTL3 could directly induce m6A methylation of Runx2 mRNA to alter its cellular stability and negatively regulate its levels. By mediating the methylation of key genes and affecting their expression, METTL3 has been reported to play functional roles in diverse diseases. For instance, METTL3 mediated m6A methylation restrained the expression of autophagy-related 7, the enzyme critical for the formation of autophagosomes, by mitigating its RNA stability in the progression of osteoarthritis47. METTL3-based m6A modification also promoted direct methylation of Runx2 mRNA in bone defect48 and osteoporosis49, which was similar to our findings. Studies have well documented the mechanism by which METTL3 regulates gene expression, such as mRNA stability, RNA splicing, and transcription50. In our study, we found that knocking down METTL3 prolonged the half-life of Runx2, revealing that METTL3 negatively regulated Runx2 expression mainly by affecting the degradation of mRNA.
Thus, a crucial question to consider is how METTL3 regulated Runx2 mRNA degradation. An intriguing point reveals that METTL3-modiated m6A methylation of genes may lead to different outcomes due to the different readers, which have different m6A localization functions18,51. In mammalian mRNA, IGF2BPs can stabilize mRNA through co-factors HuR and MATR352, while YTHDF1 and YTHDF3 are considered to active translation of target transcripts by recruiting translation initiation factors53. m6A-regulated mRNA degradation is mainly mediated by YTHDF2, which recruits methylated transcript to the mRNA decay sites54. Yu et al. reported that YTHDF2 recognized the m6A modified PER1 and TP53 mRNAs and promoted their degradation in ocular melanoma55. Similarly, our study showed that YTHDF2 deficiency was enhanced, whereas YHDF2 overexpression decreased Runx2 expression and mRNA stability. Furthermore, the degradation of Runx2 mediated by YTHDF2 could be reversed by knocking down METTL3, revealing the regulatory mechanism was dependent on m6A modification site. According to a previous study, METTL3 preferentially installs m6A methylation in a relatively conservative RRACH motif (base R is A or G; H is A, C, or U), and the methylation of m6A is usually abundant in the coding region, 3′ UTR and 5′ UTR of mRNAs34,56. In our study, a GAACU site at chr 6:45547767, the 3′ UTR region of Runx2, was shown to be recognized by YTHDF2.
Considering the crucial function of m6A in regulating gene expression, a recent study has explored its therapeutic potential in human disease. Small inhibitors of m6A methylase, demethylase, and readers, as well as activators are designed to treat diverse cancers in vitro and in mouse models57. Recently, STM2457, a small molecule inhibitor of METTL3, has been identified as a potential therapeutic strategy for the treatment of acute myeloid leukemia and underwent Phase I clinical trials58,59. In our study, we found that VSMCs treated with a METTL3 inhibitor showed enhanced osteogenic reprogramming capacity in a CKD-VC rat model and confirmed the potential function of METTL3 to negatively regulate Runx2 expression in vivo. Therefore, based on the potentially opposed effects of METTL3 on tumor cells and normal cells, the side effects on normal cells should be carefully considered if targeting METTL3 to treat tumors. In addition, the application of METTL3-specific inhibitors for in vivo experiments will be better to bolster our results. Therefore, further exploration is needed to demonstrate the relationship between STM2457 and CKD-VC.
In conclusion, as far as we know, this is the first study to identify the diagnostic and therapeutic value of METTL3 to precisely regulate osteogenic reprogramming through post-transcriptional modulation the mRNA stability of Runx2. The results support that METTL3-mediated m6A modification ameliorates VC by down-regulating the expression of Runx2 in a m6A-YTHDF2-dependent way (Fig. 8).
Materials and methods
CKD patient recruitment and samples
The case control study of CKD patients was conducted in accordance with Declaration of Helsinki principles, and was approved by the Ethics Committee of the Fourth Hospital of Hebei Medical University, Shijiazhuang, Hebei, China (No.2022ky033) with obtaining written informed consent from all the participants who were identified by number. All ethical regulations relevant to human research participants were followed. The study was performed from June 2022 to January 2023 in the Fourth Hospital of Hebei Medical University. A total of 62 CKD patients accepting maintenance hemodialysis younger than 80 years or older than 18 years were included, excluding diabetes mellitus, malignancy, severe malnutrition, uncontrolled hyperlipidemia, acute cardiovascular events, acute infections, chronic active hepatitis, and fractures occurring in the past 6 months. Clinical and biochemical parameters were assembled from the hospital’s electronic medical records after informed consent was signed. Overnight fasting median cubital venous blood was collected and centrifuged at 3000 g for 10 min at 4 °C, then the supernatant was collected as serum. Serum samples were stored in 250 μl aliquots at -80 °C for total RNA extraction. A 3 to 5 mm segment of the radial artery was resected from CKD patients who underwent an arteriovenostomy before hemodialysis (n = 12). After removing fat and adventitia, the tissue sections were placed in 4% paraformaldehyde for subsequent paraffin embedding.
Assessment of calcification
Imaging of the coronary arteries was performed with multi-detector computed tomographic scanner (Definition Flash, Siemens Healthcare, Forchheim, Germany). Agatston scores were blind-calculated by 3 independent image analysts with SmartScore software (Definition Flash, Siemens Healthcare, Forchheim, Germany) according to standard criteria. According to the studies focused on coronary artery calcium scores, patients were divided into without-VC (Agatston score = 0), mild-VC (0 <Agatston score ≤400), or severe-VC (Agatston score > 400).
Animals and treatment
The animal experiment protocols were approved by the Institutional Animal Care and Use Committee at the Fourth Hospital of Hebei Medical University, Shijiazhuang, Hebei, China (approval No. IACUC-4th Hos Hebmu-2022036). We have complied with all relevant ethical regulations for animal use. All procedures were conducted in compliance with the NIH Guide for the Care and Use of Laboratory Animals. 8-week-old male Sprague-Dawley (SD) rats weighing 200 to 220 grams were purchased from the SiPeiFu Biotechnology Co., Ltd (Beijing, China) and raised in temperature and humidity-controlled conditions on a 12-hour dark/light cycle. By adopting a simple random sampling method, rats were divided into three groups with 6 animals per group: control group, CKD model group (CKD), and CKD + SAH group. The control group was fed with standard normal chow diet, while CKD model rats were indued by feeding with special chow containing adenine (0.75%) and high levels of phosphorus (1.2%). The serum creatinine (Cr), urea, and urine protein levels were tracked to assess a rat model of CKD. In detail, blood was collected from the tail vein of fasted overnight rats every two weeks. Urine was collected by using a metabolic cage for 24 h, and the supernatant was obtained after centrifugation. The 24 h urine protein levels were obtained by multiplying the measured protein concentration by the urine volume. After 4 weeks, the Cr, urea, and urine protein levels in both the CKD group and the CKD + SAH group were significantly increased. Then, the CKD rats were administered calcitriol (active vitamin D3; 1 μg/kg; twice a week; PHR2663, Sigma) via gavage to induce media calcification, while SAH rats were given intraperitoneal injections of METTL3 inhibitor S-Adenosylhomocysteine (SAH, 5 mg/kg; twice a week; HY-19528, MedChemExpress) dissolved in normal saline (0.9% NaCl solution) additionally. For euthanasia, the rats were deeply anaesthetized with 5% isoflurane introduced via a vaporizer, followed by cervical dislocation. Blood samples were collected to evaluate the serum creatinine (C011-2-1), urea (C013-2-1), calcium(C004-2-1), and phosphate (C006-1-1) levels applying commercial test kits (Nanjing Jiancheng Bioengineering Institute) following the instructions. Parathyroid hormone (PTH) levels were detected using the Rat Parathyroid Hormone ELISA kit (CSB-E07866r; CUSABIO). Urine samples were collected to determine urine protein concentration usinga biochemistry analyzer (Beckman). Whole aorta samples were harvested for further analysis.
Cell culture
Primary VSMCs were obtained from male SD rats aged 4 weeks, weighing 80 to 100 grams. Briefly, the adventitia and intima of the blood vessels were removed from thoracic aortas, and the remaining segments were cut into 1–2 mm2 sections. Arterial tissues were then placed in cell culture dishes and immersed in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, 31600-034) containing 10% fetal bovine serum (FBS) in a 37 °C humidified incubator with 5% CO2 atmosphere. The VSMCs were migrated from the explants, and cells at passages 3 to 5 were utilized for experiments. To induce VSMCs calcification, β-glycerophosphate (Sigma, USA, G5422) was used according to previous reports60,61,62. The VSMCs were treated with concentrations of 0 mM, 1 mM, 2.8 mM, 5 mM, 7.5 mM, and 10 mM for 7 days. Then, treatment with β-glycerophosphate in the presence of 10 mM was selected for further experiments (Supplementary Fig. 3).
HEK293T (293T) cells were propagated in DMEM supplemented with 10% FBS in the same atmosphere as above.
Calcium staining
Artery calcification was identified by von Kossa staining and Alizarin red S staining, while VSMCs calcification was assessed by Alizarin red S staining. For von Kossa staining, 5-micrometer paraffin slides from the artery were dehydrated and then immersed in 5% silver nitrate solution under ultraviolet light for 1 h, and calcified spots were stained brown. For Alizarin red S staining, the calcified aortas and cells were exposed to 1% Alizarin red S solution pH 4.2 (G1452, Solarbio Technology Co.) for 15 min. Positively staining was red-orange in color, suggesting calcification.
RNA m6A quantification
The m6A modification level of total RNA was measured via performing an EpiQuik m6A RNA Methylation Quantification Kit (P-9005-96, Epigentek) following the manufacturer’s protocols. Briefly, 200 nanogram RNA accompanied with m6A standard were coated on assay wells, which was used for subsequent detection by incubating capture antibody solution and detection antibody solution. The m6A content was quantified colorimetrically at 450 nanometer wavelength and calculated based on the standard curve.
Plasmids and transfection
METTL3 overexpression (OE-METTL3) and corresponding negative control (OE-NC) plasmids were purchased from iGeneBio Co. (Rn17455, Guangzhou, China), while YTHDF2 overexpression (OE-YTHDF2) and negative control (OE-NC) plasmids were constructed from Sino Biological Inc. (HG17055-ACG, Beijing, China). The small hairpin RNA against METTL3 (sh-METTL3), YTHDF2 (sh-YTHDF2), and negative control shRNA (sh-NC) were obtained from GeneCopoeia (USA). The sequences of sh-METTL3 and sh-YTHDF2 were 5′GCTACCGTATGGGACGTTAAC3′ and 5′GGTCAAGGAAACAAAGTACAA′, respectively. VSMCs were transfected with 2.5 μg plasmids using Lipofectamine 3000 reagent (L3000015, Invitrogen) according to the manufacturer’s instructions. Further experiments were employed after 48 h, and the transfection efficiency was detected by RT-qPCR and western blot, respectively.
RT-qPCR analysis
Total RNA from clinical serum, aortic tissue, and VSMCs were extracted with Trizol Reagent (15596026, Invitrogen Carlsbad). Reverse transcription and real-time PCR were conducted to detect mRNA expression by using a FastKing one-step RT-qPCR Kit (SYBR Green) (FP313, TianGen). The primer sequences are listed in Supplementary Table 2. All experiments were performed in three biological replicates. The expression of selected mRNAs was calculated according to the 2-ΔΔCt method, with GAPDH level as an internal control.
RNA stability assays
To determine the stability of Runx2 mRNA in VSMCs under the impact of knockdown or overexpression of METTL3/YTHDF2, actinomycin D (HY-17559, MedChemExpress; 5 μg/mL) was applied to cells for 0 h, 2 h, 4 h and 6 h, and the mRNA expression at the specified time point was evaluated by performing RT-qPCR described above. GraphPad Prism was performed to calculate the decay rate and half-life of Runx2 mRNA.
m6A RIP-qPCR
The m6A RIP assay was conducted using a PureBinding®RNA Immunoprecipitation Kit (P0102, GeneSeed) and a specific anti-m6A antibody according to the manufacturer’s protocols. In brief, VSMCs were lysed and incubated with RIP buffer containing prepared Protein A/G magnetic beads conjugated with anti-m6A antibody, anti-YTHDF2, or IgG negative control overnight at 4 °C. Then, immunoprecipitated RNAs were extracted and purified for RT-qPCR assays to analyze the m6A enrichment of target genes.
Runx2 mutant plasmids construction and transfection
The Runx2 mRNA sequence generated from the NCBI reference sequence NM_001024630.4 was regarded as Runx2-WT. The adenine located in chr 6:45547738 (GRCh38) (Runx2-Mut1) and chr 6:45547767 (GRCh38) (Runx2-Mut2) was mutated to guanine. Runx2-WT or mutant plasmids were transfected into 293 T cells using Lipofectamine 3000 reagent. After 48 h, total RNA or protein was extracted to detect the expression of Runx2 using RT-qPCR or western blot.
Western blot
Protein was collected using RIPA lysis buffer (Solarbio Science & Technology Co., Beijing, China) and quantified using a BCA Protein Assay Kit (Solarbio Science & Technology Co., Beijing, China). Equal amounts of proteins were separated via SDS-PAGE gel electrophoresis and electrotransferred to polyvinylidene difluoride membranes (Merck Millipore). After being blocked with 5% BSA (Solarbio Science & Technology Co., Beijing, China) at 37 °C for 1 h, the blots were incubated overnight at 4 °C with primary antibodies. After rinsing with 1% Tween-Tris buffered saline, the blots were incubated with a peroxidase–conjugated secondary antibody (1:5000 diluted) for 1 h at room temperature. The antibody details can be found in Supplementary Table 3. Then the blots were visualized by using ECL reagent (Thermo Fisher Scientific, Waltham, MA, USA), and the mean integrated optical density of spots was quantified by ImageJ software, normalized by GAPDH. See original western blots in Supplementary Fig. 4.
Immunofluorescence
VSMCs were fixed with 4% paraformaldehyde and permeabilized with 0.3% Triton X-100 at room temperature, respectively. For artery, samples were fixed in 10% formalin and embedded in paraffin. Then, the paraffin slides were deparaffinized, rehydrated, and boiled in citrate buffer for epitope retrieval. After blocking with goat serum for 1 h, cells and slides were incubated with primary antibodies 4 °C overnight. Then, the cells and slides were observed after an incubation with the secondary antibody and DAPI.
Statistics and reproducibility
All data were presented as mean ± standard deviation from at least three replicates. GraphPad Prism 9 software was used for statistical analyses. Unpaired, two-tailed Student’s t-tests or one-way analysis of variance (ANOVA) were performed to evaluate the statistical significance of differences between two groups or among multiple sample groups. Pearson’s correlation analysis was performed to evaluate the association between two variables. Significance levels were determined as (*) p < 0.05, (**) p < 0.01, and (***) p < 0.001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The original data for western blots, as well as the prime sequence of genes, are available in the supplementary results. All source data underlying the graphs presented in the figures were available in Supplementary Data.
References
Wang, Y. et al. CCN2 deficiency in smooth muscle cells triggers cell reprogramming and aggravates aneurysm development. JCI Insight 8, e162987 (2023).
Ouyang, L. et al. ALKBH1-demethylated DNA N6-methyladenine modification triggers vascular calcification via osteogenic reprogramming in chronic kidney disease. J. Clin. Invest. 7, 131 (2021).
Jono, S. et al. Phosphate regulation of vascular smooth muscle cell calcification. Circ. Res. 87, E10–E17 (2000).
Basatemur, G.L. et al. Vascular smooth muscle cells in atherosclerosis. Nat. Rev. Cardiol. 16, 727–744 (2019).
Steitz, S.A. et al. Smooth muscle cell phenotypic transition associated with calcification: upregulation of Cbfa1 and downregulation of smooth muscle lineage markers. Circ. Res. 89, 1147–1154 (2001).
Gjorgjieva, T. et al. Loss of β-Actin Leads to Accelerated Mineralization and Dysregulation of Osteoblast-Differentiation Genes during Osteogenic Reprogramming. Adv. Sci. 7, 2002261 (2020).
Chen, N.X. et al. Phosphorus and uremic serum up-regulate osteopontin expression in vascular smooth muscle cells. Kidney Int. 62, 1724–1731 (2002).
Vervloet, M. et al. Vascular calcification in chronic kidney disease: different bricks in the wall?. Kidney Int. 91, 808–817 (2016).
Nadia, R. et al. Molecular mechanisms of vascular health: insights from vascular aging and calcification. Arterioscler. Thromb. Vasc. Biol. 43, 15–29 (2023).
London, G.M. et al. Arterial media calcification in end-stage renal disease: impact on all-cause and cardiovascular mortality. Nephrol. Dial. Transplant. 18, 1731–1740 (2003).
Cobb, A.M. et al. Runx2 (Runt-Related Transcription Factor 2) Links the DNA damage response to osteogenic reprogramming and apoptosis of vascular smooth muscle cells. Atertioscler. Thromb. Vasc. Biol. 41, 1339–1357 (2021).
Liu, H. et al. Rho A/ROCK1 signaling-mediated metabolic reprogramming of valvular interstitial cells toward Warburg effect accelerates aortic valve calcification via AMPK/RUNX2 axis. Cell Death Dis. 14, 108 (2023).
Bai, Y. et al. Manipulation of heterogeneous surface electric potential promotes osteogenesis by strengthening RGD peptide binding and cellular mechanosensing. Adv. Mater. 35, e2209769 (2023).
Dong, S. et al. RUNX2 stabilization by long non-coding RNAs contributes to hypertrophic changes in human chondrocytes. Int. J. Biol. Sci. 19, 13–33 (2023).
Towler, D.A. et al. AMPKα1: SUMO Wrestling Runx2 as a strategy to inhibit arteriosclerotic calcification. Circ. Res. 119, 398–400 (2016).
Ahn, B. et al. Cdon suppresses vascular smooth muscle calcification via repression of the Wnt/Runx2 Axis. Exp. Mol. Med. 55, 120–131 (2023).
Ouyang, L. et al. Indoleamine 2,3-Dioxygenase 1 deletion-mediated kynurenine insufficiency in vascular smooth muscle cells exacerbates arterial calcification. Circulation 145, 1784–1798 (2022).
Michaela, F. et al. RNA modifications modulate gene expression during development. Science 361, 1346–1349 (2018).
An, Y. et al. The role of m6A RNA methylation in cancer metabolism. Mol. Cancer 21, 14 (2020).
Zheng, J.R. et al. Epitranscriptomic modifications in mesenchymal stem cell differentiation: advances, mechanistic insights, and beyond. Cell Death Differ. 31, 9–27 (2024).
Luan, J. et al. N6-methyladenine RNA methylation epigenetic modification and kidney diseases. Kidney Int. Rep. 8, 36–50 (2022).
Wu, S. et al. m6A RNA methylation in cardiovascular diseases. Mol. Ther. 28, 2111–2119 (2020).
Liu, T. et al. A “reader” YTHDF1 promotes osteogenesis of bone marrow mesenchymal stem cells through translational control of ZNF839. Cell Death Dis. 12, 1078 (2021).
Zhang, M. et al. YTHDC1 inhibits osteoclast differentiation to alleviate osteoporosis by enhancing PTPN6 messenger RNA stability in an m6A-hUR-dependent manner. J. Leukoc. Biol. 115, 1154–1164 (2024).
Zhu, B. et al. Total Panax notoginseng saponin inhibits vascular smooth muscle cell proliferation and migration and intimal hyperplasia by regulating WTAP/p16 signals via m6A modulation. Biomed. Pharmacother. 124, 109935 (2020).
Wang, C. et al. Poly(ADP-ribose) polymerase 1 accelerates vascular calcification by upregulating Runx2. Nat. Commun. 10, 1203 (2019).
Shroff, R. et al. Mechanistic insights into vascular calcification in CKD. J. Am. Soc. Nephrol. 24, 179–189 (2013).
Deobrat, D.C. et al. The RNA m6A Reader YTHDF2 maintains oncogene expression and is a targetable dependency in Glioblastoma stem cells. Cancer Discov. 11, 480–499 (2021).
Seitz, F. et al. Atomic Mutagenesis of N6-Methyladenosine Reveals Distinct Recognition Modes by Human m6A Reader and Eraser Proteins. J. Am. Chem. Soc. 146, 7803–7810 (2024).
Du, H. et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 7, 12626 (2016).
Luo, X. et al. RMVar: an updated database of functional variants involved in RNA modifications. Nucleic Acids Res. 49, D1405–D1412 (2021).
Zhao, B. et al. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell. Biol. 18, 31–42 (2017).
Huang, W. et al. N6-methyladenosine methyltransferases: functions, regulation, and clinical potential. J. Hematol. Oncol. 14, 117 (2021).
Zhang, H. et al. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 48, 6251–6264 (2020).
Shao, Y. et al. CircRNA CDR1as promotes cardiomyocyte apoptosis through activating hippo signaling pathway in diabetic cardiomyopathy. Eur. J. Pharmacol. 922, 174915 (2022).
Meng, L. et al. METTL14 suppresses pyroptosis and diabetic cardiomyopathy by downregulating TINCR lncRNA. Cell Death 13, 38 (2022).
Chen, J. et al. METTL14-dependent m6A regulates vascular calcification induced by indoxyl sulfate. Life. Sci. 239, 117034 (2019).
Wang, P. et al. Structural basis for cooperative function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell. 63, 306–317 (2016).
Li, B. et al. RNA N6-methyladenosine modulates endothelial atherogenic responses to disturbed flow in mice. Elife 11, e69906 (2022).
Wan, S. et al. METTL3-dependent m6A methylation facilitates uterine receptivity and female fertility via balancing estrogen and progesterone signaling. Cell Death Dis. 14, 349 (2023).
Gatsiou, A. et al. RNA modifications in cardiovascular health and disease. Nat. Rev. Cardiol. 20, 325–346 (2023).
Wu, Y. et al. Mettl3-mediated m6A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat. Commun. 9, 4772 (2018).
Zhou, S. et al. METTL3 potentiates osteogenic differentiation of bone marrow mesenchymal stem cells via IGF2BP1/m6A/RUNX2. Oral. Dis. 30, 1313–1321 (2023).
Zhou, T. et al. Factors influencing osteogenic differentiation of human aortic valve interstitial cells. J. Thorac. Cardiovasc. Surg. 161, 163–e185 (2021).
Fang, Z. et al. Methyltransferase-like 3 suppresses phenotypic switching of vascular smooth muscle cells by activating autophagosome formation. Cell Prolif. 56, e13386 (2023).
Chen, Y. et al. Transcriptional Programming in Arteriosclerotic Disease: A Multifaceted Function of the Runx2 (Runt-Related Transcription Factor 2). Arterioscl. Thromb. Vas. 41, 20–34 (2021).
Chen, X. et al. METTL3-mediated m6A modification of ATG7 regulates autophagy-GATA4 axis to promote cellular senescence and osteoarthritis progression. Ann. Rheum. Dis. 81, 87–99 (2022).
Sun, X. et al. 3D-Printed β-Tricalcium phosphate scaffolds promote osteogenic differentiation of bone marrow-deprived mesenchymal stem cells in an N6-methyladenosine-dependent manner. Int. J. Bioprint. 8, 544 (2022).
Yan, G. et al. m6A Methylation of precursor-miR-320/RUNX2 controls osteogenic potential of bone marrow-derived mesenchymal stem cells. Mol. Ther. Nucleic Acids 19, 421–436 (2020).
Jiang, X. et al. The role of m6A modification in the biological functions and diseases. Signal. Transduct. Target. Ther. 6, 74 (2021).
Sara, Z. et al. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell. Biol. 20, 608–624 (2019).
Shi, H. et al. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell. 74, 640–650 (2019).
Wang, X. et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399 (2015).
Kumari, R. et al. mRNA modifications in cardiovascular biology and disease: with a focus on m6A modification. Cardiovasc. Res. 118, 1680–1692 (2022).
Yu, J. et al. Histone lactylation drives oncogenesis by facilitating m 6 A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 22, 85 (2021).
Yue, Y. et al. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355 (2015).
Sabrina, G. et al. m6A RNA methylation and beyond – the epigenetic machinery and potential treatment options. Drug. Discov. Today 26, 2559–2574 (2021).
Yankova, E. et al. Small molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593, 597–601 (2021).
Cully, M. Chemical inhibitors make their RNA epigenetic mark. Nat. Rev. Drug. Discov. 18, 892–894 (2019).
Li, Z. et al. Cold exposure protects against medial arterial calcification development via autophagy. J. Nanobiotechnol. 21, 226 (2023).
Alesutan, I. et al. Augmentative effects of leukemia inhibitory factor reveal a critical role for TYK2 signaling in vascular calcification. Kidney Int. 106, 611–624 (2024).
Ma, W. et al. PDK4 promotes vascular calcification by interfering with autophagic activity and metabolic reprogramming. Cell Death Dis. 11, 991 (2020).
Acknowledgements
We thank all the subjects who participated in this study. This work was supported by the Hebei Natural Science Foundation (H2023206385).
Author information
Authors and Affiliations
Contributions
M.J.C. and J.S.X. designed the research. M.J.C., J.J.J., D.X.Z., M.X., H.R.Z., and X.Y.Z. performed the follow-up experiments. J.J.J. collected data and performed calculations. M.J.C. wrote this manuscript. J.J.J. supervised the language editing. S.L.Z. and Y.L.B. provided critical ideas. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
These authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Rui Qu, Viktória Jeney, and Zhongyu Han for their contribution to the peer review of this work. Primary Handling Editors: Ashwani Gupta & Rosie Bunton-Stasyshyn.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cheng, M., Jin, J., Zhang, D. et al. METTL3 obstructs vascular smooth muscle cells osteogenic reprogramming by methylating Runx2 in chronic kidney disease. Commun Biol 8, 582 (2025). https://doi.org/10.1038/s42003-025-07972-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-07972-6
This article is cited by
-
METTL3 promotes vascular stability in intracranial aneurysm via m6A-AMPK axis
Scientific Reports (2025)