Abstract
Non-obstructive azoospermia, a severe form of male infertility caused by spermatogenic failure (SPGF), has a largely unknown genetic basis across ancestries. To our knowledge, this is the first trans-ethnic meta-analysis of genome-wide association studies on SPGF, involving 2255 men with idiopathic SPGF and 3608 controls from European and Asian populations. Using logistic regression and inverse variance methods, we identify two significant genetic associations with Sertoli cell-only (SCO) syndrome, the most extreme SPGF phenotype. The G allele of rs34915133, in the major histocompatibility complex class II region, significantly increases SCO risk (P = 5.25E-10, OR = 1.57), supporting a potential immune-related cause. Additionally, the rs10842262 variant in the SOX5 gene region is also a genetic marker of SCO (P = 5.29E-09, OR = 0.72), highlighting the key role of this gene in the male reproductive function. Our findings reveal shared genetic factors in male infertility across ancestries and provide insights into the molecular mechanisms underlying SCO.
Similar content being viewed by others
Introduction
Infertility represents a significant health concern, showing a consistent increase in global disease burden during the last 30 years1. It has been estimated that around 15% of couples attempting to conceive are affected by this problem, with male factors contributing to around half of the infertility cases2. These factors often involve issues such as low sperm counts or poor sperm quality, either due to an obstruction of the post-testicular tract or to spermatogenesis failure (SPGF)3,4. In this sense, non-obstructive azoospermia (NOA) due to SPGF, which affects approximately 1% of all adult men, represents the most severe form of male infertility, since it is characterized by a complete absence of sperm in the ejaculate due to severely impaired spermatogenesis5. SPGF can also lead to severe oligozoospermia (SO), characterized by drastically reduced sperm counts in the semen5.
Known genetic causes of SPGF include karyotype abnormalities, such as Klinefelter syndrome, microdeletions in the Y-chromosome AZF locus, and rare mutations in genes with a key role in spermatogenesis6. However, the etiology of around 70% of SPGF cases remains unknown and there is increasing evidence suggesting that this idiopathic form of male infertility may represent a complex trait6,7. The genetic architecture of complex forms of SPGF involves common variation in the human genome, mainly single nucleotide polymorphisms (SNPs), which individually have mild or moderate effects but their additive effect significantly contribute to SPGF susceptibility6.
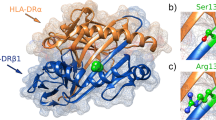
To date, only three well-powered genome-wide association studies (GWASs, in which millions of genetic variants are screened across the entire genome in a hypothesis-free manner8) have been performed in SPGF, two of them in Asian populations and a recently published study in a European population9,10,11. These studies identified several risk variants associated with SPGF susceptibility, with the major histocompatibility complex (MHC) region showing the strongest association. Notably, the European study yielded strong evidence that this human leukocyte antigen (HLA) association was specific to the most extreme histological pattern of NOA11, defined by the Sertoli cell-only (SCO) phenotype, which is characterized by a complete absence of germ cells in the testis12. At the amino acid level, the SCO-specific risk MHC allele determines the presence of a serine at position 13 of the class II HLA-DRβ1 molecule. The fact that this particular position has been identified as the top association signal for a wide spectrum of autoimmune diseases, clearly suggests that the SCO phenotype may represent an immune-mediated condition11.
Despite efforts to unravel the genetic factors contributing to severe forms of idiopathic male infertility, we are still far from a complete understanding of the intricate molecular mechanisms involved. Additionally, a substantial proportion of the heritability remains unexplained, emphasizing the need for further basic research before clinical applications can be implemented. Recent advancements in genotype imputation quality, driven by larger and more diverse reference panels, have greatly benefitted human genetic studies13. In this context, trans-ethnic meta-analyses of imputed GWAS data offer an excellent opportunity to 1) boost the statistical power by pooling together samples from different ancestries, facilitating the detection of novel genetic loci, and 2) take advantage of the comparison of distinct patterns of local linkage disequilibrium (LD) blocks observed in genetically diverse ethnic populations, which allows a more precise identification of causal variants by narrowing down the associated regions and reducing the number of putative causal variants for the different associated loci14.
Taking all the above into consideration, we aimed to shed light into the pathogenic genetic and molecular mechanisms of SPGF by conducting a trans-ethnic meta-analysis of this form of male infertility by combining the GWAS data generated by Hu et al. in Asians9 and those generated by Cerván-Martín et al. in Europeans11.
Results
Trans-ethnic genome-wide association study meta-analysis
A trans-ethnic GWAS meta-analysis was conducted to shed light into the genetic component of idiopathic SPGF. This analysis integrated previously generated GWAS data from populations of European (including both SO and NOA with a histological diagnosis) and Asian (including only NOA without a histological diagnosis) descent9,11.
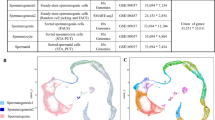
No genetic associations at the genome-wide level of significance were observed when the study of “NOA vs controls” from Asians was meta-analyzed with that of “overall SPGF vs controls” or “NOA vs controls” from Europeans (Fig. 1A, B, respectively). However, the meta-analysis between the “NOA vs controls” study from the Asian GWAS and the “SCO vs controls” study from the European GWAS revealed two significant association signals, defined by the SNPs rs34915133 (P = 5.25E−10, odds ratio [OR] = 1.57, 95% confidence interval [CI] = 1.36–1.81, MAFEUROPEANS (case/controls) = 0.20/0.11, MAFCHINESE (case/controls) = 0.16/0.11) and rs10842262 (P = 5.29E−09, OR = 0.72, 95% CI = 0.65–0.81, MAFEUROPEANS (case/controls) = 0.40/0.44, MAFCHINESE (case/controls) = 0.61/0.69) (Fig. 1C and Table 1). Although some heterogeneity in effect sizes was observed for rs34915133 (Q = 0.03), the ORs for the minor allele were consistent across studies, showing the same direction towards risk (ORIBERIANS = 2.47, ORGERMANS = 1.67, ORCHINESE = 1.42). No significant heterogeneity in ORs was observed for rs10842262 across studies (Q = 0.11), showing all of them protective effects for the minor allele (ORIBERIANS = 0.97, ORGERMANS = 0.71, ORCHINESE = 0.68). According to the variant-to-gene algorithm of Open Targets Genetics15, the genomic regions functionally implicated by these variants were the MHC class II locus (specifically the genes HLA-DQA1 and HLA-DRB1) for rs34915133, and the SOX5 gene for rs10842262 (Fig. 1C).
The inverse variance meta-analysis was performed by combining the results of the “NOA vs controls” comparison from Asians with those of the “SPGF vs controls” (A), “non-obstructive azoospermia (NOA) vs controls” (B), and “Sertoli cell-only syndrome (SCO) vs controls” (C) comparisons from Europeans. The red line represents the genome-wide level of significance (P-value < 5E−08).
Suggestive associations close to the threshold of significance (P ≤ 1E−07) were also observed in the three meta-analyses (Table 1). Regarding the meta-analysis accordingly with overall SPGF, rs7873478 emerged as the top signal (P = 2.92E−07, OR = 1.24, 95% CI = 1.14–1.35, MAFEUROPEANS (case/controls) = 0.54/0.48, MAFCHINESE (case/controls) = 0.24/0.21), with the DMRT1 gene primarily affected by this SNP. In the meta-analysis according to NOA, rs34915133 and rs13206743, impacting the loci HLA-DQA1/HLA-DRB1 and IL17A, respectively, were identified as the SNPs with the lowest P-values. However, as previously mentioned, the rs34915133 variant exhibited a stronger association when considering SCO. Finally, two additional variants, rs4492992 and rs112307255, showed a trend of association in the SCO-specific meta-analysis, influencing the genes TJP1 and TAF1B, respectively. All these suggestive signals displayed no significant heterogeneity in ORs across studies (Q > 0.05), except for the NOA-related variant IL17A-rs13206743 (Q = 0.02, I2 = 74.50) (Table 1).
Overrepresentation of Sertoli-cell only patients in the Asian cohort
The previous SPGF GWAS conducted in the European population suggested that SCO should be considered as a genetically homogeneous entity with a distinct genetic and molecular basis compared to other NOA phenotypes11. According to such study, SCO may represent an immune-mediated condition primarily associated with the MHC class II region11. Consequently, the strong association between MHC and NOA reported in the Asian GWASs for this condition led us to speculate that the NOA cohorts analyzed in those studies could have an overrepresentation of SCO patients11.
To explore this hypothesis, we calculated two polygenic risk scores (PRSs) for each participant within the Asian cohort analyzed here. These scores were derived using the summary statistics data from the comparative analyses between “SCO vs controls” and “SPGF patients without SCO vs controls”, within the European cohort. Regarding the first (PRSs based on SCO data), we observed that the best-fitting model (PRS R2 = 0.0025; P-value = 0.03) included 1077 independent SNPs with P-values < 1.87E−03. Conversely, the PRSs based on non-SCO data, including other SPGF phenotypes such as SO, hypospermatogenesis (HS, in which all stages of spermatogenesis are present in the testis but in an extremely low number), and maturation arrest (MA, with incomplete differentiation of the germline), showed a best-fitting model (PRS R2 = 0.0041; P-value = 0.005) including 12 independent SNPs with P-values < 6.71E−05 (Supplementary Fig. 1, Supplementary Table 1).
Notably, the Asian NOA cases and controls showed opposite PRS distributions when comparing scores derived from SCO data (control group mean = 0.0304, case group mean = 0.0307, t-test P-value = 1.04E−01) and from non-SCO data (control group mean = −0.0291, case group mean = −0.0356, t-test P-value = 4.63E−03), with controls manifesting higher average PRS values than the NOA group in the former case (Supplementary Fig. 1, Supplementary Table 1).
Functional relevance of Sertoli cell-only associated variants
To elucidate the functional implications of genetic associations with SCO susceptibility identified in the trans-ethnic meta-analysis, we systematically examined functional annotations for the lead variants and their proxies (R2 ≥ 0.8) utilizing publicly available databases and bioinformatics tools.
Regarding the MHC association on chromosome 6, five proxies of rs34915133 (all with R2 = 0.99) exhibited strong functional relevance. Particularly, the rs113705304 variant showed the maximum pathogenicity score in RegulomeDB (1a); while rs34831921, rs201769738, rs35001273, and rs73728618 exhibited the highest FunSeq2 values (1.30), with rs35001273 additionally displaying the highest CADD score (14.51). Furthermore, these variants affected the promoter regions of various cell types (including those from fetal thymus) and overlapped with the CCCTC-binding factor (CTCF) site in monocytes CD14+ and B cells, among others. Additionally, these SNPs were predicted to alter the binding sites of numerous proteins and transcription factors known to play pivotal roles in testicular function. Lastly, these variants overlapped with multiple histone marks in several immunological cells such as B and T cells, natural killer cells, and neutrophils. These SNPs were located between the HLA-DRB1 and HLA-DQA1 genes, acting as both expression and splicing quantitative trait loci (eQTL and sQTL, respectively) of several MHC class II genes in the testis (Fig. 2, Supplementary Data 1).
The different functional features are represented with specific colors, with color grade correlating with the probability of functional impact for each tested variant (darker colors indicate higher probability). This figure summarized the information provided in Supplementary Table 2. CADD combined annotation dependent depletion score, ChIP-seq proteins bound from chromatin immunoprecipitation flowing by sequencing experiments in the testis (using ENCODE data), eQTL expression quantitative trait locus effects in the testis, GRCh38 genome reference consortium human build 38, sQTL splicing quantitative trait locus effects in the testis, TFBS transcription factor binding sites modifications related to transcription factors involved in spermatogenesis based on protein weight matrix data.
Notably, the G allele of the lead SNP (rs34915133) was found to be completely linked (D’ = 1) with the G allele of the multiallelic polymorphism that best explained the MHC association with SCO in Europeans (rs1136759). This variant defines the presence of serine at position 13 of the HLA-DRβ1 molecule, impacting antigen presentation to CD4+ T cells11.
In relation with the SOX5 signal (rs10842262), only two proxies were identified, i.e. rs2418156 (R2 = 0.89) and rs7962638 (R2 = 0.80). Both variants were located in an intronic region of the SOX5 gene and overlapped with histone marks in multiple tissues, as well as with binding sites of several spermatogenesis-related proteins such as SMAD and HDAC. Both variants exhibited similar FunSeq2 scores (0.16); however, rs2418156 displayed a higher CADD score (7.37), while rs7962638 was assigned a more relevant score in RegulomeDB (4). Additionally, the latter variant was reported to interact with several proteins in ChIP-seq experiments (Fig. 2, Supplementary Data 1).
The suggestive association haplotypes within the TAF1B and TJP1 regions also included SNPs with strong evidence of functionality (Supplementary Data 1). Both genes were predominantly expressed in testicular tissues (Supplementary Fig. 2), and several TAF1B genetic variants were annotated in the GTEx database as eQTL and sQTL in the testis, with highly relevant RDB scores (i.e. 1b and 1f; see Supplementary Data 1).
Further functional annotation analyses using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) and the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) revealed that the proteins encoded by both genes are essential for maintaining transcriptional and cellular architecture (Supplementary Data 2 and Supplementary Fig. 3). TAF1B plays a critical role in RNA polymerase I transcription and rRNA synthesis, with key domains such as the RRN7-type zinc finger facilitating DNA binding and transcriptional regulation. TJP1, on the other hand, is crucial for cell-cell junction assembly, including tight junctions, and supports cytoskeletal organization and cell-cell barrier integrity, with PDZ and SH3 domains driving protein interactions. Interestingly, genetic regions encoding components of the molecular networks linked to TAFB1 and TJP1 harbor variations showing some evidence of genetic association with SCO (Supplementary Data 3).
Finally, a functional enrichment analysis was performed on the grey zone (i.e., signals with P-values < 1E−05) from our trans-ethnic GWAS meta-analysis to further investigate the genetic basis of this condition (Supplementary Table 2). The Multi-marker Analysis of GenoMic Annotation (MAGMA) gene-set analysis identified the “WP_SARSCOV2_B117_VARIANT_ANTAGONISES_INNATE_IMMUNE_ACTIVATION” pathway as the most significantly associated with SCO (P = 1.05E−05, β = 1.21). This pathway also emerged as the top signal in the MAGMA analyses of SPGF and NOA grey zones. However, SCO demonstrated unique enrichment for other immune-related pathways, including the NLRP3 inflammasome complex and MHC class II antigen presentation, highlighting its distinct immunological profile (Supplementary Table 2).
Phenome-wide association study
A phenome-wide association study (PheWAS) was conducted to complement the findings from our GWAS, allowing us to evaluate the roles of the most relevant risk variants (i.e. those reported in Table 1) across a broad spectrum of phenotypes. This approach provided insights into potential pleiotropic effects, where a single genetic variant may influence multiple traits. Several significant traits associated with our male infertility signals also implicated other reproductive system-related issues (Supplementary Data 4–9). For example, DMRT1-rs7873478 was linked to “bioavailable testosterone levels” (P = 0.0043, β = 0.0084), and IL17A-rs13206743 was associated with both “sex hormone-binding globulin levels” (P = 0.0040, β = 0.0036) and “benign neoplasm of male genital organs” (P = 0.0025, β = 0.2332) (Supplementary Data 4 and 5). Interestingly, the SCO-risk variants showed associations with traits related to female reproductive system dysfunction, including “endometriosis of ovary” for HLA-DQA1/HLA-DRB1-rs34915133 (P = 0.0046, β = 0.1031), “endometriosis of fallopian tube” for SOX5-rs10842262 (P = 0.0045, β = −0.3474), “pain and other conditions associated with female genital organs and menstrual cycle” for TJP1-rs4492992 (P = 0.0024, β = 0.0876), and “ovarian dysfunction” for TAF1B-rs112307255 (P = 0.0026, β = 0.1591) (Supplementary Data 6–9).
Apart from reproductive traits, the PheWAS analysis also revealed associations with immune system conditions, especially when considering the SCO variants. Those included “autoimmune diseases” for HLA-DQA1/HLA-DRB1-rs34915133 (P = 4.06E−67, β = −0.2198), “ulcerative colitis” for SOX5-rs10842262 (P = 0.0041, β = 0.1760), “circumscribed scleroderma” for TJP1-rs4492992 (P = 0.0024, β = 0.6640), and “multiple sclerosis” for TAF1B-rs112307255 (P = 0.0049, β = 0.0863) (Supplementary Data 6–9). Evaluation of previously reported associations with spermatogenic failure.
To check the association of variants previously associated with NOA and SCO in Asian and European cohorts in our trans-ethnic meta-analysis, we evaluated the statistical significance of these genomic regions (±0.5 Mbp centered in the lead SNP) within our dataset and compared them with their reported association signals in the independent European and Asian studies.
Only one SNP, rs13206743, located within the IL17A region, showed a P-value < 0.05 in both Asian (P = 1.11E−06) and European (PNOA = 2.32E−03, PSCO = 2.32E−02) studies independently, exhibiting a P-value close to the statistical significance in the meta-analysis considering either NOA (P = 4.19E−07) or SCO (P = 5.52E−07) (Supplementary Data 10). Furthermore, the OR directions remained consistent across studies, with the C allele indicating a risk effect for NOA and the SCO histological phenotype in the independent European and Asian cohorts as well as in the trans-ethnic meta-analysis (Supplementary Data 10). This same SNP emerged as the top signal in the trans-ethnic meta-analysis for both NOA and SCO phenotypes within the analyzed 1 Mbp region (Supplementary Data 11).
Discussion
Trans-ethnic meta-GWASs have proven very useful in advancing our understanding of the genetic architecture of complex conditions, such as autoimmune and cardiovascular diseases, diabetes, cancer, and COVID-1916,17,18,19,20,21. By performing a trans-ethnic meta-analysis of GWAS data in SPGF and subsequent functional annotation analyses, we have been able to advance our understanding about the genetic component of this condition. While the relevant role of the MHC class II locus in NOA development was previously recognized6, here we provide further evidence that this association is specific to the SCO phenotype by calculating PRSs predictive for SCO risk within the Asian GWAS cohort. Indeed, the strongest MHC association signal was observed when integrating data from the Chinese NOA study with that of the European SCO study. This finding reinforces our previous idea that this extreme male infertility histological pattern represents a genetically homogeneous clinical entity with an immune-mediated origin11.
In this regard, the top SCO-associated MHC allele in our meta-analysis accordingly with SCO (rs34915133*G) exhibits complete LD with a serine residue at position 13 of the HLA-DRβ1 protein, known to be crucial in the antigen presentation to T cells22,23. It has been recently reported that this site substantially increases autoimmune predisposition by influencing amino acid composition within the T-cell receptor complementarity-determining region 3 (TCR-CDR3)24. Indeed, the codon that defines HLA-DRβ1 13 is highly polymorphic, and specific residues at this position are known to drive the major risk of multiple autoimmune diseases25,26,27,28. Additionally, our in silico functional annotation analyses suggested that the pathogenic impact of this haplotype block could also extend to the modulation of gene expression in class II HLA genes through eQTL and sQTL effects in testicular immune cells, as reported in other diseases involving the immune system29.
These observations support the hypothesis that SCO is predominantly an immune-mediated condition of NOA, which may result from the loss of the immune privilege of the testis (a mechanism essential for preventing autoimmune responses against germline cells)11,30. Beyond male infertility, parallels can be drawn to inflammatory processes or autoimmune reactions targeting reproductive tissues in women. For instance, ovarian endometriosis (a condition in which endometrial tissue is present in the ovary leading to infertility) is characterized by chronic inflammation and immune activation. Notably, high-precision single-cell RNA sequencing has recently revealed elevated expression of the MHC class II complex in epithelial cells within ectopic endometrial lesions of affected women, thus activating CD4+ T cells and perpetuating chronic inflammation31. The above is consistent with our PheWAS analysis, in which endometriosis and other immune-mediated conditions were linked to the SCO-associated variants. Overall, these similarities highlight shared biological mechanisms underlying immune-mediated infertility across sexes. Future research should aim to comprehensively elucidate these molecular pathways and evaluate their potential for therapeutic intervention.
The identification of the HLA-DRβ1 site 13 as a primary risk factor for male infertility in Asians may had not been possible in the MHC fine mapping performed by Huang et al. 32 because of the inclusion of not only SCO patients but also other NOA phenotypes. Although the authors were able to establish the classical haplotype HLA-DRB1*1302 (which includes a serine at position 13) as a key MHC contributor for NOA susceptibility, the statistical noise coerced by the lack of histological phenotype information potentially hindered the analysis at the amino acid level. The data presented here strongly support this assumption.
Regarding the implication of the SOX5 region in male infertility, the lead variant of this association signal (rs10842262) has been reported to increase the genetic predisposition to NOA across various Chinese and Serbian populations9,33,34,35. This fact highlights rs10842262 as a pivotal genetic determinant in the pathogenesis of NOA, particularly within the context of SOX5 gene regulation. However, our results indicate that this is likely another SCO-specific association. We would like to note that the lack of histological phenotype information for the NOA cohorts in the Chinese and Serbian studies precludes definitive confirmation of this observation. Thus, while the rs10842262 effect on extreme patterns of male infertility is well-established, further investigation integrating detailed phenotype characterization is warranted to elucidate the precise phenotypic manifestations of this genetic association.
SOX5 encodes a member of the SRY-related HMG-box family of transcription factors, which are involved in the regulation of different developmental processes including sex determination and differentiation36. In humans, SOX5 exhibits predominant expression in the testis, particularly in spermatocytes and round spermatids37,38. Its role in the testis development and function seems to be highly conserved. In this regard, a recent study on the fish Chinese tongue sole revealed that SOX5 was mainly expressed in spermatogonia, with expression levels decreasing as differentiation progresses39. Interestingly, the authors observed that SETDB1, the predominant histone lysine methyltransferase catalyzing H3K9me3, is implicated in SOX5 transcription in gonads, which provides molecular insights into the relevance of the epigenetic control of SOX5 expression during sex differentiation and gametogenesis39.
Other genetic regions showing trends of association with SPGF in our meta-analysis included DMRT1 (particularly evident when meta-analyzing the Asian NOA study with the overall SPGF European study), IL17A (with the strongest signal observed when the Asian NOA study was meta-analyzed with the NOA European study), as well as TJP1 and TAF1B (observed in the meta-analysis accordingly with SCO).
DMRT1 plays a critical role in male fertility, with deleterious and rare variants within this gene being reported as NOA genetic markers40,41. Its encoded protein is a key player in male sex determination and differentiation. Moreover, this transcription factor is essential in both Sertoli cells and spermatogonia, preventing female reprogramming and maintaining the proliferative state, respectively, within the adult testis, highlighting its multifaceted importance in maintaining the male reproductive system42,43,44.
IL17A is a previously known NOA-associated gene that encodes a proinflammatory cytokine produced by activated T cells45. Although baseline levels of IL-17 are observed in healthy human testes, an increased IL-17A expression are usually detected in azoospermic testes with chronic inflammation, potentially leading to damage of the blood-testis barrier (BTB) and disrupting normal spermatogenesis46,47.
The BTB divides the seminiferous epithelium into two different compartments, harboring spermatogonia and preleptotene spermatocytes in the basal compartment and the remaining germ cell types in the adluminal compartment. Its primary roles encompass creating a suitable microenvironment for germ cell maturation and protecting germ cells from immune cell interaction with auto-antigens48. One of the proteins that constitute the Sertoli cell-Sertoli cell junctional complex of the BTB is TJP1, which acts as both a tight junction adaptor and a regulator of adherens junctions49,50. This aligns with the results of our in silico analysis, which revealed high expression of TJP1 in testicular tissues. Consequently, deregulation of the TJP1 gene could compromise the structural integrity of this protective barrier, potentially resulting in germ cell loss. Notably, delocalization of TJP1 has been identified in the testes of rats with experimental autoimmune orchitis51. The observed trend of association of this gene with the SCO phenotype in our study aligns with the aforementioned findings.
Regarding TAF1B, although there is no reported evidence that this gene is directly implicated in human fertility thus far, recent studies have revealed an association between TAF1B and the incidence of stillborn births in large white pigs52. Interestingly, the functional characterization of the TAF1B signal from our meta-analysis based on SCO revealed that several risk variants function as sQTL for this gene in the testis. Given the critical role of TAF1B in rRNA transcription53, these findings suggest that alterations in the transcript isoform ratio may disrupt ribosomal function and cellular metabolism, potentially contributing to the SCO phenotype.
In conclusion, this is a pioneering study that integrates genomic data from SPGF patients of different ancestries, allowing a deeper exploration of the molecular mechanisms underlying idiopathic forms of male infertility. The identification of shared risk polymorphisms across European and Asian populations, specially under the context of SCO, brings us closer to establishing genetic panels predictive of the histological affectation of the testis in NOA patients, thus preventing unnecessary surgeries for testicular sperm extraction54. However, the lack of detailed phenotype data in the analyzed Chinese cohort represent an important limitation of this study. While consistent diagnostic criteria were applied for inclusion, differences in sample composition across the study cohorts may have contributed to heterogeneity in the dataset. This heterogeneity, along with the limited sample size, weakened the ability to draw definitive conclusions about genetic markers across ancestries. Although the identification of shared variants is promising, it does not yet provide definitive evidence of consistent genetic effects between populations at this time. Establishing large and well-characterized SCO cohorts from Asian populations will be essential to confirm our findings. Moreover, additional post-GWAS analyses, such as LD score regression, could provide insights into the heritability and genetic correlations of SCO with related traits, including immune-mediated disorders. While beyond the scope of this study, we are pursuing a complementary analysis to explore these questions, which may further shed light on the immune-related pathways underlying SCO.
Methods
Study population
In this study, we analyzed two independent case-control cohorts comprising individuals of European and Asian ancestry. The dataset included whole-genome genotype information from 2255 individuals diagnosed with idiopathic SPGF and 3608 unaffected controls, extracted from two previously published GWASs of this condition9,11.
In brief, a total of 1274 infertile men diagnosed with idiopathic SPGF from Iberian Peninsula and Germany were recruited in different public health centers and private fertility clinics from Spain and Portugal, and at the Centre of Reproductive Medicine and Andrology (University Hospital Münster, Germany), respectively. Additionally, 981 Han Chinese men diagnosed with idiopathic NOA were recruited from the Centre of Reproductive Medicine in Nanjing. The European SPGF cohort comprised 494 SO cases (defined as having fewer than 5 million spermatozoa per milliliter of semen due to SPGF) and 780 NOA cases (showing total absence of seminal spermatozoa). Amongst these NOA patients, 120 also had a histological diagnosis of HS, 98 of MA, and 214 of SCO following testicular biopsy. The biopsy results also allowed their classification based on whether testicular sperm retrieval was successful or unsuccessful (Table 2). No histological data was available for the Asian cohort.
The recruitment process adhered strictly to predefined criteria as previously described9,11, in accordance with approved guidelines from the American Urological Association (AUA)/American Society for Reproductive Medicine (ASRM), the Canadian Urological Association (CUA), and the World Health Organization (WHO, 2010)55,56,57. Patients were included in the study based on comprehensive andrological evaluations, which encompassed a review of medical history, physical examination, semen analysis, hormone profiling (FSH, LH, and testosterone), karyotyping, and Y-chromosome microdeletion screening. To ensure diagnostic accuracy, azoospermia was confirmed by the absence of spermatozoa in two semen samples after high-speed centrifugation. Patients with known causes of male infertility, including cryptorchidism, orchitis, testicular malformations, obstruction of the vas deferens, chromosomal abnormalities, or Y-chromosome microdeletions, were excluded. All enrolled participants provided informed written consent prior to inclusion in the study. The procedures followed were in accordance with the tenets of the Declaration of Helsinki and were approved by the Ethics Committee ‘CEIM/CEI Provincial de Granada’ (Andalusia, Spain) in a session held on January 26, 2021 (approval number: 1/21). All ethical regulations relevant to human research participants were followed. The research involved collaboration with local researchers in Spain, Portugal, and China, who contributed to study design, data collection, and authorship. Recruitment followed international clinical guidelines (AUA, ASRM, CUA, WHO), and patient safety and confidentiality were ensured. No biological materials or traditional knowledge were transferred across borders, and all relevant local studies were cited.
Genotype imputation and quality control
Sample genotyping and quality controls of raw data from both analyzed cohorts were described elsewhere9,11.
Given the extensive reference panels currently available for SNP genotype imputation, we decided to maximize the genetic coverage of both original datasets by performing an imputation process with Minimac458, using the NHLBI Trans-OMICs for Precision Medicine’ (TOPMed) program (freeze 5)59 as reference for Europeans and the Nanjing Lung Cancer Cohort (NJLCC)60 for Asians. Haplotype phasing was conducted with Eagle v.2.4.61 and SHAPEIT462 for European and Asian data, respectively.
To ensure the high quality of the imputed data, variants with estimated imputation quality Rsq < 0.9 and rare variants with minor allele frequency (MAF) < 0.005 were set to missing. Additionally, the imputed dataset underwent thorough quality control (QC) procedures utilizing R and PLINK 1.9 software63. These procedures included the removal of SNPs with call rates below 98% and those exhibiting genotype frequencies deviating from Hardy−Weinberg equilibrium (HWE) at a significance level of P-value < 0.001. In order to control for population stratification, we performed a principal component (PC) analysis using R, PLINK 1.9, and EIGENSTRAT 3.0 software64.
Statistics and reproducibility
Case-control comparisons for each analyzed population, including two European cohorts (Iberian and German) and one Chinese cohort, were carried out employing PLINK 1.9 through logistic regression on the best-guess genotypes (Rsq > 0.9) and assuming additive effects. The covariates included in the logistic regression models were the 10 first principal components (PCs) and country of origin for Europeans, as well as the first PC and age for Asians. Specifically, three comparisons were performed with the European dataset: (1) SPGF (NOA + SO) vs controls, (2) NOA vs controls, and (3) SCO vs controls. Conversely, due to the lack of histological phenotype information for the Asian cohort, only the NOA vs controls comparison was feasible. For each variant in all comparisons, ORs, along with their corresponding 95% CIs, standard errors (SE), and P-values, were computed.
To identify trans-ethnic genetic associations with SPGF, combined analyses between the Asian NOA group and the European SPGF, NOA, and SCO groups were conducted by meta-analyzing the summary statistics derived from the individual logistic regression analyses of each population (Fig. 3). Meta-analyses were conducted using the inverse variance method under a fixed effects model using PLINK 1.9. To assess the potential heterogeneity in effect sizes between the different studies, I2 and Cochran’s Q tests were employed. Genome-wide statistical significance for the meta-analysis was established at a P-value < 5E−08. Manhattan plots were generated using a custom R script. Only variants showing statistical significance in the meta-analysis, displaying consistent ORs across all populations (towards the same direction), and with P-values lower than 0.05 in the Chinese study and in any of the two European cohorts (Iberian or German), were considered associated.
Enrichment analysis of Sertoli cell-only phenotype using a polygenic risk score
We previously hypothesized that the Asian NOA cohort examined in this study may exhibit a significant enrichment of SCO patients based on the reported HLA associations11. To evaluate this assumption, we used the PRSice-2 software65 to compute two distinct PRSs for each participant within the Asian NOA cohort: (1) The first PRS was derived by adding the number of risk alleles (0, 1, or 2) per SNP in the summary statistics corresponding to the European “SCO patients vs controls” analysis, each multiplied by the natural logarithm of their respective ORs; (2) The second PRS was computed similarly, but utilizing a new summary statistics obtained from the analysis of European “SPGF patients not exhibiting SCO (non-SCO SPGF patients) vs controls”, that is, those infertile men showing histological phenotypes distinct from SCO, such as MA, HS, or SO). The main goal was to ascertain whether the genetic architecture of the Asian NOA cohort more closely resembled that of European SCO patients than non-SCO SPGF patients or controls.
LD clumping was conducted with a cutoff of R2 = 0.1 to identify a set of independent SNPs that retained as much association signal as possible from the European discovery GWAS. Various models were then fitted by selecting clumped variants surpassing different P-value thresholds in the European GWAS summary statistics (defined by the argument—bar-levels 5e−11, 5e−10, 5e−09, 5e−08, 5e−07, 5e−06, 5e−05, 0.0001, 0.001, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 1), using the first PC and the age as covariates.
In silico characterization of associated variants
To understand the functional implications of suggested associated variants with NOA, and their proxy variants (R2 ≥ 0.8, identified via LDlink [30]), we conducted a comprehensive in silico functional characterization of the lead SNPs and their proxies (R2 ≥ 0.8) leveraging annotated evidence from public online databases and diverse bioinformatics resources.
The potential role of these variants as eQTL and sQTL in the testis was explored using the GTEx database66. Additionally, relevant information regarding variant functionality and the genes in which associated variants were implicated functionally was extracted from SNPnexus67 and Haploreg v.4.168, which integrate datasets from Ensembl, SIFT, Polyphen, CpG, Vista enhancers, miRbase, TarBase, TargetScan, Roadmap, Ensembl regulatory build, CADD, DeepSEA, EIGEN, FATHMM, fitCons, FunSeq2, GWAVA, and REMM.
Furthermore, RegulomeDB69 was consulted to identify protein and transcription factor binding sites overlapping with the analyzed variants, along with their respective pathogenicity scores. The used scores provide insights into the likelihood of each variant being functional or potentially pathogenic. Specifically, the CADD and FunSeq2 scores range from 1 to 99 and 0 to 6, respectively, with higher scores indicating a greater likelihood of functionality or pathogenicity. The RegulomeDB score ranges from 1 to 7, with 1 indicating higher and 7 lower pathogenicity likelihood.
To place the putative NOA genes within a broader biological network, we further explored their potential biological functions using the MAGMA tool integrated into Functional Mapping and Annotation of Genome-Wide Association Studies (FUMA)70, as well as DAVID71. Additionally, we conducted pathway analyses with STRING72 to examine protein-protein interaction networks, aiming at identifying potential interaction partners for these genes.
Finally, a PheWAS analysis was conducted to identify pleiotropic effects of the top signals using the tool for that purpose of the Open Targets Genetics platform15. This approach aimed to uncover the broader implications of the most relevant genetic variants associated with male infertility phenotypes, maximizing the insights gained from our GWAS findings.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The summary statistics of this study are available through the NHGRI-EBI GWAS Catalog (https://www.ebi.ac.uk/gwas/downloads/summary-statistics) through the accession numbers GCST90558278, GCST90558279, and GCST90558280. The accession numbers for the Chinese (NOA) and the three European original studies (SPGF, NOA, and SCO) are GCST001362, GCST90239716, GCST90239717, and GCST90239721, respectively. Individual-level genotype data will not be publicly available because they could compromise the privacy of participants and the informed consent. The source data for the plots in Supplementary Fig. 1 are included in Supplementary Data files 12–15. All other data are contained either in the article file and its supplementary information or available upon reasonable request to the corresponding authors, considering legal restrictions on transferring individual-level data under both European and Chinese law at the time of the inquiry.
Change history
16 April 2025
In the Acknowledgement section, the funding statement has been rephrased.
References
Sun, H. et al. Global, regional, and national prevalence and disability-adjusted life-years for infertility in 195 countries and territories, 1990-2017: results from a global burden of disease study, 2017. Aging 11, 10952–10991 (2019).
Cox, C. M. et al. Infertility prevalence and the methods of estimation from 1990 to 2021: a systematic review and meta-analysis. Hum. Reprod. open 2022, hoac051 (2022).
Pan, M. M., Hockenberry, M. S., Kirby, E. W. & Lipshultz, L. I. Male infertility diagnosis and treatment in the era of in vitro fertilization and intracytoplasmic sperm injection. Med. Clin. North Am. 102, 337–347 (2018).
Krausz, C. & Riera-Escamilla, A. Genetics of male infertility. Nat. Rev. Urol. 15, 369–384 (2018).
Tournaye, H., Krausz, C. & Oates, R. D. Novel concepts in the aetiology of male reproductive impairment. Lancet Diab. Endocrinol. 5, 544–553 (2017).
Cervan-Martin M., Castilla J. A., Palomino-Morales R. J. & Carmona F. D. Genetic landscape of nonobstructive azoospermia and new perspectives for the clinic. J. Clin. Med. 9, 300 (2020).
Cervan-Martin, M. et al. Changes in environmental exposures over decades may influence the genetic architecture of severe spermatogenic failure. Hum. Reprod. 39, 612–622 (2024).
Loos, R. J. F. 15 years of genome-wide association studies and no signs of slowing down. Nat. Commun. 11, 5900 (2020).
Hu, Z. et al. A genome-wide association study in Chinese men identifies three risk loci for non-obstructive azoospermia. Nat. Genet. 44, 183–186 (2011).
Zhao, H. et al. A genome-wide association study reveals that variants within the HLA region are associated with risk for nonobstructive azoospermia. Am. J. Hum. Genet. 90, 900–906 (2012).
Cervan-Martin, M. et al. Immune and spermatogenesis-related loci are involved in the development of extreme patterns of male infertility. Commun. Biol. 5, 1220 (2022).
Ghanami Gashti, N., Sadighi Gilani, M. A. & Abbasi, M. Sertoli cell-only syndrome: etiology and clinical management. J. Assist. Reprod. Genet. 38, 559–572 (2021).
Das, S., Abecasis, G. R. & Browning, B. L. Genotype imputation from large reference panels. Annu. Rev. Genomics Hum. Genet. 19, 73–96 (2018).
Li, Y. R. & Keating, B. J. Trans-ethnic genome-wide association studies: advantages and challenges of mapping in diverse populations. Genome Med. 6, 91 (2014).
Ghoussaini, M. et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 49, D1311–D1320 (2021).
Okada, Y. et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506, 376–381 (2014).
Ha, E., Bae, S. C. & Kim, K. Large-scale meta-analysis across East Asian and European populations updated genetic architecture and variant-driven biology of rheumatoid arthritis, identifying 11 novel susceptibility loci. Ann. Rheum. Dis. 80, 558–565 (2021).
Matsunaga, H. et al. Transethnic meta-analysis of genome-wide association studies identifies three new loci and characterizes population-specific differences for coronary artery disease. Circ. Genom. Precis. Med. 13, e002670 (2020).
Mahajan, A. et al. Multi-ancestry genetic study of type 2 diabetes highlights the power of diverse populations for discovery and translation. Nat. Genet. 54, 560–572 (2022).
Byun, J. et al. Cross-ancestry genome-wide meta-analysis of 61,047 cases and 947,237 controls identifies new susceptibility loci contributing to lung cancer. Nat. Genet. 54, 1167–1177 (2022).
Wu, P. et al. Trans-ethnic genome-wide association study of severe COVID-19. Commun. Biol. 4, 1034 (2021).
Dimitrov, I., Garnev, P., Flower, D. R. & Doytchinova, I. Peptide binding to the HLA-DRB1 supertype: a proteochemometrics analysis. Eur. J. Med. Chem. 45, 236–243 (2010).
Rossjohn, J. et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu. Rev. Immunol. 33, 169–200 (2015).
Ishigaki, K. et al. HLA autoimmune risk alleles restrict the hypervariable region of T cell receptors. Nat. Genet. 54, 393–402 (2022).
Carmona, F. D. et al. A large-scale genetic analysis reveals a strong contribution of the HLA class II region to giant cell arteritis susceptibility. Am. J. Hum. Genet. 96, 565–580 (2015).
Kim, K. et al. The HLA-DRbeta1 amino acid positions 11-13-26 explain the majority of SLE-MHC associations. Nat. Commun. 5, 5902 (2014).
Hu, X. et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat. Genet. 47, 898–905 (2015).
Raychaudhuri, S. et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat. Genet. 44, 291–296 (2012).
Yazar, S. et al. Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease. Science 376, eabf3041 (2022).
Gong, J., Zeng, Q., Yu, D. & Duan, Y. G. T Lymphocytes and testicular immunity: a new insight into immune regulation in testes. Int. J. Mol. Sci. 22, 57 (2020).
Yan, J. et al. Single-cell analysis reveals insights into epithelial abnormalities in ovarian endometriosis. Cell Rep. 43, 113716 (2024).
Huang, M. et al. Fine mapping the MHC region identified rs4997052 as a new variant associated with nonobstructive azoospermia in Han Chinese males. Fertil. Steril. 111, 61–68 (2019).
Gu, X. et al. PEX10, SIRPA-SIRPG, and SOX5 gene polymorphisms are strongly associated with nonobstructive azoospermia susceptibility. J. Assist. Reprod. Genet. 36, 759–768 (2019).
Zou, S. et al. Association study between polymorphisms of PRMT6, PEX10, SOX5, and nonobstructive azoospermia in the Han Chinese population. Biol. Reprod. 90, 96 (2014).
Nemanja, V. et al. Association study between single-nucleotide variants rs12097821, rs2477686, and rs10842262 and idiopathic male infertility risk in Serbian population with meta-analysis. J. Assist. Reprod. Genet. 37, 2839–2852 (2020).
Sreenivasan, R., Gonen, N. & Sinclair, A. SOX Genes and Their Role in Disorders of Sex Development. Sex Dev. 16, 80–91 (2022).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Karlsson, M. et al. A single-cell type transcriptomics map of human tissues. Science Adv. 7, eabh2169 (2021).
Yue, B. et al. Molecular functional characterization of the setdb1 and its potential target gene sox5 illuminate the histone modification-mediated orchestration of gonadal development in Chinese tongue sole (Cynoglossus semilaevis). Gene 901, 148199 (2024).
Lima, A. C. et al. Rare double sex and mab-3-related transcription factor 1 regulatory variants in severe spermatogenic failure. Andrology 3, 825–833 (2015).
Tang, D. et al. Identification of deleterious variants in patients with male infertility due to idiopathic non-obstructive azoospermia. Reprod. Biol. Endocrinol. 20, 63 (2022).
Matson, C. K. et al. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476, 101–104 (2011).
Zarkower, D. DMRT genes in vertebrate gametogenesis. Curr. Top. Develop. Biol. 102, 327–356 (2013).
Macdonald, J. et al. DMRT1 repression using a novel approach to genetic manipulation induces testicular dysgenesis in human fetal gonads. Hum. Reprod. 33, 2107–2121 (2018).
Hu, Z. et al. Association analysis identifies new risk loci for non-obstructive azoospermia in Chinese men. Nat. Commun. 5, 3857 (2014).
Duan, Y. G. et al. Immunodeviation towards a Th17 immune response associated with testicular damage in azoospermic men. Int. J. Androl. 34, e536–e545 (2011).
Perez, C. V. et al. IL17A impairs blood-testis barrier integrity and induces testicular inflammation. Cell Tissue Res. 358, 885–898 (2014).
Mruk, D. D. & Cheng, C. Y. The Mammalian blood-testis barrier: its biology and regulation. Endocr. Rev. 36, 564–591 (2015).
Fanning, A. S., Jameson, B. J., Jesaitis, L. A. & Anderson, J. M. The tight junction protein ZO-1 establishes a link between the transmembrane protein occludin and the actin cytoskeleton. J. Biol. Chem. 273, 29745–29753 (1998).
Pu, J. et al. TJP1, a membrane-expressed protein, is a potential therapeutic and prognostic target for lung cancer. Technol. cancer Res. Treat. 21, 15330338221106855 (2022).
Perez, C. V. et al. Loss of occludin expression and impairment of blood-testis barrier permeability in rats with autoimmune orchitis: effect of interleukin 6 on Sertoli cell tight junctions. Biol. Reprod. 87, 122 (2012).
Wang, X. et al. GWAS of reproductive traits in large white pigs on chip and imputed whole-genome sequencing data. Int. J. Mol. Sci. 23, 13338 (2022).
Hahn, S. The role of TAFs in RNA polymerase II transcription. Cell 95, 579–582 (1998).
Corona, G. et al. Sperm recovery and ICSI outcomes in men with non-obstructive azoospermia: a systematic review and meta-analysis. Hum. Reprod. Update 25, 733–757 (2019).
Schlegel, P. N. et al. Diagnosis and treatment of infertility in men: AUA/ASRM guideline part I. Fertil. Steril. 115, 54–61 (2021).
Jarvi, K. et al. CUA Guideline: the workup of azoospermic males. Can. Urol. Assoc. J. 4, 163–167 (2010).
Cooper, T. G. et al. World Health Organization reference values for human semen characteristics. Hum. Reprod. Update 16, 231–245 (2010).
Fuchsberger, C., Abecasis, G. R. & Hinds, D. A. minimac2: faster genotype imputation. Bioinformatics 31, 782–784 (2015).
Taliun, D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021).
Wang, C. et al. Analyses of rare predisposing variants of lung cancer in 6,004 whole genomes in Chinese. Cancer cell 40, 1223–1239.e1226 (2022).
Loh, P. R. et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 48, 1443–1448 (2016).
Delaneau, O., Zagury, J. F., Robinson, M. R., Marchini, J. L. & Dermitzakis, E. T. Accurate, scalable and integrative haplotype estimation. Nat. Commun. 10, 5436 (2019).
Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience 4, 7 (2015).
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Choi, S. W. & O’Reilly, P. F. PRSice-2: polygenic risk score software for biobank-scale data. GigaScience 8, giz082 (2019).
The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Oscanoa, J. et al. SNPnexus: a web server for functional annotation of human genome sequence variation (2020 update). Nucleic Acids Res. 48, W185–W192 (2020).
Ward, L. D. & Kellis, M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 44, D877–D881 (2016).
Dong, S. & Boyle, A. P. Predicting functional variants in enhancer and promoter elements using RegulomeDB. Hum. Mutat. 40, 1292–1298 (2019).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Sherman, B. T. et al. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 50, W216–W221 (2022).
Szklarczyk, D. et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646 (2023).
Acknowledgements
This work was supported by the Spanish Ministry of Economy and Competitiveness through the Spanish National Plan for Scientific and Technical Research and Innovation (grant: PID2023-152215OB-I00 funded by MICIU/AEI/10.13039/501100011033 and ERDF, EU; and grant: PID2020-120157RB-I00 funded by MICIU/AEI/10.13039/501100011033) and the National Key Research and Development Program of China (ref. 2021YFC2700201 and 2021YFC2700601). S.G.-M. was funded by grant ref. FPU23/02674. S. La received support from “Instituto de Salud Carlos III” (grant: DTS18/00101], co-funded by FEDER funds/European Regional Development Fund (ERDF)- a way to build Europe-), and from “Generalitat de Catalunya” (grant 2021SGR052). S. La is sponsored by the “Researchers Consolidation Program” from the SNS-Dpt. Salut Generalitat de Catalunya (Exp. CES09/020). The authors thank the support of the Unit of Excellence ‘UNETE’ from the University of Granada (reference UCE-PP2017-05). This article is related to the PhD Doctoral Thesis of Sara González-Muñoz.
Author information
Authors and Affiliations
Contributions
F.D.C., R.P.-M., Z.H., and C.W. were involved in the conception, design, and supervision of the study. S.G.-M., Y.L., A.G.-J., M.C.-M., I.H.-S., L.B.-C., and R.P.-M. participated in the methodology. S.G.-M. and Y.L. performed the formal analysis. S.G.-M., Y.L., A.G.-J., L.B.-C., R.P.-M., C.W., Z.H., and F.D.C. were involved in the interpretation of the data. J.A.C., A.C., N.G., S. Lu, X.Y., X.G., J.L., L.B., S.S., J.G., A.M.L., and S. La, were responsible for study subject and data recruitment. S.G.-M., Y.L., L.B.-C., R.P.-M., C.W., and F.D.C. were involved in the original draft preparation. All authors revised critically and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Prafulla S Ambulkar and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Hélène Choquet and Rosie Bunton-Stasyshyn. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
González-Muñoz, S., Long, Y., Guzmán-Jiménez, A. et al. Trans-ethnic GWAS meta-analysis of idiopathic spermatogenic failure highlights the immune-mediated nature of Sertoli cell-only syndrome. Commun Biol 8, 571 (2025). https://doi.org/10.1038/s42003-025-08001-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08001-2
This article is cited by
-
Genetic and epigenetic insights into non-obstructive azoospermia: mechanisms, biomarkers, and clinical perspectives
Reproductive Biology and Endocrinology (2025)