Abstract
Wolbachia, a widespread endosymbiotic bacterium, profoundly impacts insect hosts by distorting reproduction and population dynamics. Despite extensive laboratory research, its long-term effects on host evolution in nature remain poorly understood, especially the genomic consequences linked to disruptions in sex determination and reproductive processes. We present the first telomere-to-telomere (T2T) genome assembly of the sawfly Analcellicampa danfengensis and the complete genome of its symbiotic Wolbachia. Comparative population genomics across six Analcellicampa species revealed that Wolbachia-infected populations show starkly different demographic signals. While uninfected populations show similar demographic signals for both sexes, infected populations exhibit a lower apparent effective population size (Ne) in males, which may reflect a recurrent male bottleneck effect driven by Wolbachia-induced male scarcity. Genomic scans identified positively selected genes associated with reproductive functions, sensory perception, neural development, and longevity, suggesting that Wolbachia likely manipulates critical host pathways to promote its transmission. These findings provide direct genomic insights into Wolbachia as an evolutionary force, highlighting specific host genes and regions under selection resulting from these altered evolutionary dynamics. This work provides deeper insights into host–endosymbiont coevolution and has important implications for evolutionary theory and pest management strategies.
Similar content being viewed by others
Introduction
Symbiosis in nature showcases the intricate and essential relationships between species. These interactions not only play a pivotal role in biological evolution but also significantly impact modern agriculture and ecosystem diversity1,2,3,4. Among the most extensively studied and evolutionarily remarkable examples of symbiosis is that of Wolbachia pipientis, a bacterium belonging to the order Rickettsiales within the α-proteobacteria5. This obligate intracellular symbiont infects millions of insects worldwide, with estimates suggesting that up to 66% of insect species harboring the infection6. Consequently, Wolbachia infections are considered one of the most widespread symbioses in the history of life, considering the sheer number and diversity of species infected7. By exerting complex effects on the reproductive systems and population dynamics of its hosts, Wolbachia has become a focal point in contemporary biological research.
Wolbachia is well-known for its profound effects on the reproductive systems of insects, often employing mechanisms like cytoplasmic incompatibility, parthenogenesis induction, feminization, and male-killing8. By inducing mechanisms such as cytoplasmic incompatibility, male killing, feminization, and parthenogenesis, Wolbachia significantly enhances its transmission and persistence within host populations8. These characteristics have made Wolbachia a promising candidate for pest control9 and for reducing the impact of arthropod-borne diseases10. While Wolbachia is primarily transmitted maternally through vertical transmission, it can also spread horizontally between species via predation, shared habitats, or symbiotic interactions11. These horizontal transfer between different host species, along with occasional infection loss, frequently cause discordance between Wolbachia and host phylogenies.
The microbiome frequently influences the range of host phenotypes across different taxa and environmental conditions, potentially playing a significant role in the evolution of hosts12. However, the full spectrum of the microbiome in shaping host phenotypic variance, evolutionary dynamics and demographic history is not yet fully understood. This gap in knowledge is also evident in Wolbachia research. Although the reproductive manipulation of Wolbachia in hosts, including its ability to reduce the census population size, has been well-documented1,8,13,14, our genetic understanding of how it dynamically drives evolutionary changes at the population level, or alters the evolutionary trajectory of its hosts during its symbiotic relationship in nature, remain limited, which limits our full comprehension of Wolbachia dynamics in natural ecosystems. Moreover, while Wolbachia’s ability to manipulate host reproduction and skew sex ratios is well-documented8, the long-term evolutionary consequences of such disruptions for the host genome in natural populations remain largely unexplored. However, examples of host-Wolbachia interactions across different symbiotic stages within same or closely related species, especially in the wild, remain scarce.
In this study, we report the parasitism of Wolbachia in the sawfly Analcellicampa spp. (Hymenoptera, Tenthredinidae), whose larvae are fruit borers of Cerasus spp. (Rosaceae)15. These sawflies exhibit haplodiploidy, with males developing from unfertilized eggs (haploid) and females from fertilized eggs (diploid). Notably, Wolbachia infections in this group are associated with extreme male rarity, providing a unique opportunity to explore in detail how Wolbachia parasitism creates differential evolutionary pressures and causes shared population history to manifest differently in male and female genome sequences, and alters host genome evolution. We collected 89 individuals from six species across China and established a pipeline based on next-generation sequencing data to identify Wolbachia infections, discovering that three of these species were infected. We successfully assembled the first telomere-to-telomere (T2T) genome for Analcellicampa danfengensis (AD) along with the complete genome of its symbiotic Wolbachia. Utilizing these genomic resources, we investigated Wolbachia transmission characteristics in sawflies, applying an epidemiological perspective alongside genomic and population genetic approaches to assess its evolutionary impact. While previous research has predominantly focused on the origin, classification and reproductive manipulation of Wolbachia13,16,17, few studies have addressed its demographic effects and population evolutionary dynamics from a population genetics perspective. This study leverages a unique sawfly system and a T2T genome assembly to investigate how Wolbachia-driven male rarity shapes the demographic signals inferred from male and female genomes and to identify the resulting signatures of selection on the host genome. These insights will benefit the fields of ecology, evolutionary biology, and biological control. Furthermore, our findings may contribute to refining strategies for utilizing Wolbachia in the control of vector insect reproduction and transmission, ultimately enhancing pest management efforts.
Results
Assembly and annotation of the Analcellicampa danfengensis genome
Initially, we generated approximately 14.69 GB of Pacific Biosciences (PacBio) HiFi reads ( ~ 68.89×), 54.80 GB of Oxford Nanopore (ONT) long reads ( ~ 256.99×), 47.21 GB of Hi-C reads ( ~ 221.39×) and 34.54 GB illumina paired-end reads ( ~ 161.98×) from pools of five adult females (Supplementary Data 1). Heterozygous k-mer pair coverage distributions from Smudgeplot18 revealed signals of diploidy (Supplementary Fig. 1a), consistent with previous studies19. The genome size of AD was estimated to be 213.24 Mb using GenomeScope20 (Supplementary Fig. 1b), close to the previously reported genome size of another sawfly species, Orussidae abietinus, at 201 Mb19.
We utilized various cutting-edge assembly software tools to construct the genome assembly. By almost every metric, Flye21, using ONT reads, produced the best draft genome assembly, showing the most contiguous (N50) and high per-base quality among all assemblies, as indicated by its superior Benchmarking Universal Single-Copy Orthologs (BUSCO)22 score (Supplementary Data 2). In contrast, LJA23 produced the lowest-quality assembly across all metrics, while Hifiasm24 in HiFi + ONT mode generated an intermediate-quality assembly. Therefore, we selected the Flye assembly for further scaffolding with Hi-C data and gap closure.
We used Hi-C data for scaffolding and then extended both ends of the scaffolds using HiFi reads to further elongate and link associated scaffolds (see Materials and Methods). This approach ultimately scaffolded the genome into 10 pseudo-chromosomes (Fig. 1a, b), during which we also manually closed 77 gaps using the sequence-extension method. The final genome assembly was 211.14 Mb, featuring a contig N50 size of 22.39 Mb and a GC content of 41.99% (Supplementary Data 2). The Hi-C contact map showed stronger interaction intensity along the diagonal compared to non-diagonal regions, with no significant noise outside the diagonal, suggesting a high-quality chromosome assembly (Fig. 1a). The largest scaffold reached 57.04 Mb (Chr1), while the smallest was 13.31 Mb (Chr6). Seven scaffolds achieved telomere-to-telomere (T2T) assembly, though two scaffolds each still contained one unresolved gap each (Chr4:10537477-10537560 and Chr9:7931267-7931340) (Fig. 1a). Assembly completeness was evaluated using the insecta-specific and hymenoptera-specific BUSCO database, showing 99.85% and 97.91% completeness, respectively, indicating robust genome integrity (Fig. 1c and Supplementary Data 2). Quality-control mapping of Illumina reads to the final assembly yielded an exceptionally high mapping rate of 99.83%. HiFi reads achieved a mapping rate of 99.93%. ONT reads achieved 98.83%, and RNA-seq data showed a mapping rate of 91.76%. These results further indicate the high quality of our genome assembly. Comparative assessments with AD and Orussidae abietinus19 genomes showed significant enhancements in the AD assembly, with a contig N50 approximately 17 times that of Orussidae abietinus and almost all gaps from previous versions filled (Supplementary Fig. 2). In summary, the assembly resulted in seven gap-free chromosomes, establishing AD as a high-quality chromosomal-level reference genome.
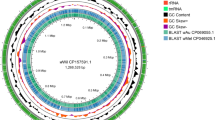
a Genome-wide Hi-C contact matrix of A.danfengensis. Red color intensity in the heatmap shows frequency of interaction between two loci at 25 kb resolution. b Distribution of genomic features of the A.danfengensis genome. Tracks are aggregated in 100-kb bins as follows: I, gene density; II, TE density; III, TR density; IV, GC content; V, SNPs density; VI, the 9 chromosomes (chr1–chr9). c BUSCO completeness scores for the host genome using insecta and hymenoptera databases, and for Wolbachia genome using rickettsiales database. d Features of the three largest centromeres: ① Chr1 (~4 Mb); ② Chr3 (~4 Mb); ③ Chr8 (~3.5 Mb). e Circular map of the wAnd genome. The outermost tracks 1 and 2 represent the positions of the CDS, tRNA, rRNA, and tmRNA genes on the positive and negative strands. Tracks 3 and 4 tracks show the GC content and GC skew, respectively. f Features of the mitochondrial genome.
Moreover, we examined whether telomeres and centromeres were present in our assembled genome. The results showed that telomeric regions could be detected on both ends of nine chromosomes. The analysis predicted nine centromeric regions and identified 18 telomeres by recognizing the five-base telomeric repeat (CCTAA/TTAGG)25. Centromeric and telomeric regions are detailed in Supplementary Data 3 and 4, respectively. The approximate locations of the nine centromeric regions was estimated based on repeat density and the Hi-C interaction heatmap. We observed that most centromeres were positioned near to the middle parts of the chromosomes, with a few located at the chromosomes’ terminals, including Chr5 and Chr6 (Fig. 1a, b and d, and Supplementary Data 4). The centromeric regions ranged in size from 124.98 kb to 4.54 Mb, with an average length of 1.96 Mb (Supplementary Data 4). The shortest centromeres are the telocentric, found on Chr5 and Chr6. Notably, “blank regions” observed in the Hi-C interaction map (Fig. 1a) corresponded with a dense accumulation of tandem repeats, especially in the centromeric regions (Fig. 1b, d), which is consistent with other T2T genome assemblies26,27. Centromeres vary greatly in their sequence organization among species28. Additionally, centromeric regions exhibit several notable common features. In AD, the centromeric regions are primarily composed of tandem repeats, accompanied by a reduction in GC content, a decrease in transposable element (TE) content, and a significantly lower gene density (Fig. 1b). These distinct characteristics, compared to other genomic regions, further support the identification of centromeres.
A total of 9569 protein-coding genes were predicted from the chromosomal-level genome of Analcellicampa. While this number is lower than that found in some model insects like Drosophila, it is comparable to gene counts reported for other sawfly species (e.g., Orussus abietinus19 and falls within the range observed across basal Hymenoptera. Of these, 4716 genes were assigned Gene Ontology (GO) terms, 4282 genes were assigned Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, and 6787 genes contained Pfam domains. Additionally, 6478 genes had BLASTN matches in the Swiss-Prot protein database. Repeat sequences constituted 32.42% of the AD genome, of which retrotransposons accounted for 12.29%, DNA transposons for 5.73%, and unclassified elements for 12.59%.
Assembly and annotation of the endosymbiotic Wolbachia in sawfly
We generated a circular contig in our assembly (Supplementary Fig. 3), and sequence homology analysis confirmed it was Wolbachia. The circular structure indicates the completeness of the assembled sequence. The Wolbachia strain was detected in AD, thus we named it wAnd. The wAnd genome is 1,236,102 bp in size and has a GC content of 35.14% (Fig. 1e and Supplementary Data 5). Using Prokka software29, we annotated the genome, identifying a total of 1166 genes were identified, 3 rRNA genes, 34 tRNA genes, and 1 tmRNA (Fig. 1e). The BUSCO completeness scores of the wAnd genome indicated that it contained 361 complete and single-copy BUSCO groups, 1 complete and duplicated BUSCO groups, 0 fragmented BUSCO groups, and 2 missing BUSCO groups (Fig. 1c). The BUSCO score of 99.5% for the wAnd genome was comparable to that of other circular, chromosome-level Wolbachia genomes in supergroup A30 (Supplementary Data 6). The genomic features of wAnd, including coding sequences (CDS), tRNA, rRNA, and others, were visualized using Proksee31 (Fig. 1e). The genome exhibits the GC skew pattern typical of Wolbachia genomes32,33. The presence of an irregular GC skew in wAnd masks the identification of a specific origin of replication, indicating potential frequent genomic rearrangements34. In summary, we discovered Wolbachia parasitism in the sawfly and successfully assembled a high-quality Wolbachia genome.
We assembled a complete mitochondrial genome of 199,511 bp, encompassing all 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a control region (Fig. 1f and Supplementary Data 7). The control region is situated between trnQ and trnI and measures 4064 bp in length.
Resequencing data collection and high-quality SNPs construction
In this study, we collected 89 high-quality whole-genome resequencing samples (including males and females where available, see Supplementary Data 8) from across China (Fig. 2a). Specifically, we obtained 30 AD, 26 A.xanthosoma (AX), 7 A.acutiserrula (AA), 13 A.maculidorsatus (AM), 4 A.wui (AW) and 9 A.emei (AE) for whole-genome sequencing and analysis (Supplementary Data 8). We used two species Monocellicampa pruni and Monocellicampa yangae in closely related genus, as an outgroup. All 91 genomes were aligned against the AD reference genome. The lowest alignment rate was observed in the outgroup at 69.67%, while the average mapping rate for the Analcellicampa species was 89.07% (see Supplementary Data 8 for alignment rate per species). Among the Analcellicampa species, the lowest alignment rate was observed in AE at 78.87%, while the highest alignment rates were observed in AD at 99.90%, followed by AX, with rates of 99.90% and 93.65%, respectively (Supplementary Data 8). A total of 20,292,172 putative bi-allelic single-nucleotide polymorphisms (SNPs) were identified and passed the filtering criteria across the 91 genomes for subsequent analyses, of which 11.83 million (39.62%) were intergenic, 6.83 million (22.86%) were intronic and 1.53 million (5.13%) were exonic (Supplementary Data 9). We observed the highest number of SNPs per individual in the AE, with 3.39 million SNPs per individual, corresponding to ~24.51% more SNPs than the average of 2.73 million (Supplementary Data 8). To further assess the quality of the genetic variants, we calculated the transition-to-transversion (Ts/Tv) ratio, an indicator of potential sequencing error35. In our study, the Ts/Tv ratio for global populations SNPs was found to be 1.99, close to 2, indicating that the genetic variants identified are of high quality and their distribution is relatively balanced.
a Sampling sites. The bar charts indicate the species and number of samples collected at each sampling location. Colors correspond to those in (d). b Maximum-likelihood tree depicting the evolutionary relationships among genus Analcellicampa and outgroup. c ADMIXTURE analysis with K = 3, 7 and 8. Colors in each column represent ancestry proportion. d Nucleotide diversity (π), nucleotide differences (dxy) across the six species. The value in each circle represents a measure of nucleotide diversity for each species; values in red on each line indicate pairwise population nucleotide differences between species. e Patterns of LD (linkage disequilibrium) decay across the genome in different geographic populations. r2, Pearson’s correlation coefficient. f Genomic similarity of six species of Analcellicampa to the AD reference genome. Chromosomes are indicated by different colors along the left y axis. Identical score (IS) values are shown for SNPs within each 50-kb window across the genome. g Estimated split times between each species after 50 bootstraps. The widths indicate probability densities. h Dynamic of Ne inferred by PSMC. The vertical lines indicate divergence times corresponding to those shown in (g). The marked pentagrams indicate species fully infected with Wolbachia.
Population characterization and evolutionary history of sawfly
To decipher genetic relationships among Analcellicampa sawfly populations, we constructed a Maximum Likelihood (ML) tree and performed principal component analysis (PCA) on the above 91 individuals using host nuclear genome SNPs (hereafter referred to as autosomal SNPs, in contrast to symbiont or mitochondrial SNPs). Our phylogenetic and genetic clustering analyses resolved six genetic groups (Fig. 2b and Supplementary Fig. 4), which corresponded to species classifications based on morphological characteristics (Supplementary Fig. 5 and 6). Using Monocellicampa species as outgroup, the phylogenetic analysis revealed that individuals from the same population clustered together, with AE and AM forming a clade distinct from the other genetic lineages in the ML tree (Fig. 2b). Higher π values observed in AE and AM (with the exception of AX) reflected their rich genetic diversity (Fig. 2d), potentially indicating that species from South and East China have more ancient origins compared to other populations (Fig. 2a). Species from the northern and western regions clustered together, with each species forming stable genetic clusters (Fig. 2a, b). Population structure inferred by ADMIXTURE36 also aligned with the six genetic groups when the optimal K = 7, with AD further subdivided into two subgroups corresponding to their geographical distribution (Fig. 2c and Supplementary Fig. 7). Additionally, when K values ranged from 2 to 6, the ancestry component of AE and AM accounted for the largest proportion in the outgroup (Supplementary Fig. 7), supporting the previous finding suggested by π values that populations from South and East China have more ancient origins compared to other populations.
Notably, AX and AD formed a sister clade (Fig. 2b), with their dxy value being the lowest among all populations (0.017, Fig. 2d). The geographic ranges of the two species overlapped, though AX had a broader distribution, with a few individuals still found in Hunan and eastern regions (Fig. 2a and Supplementary Data 7). AX exhibited the second-highest nucleotide diversity (π = 7.61 × 10−3), following AM (π = 8.97 × 10−3), while AD and AW showed the lowest nucleotide diversity (Fig. 2d). Linkage disequilibrium (LD) decay analysis showed slower decline in LD for AW and AD (Fig. 2e), suggesting smaller population sizes and further supporting their lower nucleotide diversity. Identity Score (IS) analysis demonstrated a high level of genomic similarity between AX and AD (Fig. 2f). The dxy between AD and other species was approximately 0.03, with genomic IS reaching at least 0.6, indicating a close genetic similarity among populations and providing a solid foundation for our subsequent population genetic analyses using AD as the reference genome.
We also examined possible gene flow between species using TreeMix (Supplementary Figs. 8 and 9), which calculates a phylogeny of populations based on shared drift and tests whether migration edges (i.e., introgression) improve the model fit. The overall topology of the tree was consistent with that of the ML tree (Fig. 2b), confirming the reliability of this tree’s topological structure. Likelihood improvements declined after four migration edges (Supplementary Fig. 8), so we presented results using this value (Supplementary Fig. 9), with 100 bootstraps performed. At four migration edges, TreeMix identified evidence for low levels of gene flow (migration weight < 0.05) from AA into AX, AE into AD, and a higher level of gene flow ( ~ 0.2) from AD into outgroup (Supplementary Fig. 9).
Furthermore, we explored the demographic history of the sawflies by using site-frequency spectrum (SFS) via momi237 and inferred changes in effective population size (Ne) over time for each population with the pairwise sequential Markovian coalescent (PSMC) model38. Since low levels of gene flow have negligible impact on timing inference39,40 and our study primarily focused on changes in Ne across species, we tested two models—one assuming constant population size and another allowing for variable population size—to seek the best one with lowest AIC value (see Materials and Methods and Supplementary Fig. 10). Based on this best-fit model (Supplementary Fig. 10), we inferred that the most recent common ancestor of the genus Analcellicampa diverged approximately 857,445 years ago (95% CI, 844.17 to 870.72 Kya). The basal clade within sawflies, containing AE and AM, separated from other lineages 368,609 years ago (95% CI, 313.15 to 384.07 Kya). Within Analcellicampa, the most recent divergence occurred between AE and AM at 79,678 years ago (95% CI, 74.39 to 84.97 Kya), followed by AD and AX at 185,866 years ago (95% CI, 174.66 to 197.07 Kya) (Fig. 2g and Supplementary Data 10). In the optimal model, we allowed population size to vary from the Last Glacial Maximum to the present. All populations showed a decline in Ne, though AW exhibited the fastest decline, followed by AD. This result aligns with the previous LD decay analysis, where these two populations demonstrated the slowest LD decay (Fig. 2e).
The demographic history of the six species in our study was first inferred by analyzing the whole-genome sequence using Pairwise Sequentially Markovian Coalescent (PSMC) model38 (Fig. 2h). The inferred demographic histories of these six species spanned from approximately 10 million years ago (Mya) to 10,000 years ago (Kya). Since all species analyzed here are younger than 10 million years, the inferred demographic dynamics likely reflect an ancestral form with a potentially different geographic distribution (Fig. 2h). All sawfly populations experienced two population bottlenecks, one around 0.4 Mya (Bottleneck 1, B1) and another near 20 Kya (B2) (Fig. 2h). Correlating species divergence times with Ne dynamics, we observed that the formation of the Analcellicampa genus occurred approximately 857 Kya, coinciding with the early phase of B1 (Fig. 2g, h). The sawfly Analcellicampa, being monophagous with their larvae that exclusively parasitize cherry fruits (Cerasus spp.), likely experienced a founder effect at this bottleneck stage. Following B1, Analcellicampa underwent rapid radiation, accompanied by an increase in Ne. Over a span of approximately 300,000 years, six distinct species emerged, among which AD experienced a notable Ne expansion post-divergence. During the Last Glacial Maximum (LGM, ~ 20 Kya), all populations experienced a decline in Ne, leading to the second bottleneck (B2).
To sum up, our results provided a comprehensive overview of the evolutionary history of Analcellicampa. The earliest emergence of the genus was around 857 Kya, originating in the southwestern region and subsequently spreading northeastward, resulting in six distinct Analcellicampa species in China. The most widely distributed species was AX, which diverged from its sibling species AD approximately 185 Kya. We did not observe any introgression events from AD, the species for which we constructed the reference genome, into other sawfly species. Overall, despite ~857 thousand years of divergence, these populations maintain relatively high similarity, with IS values above 0.6.
Discordance between phylogenetic relationships of the sawfly and its symbiotic Wolbachia
The transmission modes of Wolbachia play a crucial role in its spread and coevolution with hosts41,42. Our comprehensive genomic dataset allowed us to explore Wolbachia evolutionary dynamics within host populations, particularly to assess the extent of intraspecific horizontal transmission. Through our Wolbachia detection pipeline (See “Materials and Methods”), we observed infections in four sawfly species: AW, AM, AX and AD (Supplementary Data 8). AW and AD were fully infected, while AM and AX showed partial infection, with AM having an infection rate of 3/13 among sampled individuals and AX exhibiting an infection rate of 1/26. Maximum likelihood phylogenetic trees of these Wolbachia strains classified them into three genetic groups: the first group parasitized only AW; the second included two individuals from AM; and the third encompassed all AD, one individual from AM, and the single Wolbachia strain found in AX (Fig. 3). The Wolbachia strains found in AW form a distinct early-diverging lineage in the phylogenetic tree, suggesting a long independent evolutionary history of this host-symbiont partnership (Fig. 3). The Wolbachia strains from AM were resolved into two distinct clades, while those from AD formed several closely related terminal clades with a lower genetic diversity (Type 1: 4.95 × 10-3 vs. Type 3: 2.05 × 10-4) and relatively fast LD decay (Supplementary Fig. 11). Two Wolbachia strains from AM formed a sister group relationship with the AD-associated strains, suggesting a shared evolutionary history between these infections. The single Wolbachia strain detected in AX was nested within the AD-associated clade, with both species sharing overlapping habitats. This relationship suggests a potential recent horizontal transfer from AD to AX, likely facilitated by ecological factors such as shared habitats and host density, aligning with the broader understanding that environmental context plays a critical role in host-symbiont evolution43.
When comparing host phylogeny with that of symbiotic Wolbachia, we found that the overall topologies of the host and Wolbachia trees were not fully congruent. However, some clusters maintained congruent phylogenetic relationships. For example, the phylogenetic patterns between AW and its associated Wolbachia showed strong congruence, suggesting stable host-symbiont associations likely due to long-term coevolution. The distribution of Wolbachia in AD showed partial congruence with host phylogeny, with both host and symbiont phylogenies revealing two main clusters that correspond to geographical distribution. However, within these clusters, the detailed phylogenetic relationships between hosts and their Wolbachia were discordant; For instance, while A.danfengensis_MC0301 and A.danfengensis_MC0303 formed sister clades in the host phylogeny (Fig. 3), their associated Wolbachia strains showed greater phylogenetic divergence–this pattern was commonly observed within AD. These phylogenetic patterns are consistent with multiple modes of Wolbachia acquisition described in previous studies11, including vertical inheritance during host speciation, hybridization and gene flow between closely related hosts, and horizontal transmission events. In our study, these phenomena also reflect the existence of significant hybridization and horizontal transfer events.
Divergent demographic signals in male and female genomes reflect a Wolbachia-driven male bottleneck
PSMC38 was used to infer historical changes in effective population size (Ne). The most dramatic demographic changes occurred between 20 and 200 Kya (Fig. 2h), although the timing of population expansions and contractions varied among species. Interestingly, inferred Ne trajectories differed dramatically between male and female AD during this period (Fig. 4). To confirm this result, we inferred the demographic history of AD from three geographically distinct populations and performed 100 bootstrap replicates. The results remained robust, consistently showing significant differences in Ne between males and females during the 20–200 Kya period. Specifically, female Ne increased sharply around 200 Kya, peaking around 70–80 Kya, where Ne inferred from female genomes was 13 times larger than the apparent Ne inferred from male genomes (13 × 105 vs. 1 × 105), before declining during the LGM to approximately 1 × 105 (Fig. 4a). In contrast, the male Ne remained relatively stable throughout this period, with a slight decline during the LGM.
PSMC analyses based on re-sequenced data (Fig. 4) enabled us to compare the demographic histories across the different populations. We included AX and AM as comparison groups (Fig. 4b, c), as these populations contained predominantly uninfected individuals with only a small proportion showing recent Wolbachia infection. Using data from uninfected individuals, our analyses of these uninfected populations revealed nearly identical Ne trajectories inferred from both male and female genomes (Fig. 4b, c). This demonstrates that our analytical approach does not inherently create a sex-biased signal, and the stark differences observed in infected populations (Fig. 4a) are indeed a consequence of Wolbachia infection.
PSMC results become less reliable for recent timeframes (within the last 10,000 years) due to reduced inference power38. To address this limitation, we employed the SMC + + 44, which enable more precise inference of population history within the last 10,000 years. The demographic trajectories inferred by SMC + + prior to 10,000 years ago were largely consistent with the PSMC results, confirming an increased Ne of AD females around 70–80 Kya. However, SMC + + revealed that within the past 10,000 years, Ne inferred from female genomes remained stable, while the apparent Ne inferred from male genomes experienced a dramatic decline, decreasing from approximately 10,000 to fewer than 1000 individuals, a signal consistent with an intensifying male bottleneck. In contrast, analyses of uninfected populations (AX and AM) exhibited consistent demographic trajectories from both male and female genomes throughout their entire evolutionary history, despite differences in the range of Ne changes between the two species.
The momi2 results, which model the demography of the population as a whole, further supported the overall population declines observed in AD and AW. We tested two models: one assuming constant Ne and another allowing for variable Ne. The variable Ne model, yielding the lowest AIC, demonstrated that Wolbachia-infected AD and AW experienced the fastest declines in overall population Ne, and their current population sizes remain at low levels (Supplementary Fig. 10 and Supplementary Data 10).
In summary, our analyses reveal complex temporal changes in host population dynamics associated with Wolbachia infection. While previous studies have largely focused on Wolbachia’s male-killing effects and their role in causing sex ratio imbalances, our findings revealed a more complex picture of how these effects are reflected in the genome. In AD, Ne inferred from female genomes increased sharply around 70-200 Kya, followed by a decline during the LGM, whereas the apparent Ne inferred from male genomes showed a prolonged and significant decline (Fig. 4). This divergence in signals indicates that the observed bias is not solely a prolonged intensification of the male bottleneck but reflects complex host-symbiont interactions over time.
Genomic signatures of Wolbachia-induced changes in AD
Leveraging the unique system of sympatric sibling species—AD (fully infected) and AX (nearly uninfected)—with overlapping distributions and shared ancestry, we had a rare opportunity to explore the evolutionary consequences for the host genome resulting from Wolbachia’s modification of host reproduction. By focusing on these populations, we minimized confounding variables, such as environmental effects, which are known to influence Wolbachia’s impact on sex determination45,46. This system, with its genomic similarity and robust sample sizes, allowed us to investigate Wolbachia-induced genetic modifications in an ecologically controlled context.
Specifically, we analyzed whole-genome sequences of AD (n = 30) and AX (n = 26) to investigate Wolbachia-induced modifications in the host genome. To identify the genomic region that presents signatures of natural selection associated with Wolbachia-coevolution, we performed whole-genome genetic differentiation analysis by estimating the cross-population composite likelihood ratio (XP-CLR) and differences in nucleotide diversity (π log2-ratio Wolbachia_positive/Wolbachia_negative) between AD and AX in 10-kb windows with a 5-kb sliding window along the genome. Applying a 5% threshold for maximum haplotype frequency difference (XP-CLR) and nucleotide diversity (log2-π ratio), we identified a total of 3.94 Mb genomic regions that covered all outliers (Fig. 5a and Supplementary Fig. 12). These regions contained 287 annotated positively selected gene (PSG) candidates (Supplementary Data 11).
a Distribution of the log2(π-AD/π-AX) and XP-CLR values were calculated in 10 kb sliding windows with 5 kb steps. The horizontal and vertical lines represent threshold lines of the top 5% of the XP-CLR and π ratio values, respectively. Points (red) located in the top right sector represent selective signatures in AD. Yellow and black bins in the histograms of XP-CLR (right) and π ratio (top) represent levels respectively higher and lower than the threshold line. b Significant GO enrichment of PSGs associated with Wolbachia-induced reproductive manipulations.
Gene Ontology (GO) analysis of the PSGs revealed significant enrichment in categories associated with reproductive processes, sensory and neural development, among others (Supplementary Data 13). Notably, the most highly enriched Gene Ontology (GO) terms were those associated with sensory system development (P = 3.0 × 10-4, Benjamini-Hochberg corrected) and compound eye development (P = 4.0 × 10-4) (Fig. 5b). Enrichment was also observed in reproductive development categories, such as germ-line stem cell population maintenance (P = 1.3 × 10-3), germ-line cyst formation (P = 1.4 × 10-2), male sex differentiation (P = 1.4 × 10-2) and sex differentiation (P = 3.8 × 10-2) (Fig. 5b). Additionally, neural development terms like synaptic signaling (2.7 × 10-3), synapse assembly (P = 9.9 × 10-4), synapse organization (P = 1.3 × 10-2), regulation of synapse organization (P = 1.2 × 10-2), regulation of synapse structure or activity (P = 6.9 × 10-3), synapse organization (P = 1.3 × 10-2), neuron fate commitment (P = 2.7 × 10-3) and learning or memory (P = 1.2 × 10-2) were significantly enriched (Fig. 5b). Notably, we also observed significant enrichment in the category of determination of adult lifespan (P = 4.4 × 10⁻2) (Fig. 5b), suggesting potential selective pressures related to longevity and its influence on population dynamics under Wolbachia infection.
We identified several reproduction-related genes potentially linked to Wolbachia’s sex-manipulation mechanisms, corresponding to the significant demographic differences observed between males and females in our study. Among the reproductive GO terms we identified, the following genes were included: mys, otk2, puc, Six4, alien, Cbl, Inx2, dally, Rap1, and Rtel1. Among these, Inx2 exhibited the strongest selection signal with XP-CLR = 1297.74 and log2(π ratio)=4.85, while puc showed the lowest signal with XP-CLR = 700.75 and log2(π ratio)=3.69 (Supplementary Data 11).
Notably, Six4, which encodes a transcription factor essential for embryonic gonad formation, has been implicated in sex determination pathways. Its mammalian homolog regulates Sry expression, a gene essential for male development, and the loss of Six4 function can result in the reversal of sex from male to female in mice47. This gene similarly contributes to Wolbachia-induced feminization of genetic males, as observed in some isopod (Crustacean) species and lepidopteran species5, where Wolbachia cause male embryos to develop as functional females. This process may underlie the increased female population numbers observed in our study.
Several of the above genes are associated with sperm formation, highlighting Wolbachia’s potential influence on male reproductive capabilities. For instance, Rtel1, a regulator of telomere elongation helicase 1, plays a role in male germline stem cells regulation48. asun is essential for spermatogenesis in Drosophila, with mutations leading to male sterility49. Dcst2 is required for sperm-egg fusion, with knockout resulting in impaired fertilization in zebrafish50. Additionally, genes such as Otk2, linked to male genitalia development51, and Inx2, which encodes an innexin protein critical for gap junction communication during oogenesis, may influence oocyte development and fecundity52, underscoring their importance in the development of the male reproductive system.
Beyond the genes enriched in the aforementioned GO terms, we identified three additional reproduction-related genes, kl-2 (XP-CLR = 978.52, log2(π ratio)=4.03), 5-HT7 (XP-CLR = 868.21, log2(π ratio)=4.88) and gb (XP-CLR = 810.20, log2(π ratio)=4.71). kl-2 is a male fertility factor, is involved in flagella movements53. The remaining two genes are linked to mating behavior. 5-HT7, a serotonin receptor, regulates sexual receptivity in virgin female Drosophila melanogaster, and its absence significantly reduces the mating rate of female flies54. Meanwhile, gb (genderblind), encoding an amino acid transporter involved in glutamate secretion and synaptic transmission, was identified as a target of selection and is known to affect male courtship behavior in Drosophila55. Notably, a majority of these reproduction-related genes (8 out of 13) are located on Chr1 (Supplementary Data 11 and Supplementary Fig. 12), suggesting potential chromosomal clustering of selection signals.
To gain deeper insights into the specific genes potentially driving this adaptation, we further annotated variants within the key reproduction-related candidate genes (e.g., mys, otk2, puc, Six4, alien, Cbl, Inx2, dally, Rap1, Rtel1, kl-2, 5-HT7 and gb) using snpEff56. This analysis revealed striking allelic differentiation between the Wolbachia-infected (AD and AW) and uninfected (AX) populations for six of these 13 genes (Supplementary Data 12), driven largely by missense mutations suggesting potential alterations in protein function. To assess the likely functional impact of these amino acid changes, we employed the SIFT algorithm57. This predictive analysis identified one missense SNP within the Cbl gene as “deleterious”. As a known negative regulator of EGFR signaling critical for Drosophila oogenesis58, and given that EGFR signaling is also implicated in male germline stem cell regulation and spermatogenesis59, this deleterious Cbl variant likely impacts male reproduction by dysregulating this pathway. Its signature of positive selection suggests Cbl is a target of host adaptation in response to the male-specific pressures imposed by Wolbachia. A potentially deleterious mutation fixed or at high frequency in the infected population suggests strong selection acting on Cbl function, possibly as a direct response to Wolbachia infection or as an adaptation to the altered reproductive environment caused by male scarcity.
This finding pinpoints Cbl as a specific gene, carrying a potentially deleterious mutation under strong selection, that warrants future investigation into host-symbiont coevolution in this system. More broadly, the presence of distinct, functionally relevant allele states (including deleterious variants like the one in Cbl) within key reproductive genes distinguishing infected and uninfected populations provides tangible evidence for the profound genomic reshaping driven by Wolbachia. These findings underscore the potential for Wolbachia infection to drive significant genomic changes in host genes related to reproduction, development, and behavior, highlighting the multifaceted impact of Wolbachia on host biology. By exerting selective pressures on critical biological processes, Wolbachia likely enhances its own transmission success60, while simultaneously altering host sex ratios and driving male extinction through complex, symbiont-mediated genetic changes.
Discussion
Our findings reveal compelling evidence linking Wolbachia infection status to shifts in host reproductive and population dynamics. Specifically, the observed female-biased sex ratios and the scarcity of males in infected populations correspond closely to established outcomes of Wolbachia-induced reproductive manipulation. Supporting evidence from controlled laboratory investigations has extensively documented Wolbachia’s capacity to reshape host reproductive biology through mechanisms such as male-killing, parthenogenesis induction, and behavioral alterations in mating systems13,61. Particularly compelling is our detection of strong selective signatures in genetic loci associated with reproduction, neural development, and sperm formation. These genomic signatures, corresponding to phenotypic traits known to be targeted by Wolbachia, provide a direct window into the host’s evolutionary response to this manipulation. The consistency between molecular signatures of selection and experimentally verified Wolbachia-host interactions13 strengthens the foundational understanding of these host-symbiont interactions. This integration of genomic and phenotypic evidence provides a robust framework to investigate how Wolbachia fundamentally shapes host evolutionary trajectories, including how a shared population history is differentially reflected in male and female genomes.
Moreover, alternative explanations, such as neutral demographic shifts or environmental factors, are less consistent with the observed patterns. Neutral demographic processes, such as genetic drift or random migrations, typically result in stochastic genomic changes that are distributed randomly across the genome62. In contrast, we observe concentrated selection signals specifically enriched in functional categories tied to Wolbachia’s reproductive manipulations, such as genes associated with reproduction and mating behavior. This discrepancy suggests that the observed genomic patterns are more likely driven by directional selective pressures than by random demographic events. Similarly, environmental factors alone are insufficient to account for the congruence between Wolbachia infection rates and the persistent female-biased demography observed in infected populations. Environmental variation typically produces diffuse and temporally variable effects, often fluctuating with changing ecological conditions63,64. By contrast, the observed patterns exhibit a more targeted and consistent influence, consistent with an endosymbiont-imposed selective pressure rather than stochastic or transient factors. Collectively, these findings provide compelling evidence that Wolbachia acts as a major selective force, reshaping host reproductive dynamics over evolutionary timescales.
Our findings illustrate how microbial symbionts can fundamentally alter host evolutionary trajectories by driving profound shifts in reproductive dynamics. In this haplodiploid system, the population’s shared demographic history is reflected differently in male and female genomes. To explain the starkly different demographic signals observed (Fig. 4), we propose a “recurrent male bottleneck” model. Although male genomes are resampled from the female gene pool each generation65, the Wolbachia-induced rarity of males creates an intense bottleneck for any gene lineage passing through the male stage. This bottleneck accelerates coalescence, causing analyses of male genomes to reveal a lower apparent Ne compared to female genomes. The fact that this divergence is absent in uninfected populations (Fig. 4b, c) confirms this phenomenon is a direct consequence of Wolbachia’s impact, not an analytical artifact.
The intense selective pressures created by this symbiont-driven male rarity are further evident in our genomic scans (Fig. 5). Interestingly, the process of resampling male genomes from the female pool introduces another layer of potential evolutionary conflict. This transmission from a diploid mother to a haploid son is typically assumed to be Mendelian (i.e., random segregation of alleles). However, this assumption may not hold true. Biological mechanisms such as meiotic drive or other forms of intragenomic conflict—potentially even mediated by Wolbachia itself—could lead to a non-random, preferential transmission of certain driving alleles into the eggs destined to become males66,67. Such a meiotic drive allele would rapidly increase in frequency, creating a strong, localized signature of positive selection that could be mistaken for or interact with demographic signals68. This provides a fascinating potential link between our two key findings: it is tantalizing to speculate that some of the genomic regions we identified as being under strong selection (Fig. 5) might not only be responding to the downstream consequences of male rarity but could themselves house meiotic drive elements elements shaping male inheritance.
Given these interacting evolutionary forces, interpreting the signatures of selection on male-function genes requires considering multiple, potentially competing, hypotheses. For instance, the selection we observe could be explained by at least two competing hypotheses. First, intense sexual selection on the few remaining males could drive rapid adaptive evolution of reproduction-related genes. This scenario represents an extreme form of sexual conflict, a force that can be particularly potent in haplodiploid systems where male alleles are always exposed to selection due to hemizygosity69. Alternatively, if the system is evolving toward parthenogenesis, relaxed purifying selection on male-specific functions could allow accumulation of deleterious mutations. Our discovery of a potentially deleterious mutation in the Cbl gene provides intriguing, though not conclusive, support for the latter scenario. Thus, our findings present a complex evolutionary picture, suggesting that male genes might be simultaneously subject to pressures from sexual selection, relaxed functional constraint, and potentially intragenomic conflict over their very transmission.
While our current data cannot definitively distinguish between male-killing and incomplete parthenogenesis as the underlying mechanism—especially as the latter is also known to produce rare males70—the genomic signatures clearly indicate a system in profound evolutionary flux. Regardless of whether the underlying mechanism is male-killing or incomplete parthenogenesis—both of which can produce similar patterns of male rarity—the demographic bottleneck effect we observe provides a unifying framework for understanding how Wolbachia reshapes host genomic evolution. The long-term evolutionary outcome of this ongoing selection is not predetermined. The host lineage could potentially evolve dependence on the Wolbachia-induced state, similar to scenarios observed in some parthenogenesis-inducing interactions71. Alternatively, host populations might evolve resistance mechanisms that counteract the manipulation, potentially leading to a restoration of a more balanced sex ratio, as documented in other systems72. Our data reveal the strong selective pressures currently acting on the Analcellicampa genome, but distinguishing between these potential future trajectories (dependence, resistance, or continued conflict) requires further investigation.
Beyond illuminating the fate of host reproduction, our results highlight Wolbachia’s remarkable ecological and evolutionary adaptability. Although vertical, maternal inheritance has long been considered the primary transmission route, the broad phylogenetic distribution of Wolbachia suggests that horizontal transfer also plays a pivotal role43,73,74. Our phylogenetic analyses reveal incongruence between host and Wolbachia lineages, indicating inter- and intra-specific transfers likely facilitated by ecological interactions such as shared habitats and predation11,75. For example, the close phylogenetic relationship between Wolbachia strains in AD and AX, which overlap geographically, suggests recent horizontal transfer. Such horizontal transmission strategies enable Wolbachia to colonize new host species, fostering novel coevolutionary relationships. Additionally, contrasting patterns of genetic diversity highlight different transmission dynamics: in long-established associations like AW, stable coevolution has produced high genetic diversity and slow LD decay, signaling a tightly integrated partnership, whereas in more recent infections, as seen in AD, AM, and AX, lower genetic diversity and frequent horizontal transfers mirror a dynamic phase of host-symbiont negotiation. These findings demonstrate Wolbachia’s diverse and adaptable transmission strategies, enabling its persistence across varied ecological contexts.
From a practical perspective, these findings carry substantial implications for the use of Wolbachia-based strategies in vector control and pest management. While the short-term goal of suppressing disease vectors or agricultural pests through mechanisms like cytoplasmic incompatibility is promising76,77, our data highlight the need for caution when considering the long-term evolutionary and ecological consequences. Persistent female bias in target populations could trap them in demographic cul-de-sacs, reducing overall genetic diversity and constraining their adaptive capacity in changing environments. Such genetic bottlenecks might initially seem beneficial for pest suppression but could lead to broader ecological disruptions, including niche vacuums that might be filled by more invasive or harmful species78. Furthermore, horizontal transmission of Wolbachia to non-target species—such as pollinators or other beneficial insects—poses additional risks, potentially destabilizing ecosystems. In an era of rapid environmental shifts and emerging pest challenges79, it is crucial to anticipate these long-term outcomes. A deeper understanding of Wolbachia’s genetic, demographic, and ecological impacts will help design more sustainable and responsible pest management strategies, minimizing risks while maximizing benefits. Recognizing the potential for unintended consequences in non-target populations is essential to ensure that these interventions achieve their goals without compromising broader ecosystem stability.
While this study highlights the profound impact of Wolbachia infection on host sex ratios and population dynamics, investigating such complex host-symbiont interactions in natural environments inevitably presents certain limitations. Although candidate genes associated with reproduction and sex determination were identified through genomic and population genetic analyses, their specific functions and causal relationships with Wolbachia infection remain experimentally unverified. In natural settings, directly manipulating gene functions or isolating environmental confounding factors is challenging, which restricts a deeper understanding of the mechanisms underlying Wolbachia’s effects. Despite these challenges, our study provides critical insights at both the genomic and population levels. It demonstrates how Wolbachia-driven male rarity creates intense, selective pressures and results in divergent demographic signals within male and female genomes. These findings lay the groundwork for future experimental investigations80,81, particularly in functional validation and the molecular mechanisms underpinning host-symbiont interactions. Furthermore, by capturing the long-term evolutionary dynamics of natural populations, this study complements the shorter temporal focus of laboratory-based research, offering a broader perspective on Wolbachia’s ecological and evolutionary roles under field conditions.
In sum, our findings highlight Wolbachia as a potent evolutionary force capable of sculpting host genomic landscapes and fundamentally altering host evolutionary dynamics. Far from being an evolutionary anomaly, this scenario underscores the pervasive influence of microbial symbionts. Our proposed “recurrent male bottleneck” model demonstrates how endosymbiont-driven ecological pressures can create complex evolutionary signatures. By causing shared population history to manifest differently in male and female genomes, Wolbachia illustrates the profound ways that microbial symbionts can reshape host evolutionary trajectories. Furthermore, our results reveal a complex selective landscape acting on the host. The selection signatures on male-function genes suggest two competing hypotheses: one of intense sexual selection on the few remaining males, and an alternative of relaxed selection reflecting a trajectory towards male obsolescence and parthenogenesis. The discovery of potentially deleterious mutations in key male-function genes lends intriguing, though not conclusive, support to this latter possibility. Ultimately, this work highlights how microbial symbionts create profound co-evolutionary conflicts and emphasize the need to integrate these intricate dynamics into our understanding of evolution.
Materials and methods
Sample collection and sequencing
We collected samples from six species of the genus Analcellicampa (AD, AX, AA, AM, AE, AW). Two previously sequenced and published species from the sister genus Monocellicampa82 were used as outgroup references. For AD, we performed PacBio SMRTbell library construction and sequencing using the PacBio Sequel II platform, with data quality evaluated through CCS v6.0.0 software (https://github.com/pacificbiosciences/unanimity). High-accuracy circular consensus sequencing (CCS) reads were obtained for downstream analysis, yielding a total of 14.69 GB of HiFi data. Hi-C data was generated on MGI sequencing platform to assist in genome assembly. Additionally, 54.80 GB of ultra-long read data was obtained from Oxford Nanopore Technology (ONT) sequencing. The remaining samples were sequenced using Illumina paired-end sequencing, with achieving depths exceeding 20× (Supplementary Data 8).
Genome survey and assembly
Genomic sequences were characterized for size, heterozygosity and repetitiveness through k-mer frequency analysis (k = 21) using KMC3 [v3.2.1]83 and GenomeScope 2.018. To ensure reads are reliable, Illumina paired-end sequenced raw reads for the genomic survey were first filtered using the Fastp v.0.20.0 preprocessor (set to default parameters)84. Heterozygous k-mer pairs were then analyzed using Smudgeplot18 to estimate ploidy levels and infer genomic complexity.
The Nanopore sequencing platform was used for the Ultra-long sequencing of DNA samples. The failed reads were removed from the raw data and We quality-filtered male and female ONT reads using NanoFilt [v2.5.0]85 software was used to filter the fragments <10 kb. The obtained pass reads were used for subsequent analyses. The joint sequence was filtered using Porechop v0.2.4 software (https://github.com/rrwick/Porechop), reads with retention lengths ≥30 kb and mean read quality scores >90% were used for assembly.
To maximize the quality of the final assembly with these data, we generated and compared several draft genome assemblies using four methods: Flye [v2.9.2]21, hifiasm [v0.18.2]24 and LJA [v0.2]23. Each assembly was assessed the contiguity and completeness using the program compleasm86. against the insecta ortholog database (Supplementary Data 2). Based on these evaluations, we selected the highest quality genome assembly (the Flye assembly; Supplementary Data 2) to proceed with Hi-C scaffolding. we mapped Hi-C reads to the draft assembly using BWA [v0.7.17]87 and scaffolds were constructed using YaHS [v1.2a.1]88, producing 232 scaffolds. HiFi reads were compressed following a process similar to Canu89 and used to extend scaffolds larger than 1 Mb with custom scripts. During each round of extension, we performed a BLAST search of the extended sequences against the assembly to check for overlaps with the assembled scaffolds. If an overlap was consistently and stably detected at the ends of two scaffolds, the two scaffolds were connected; otherwise, the extension continued until it could no longer proceed. Telomere sequences were also extended as much as possible using this method. Then, assembly underwent two rounds of additional polishing with NextPolish90 using resequencing data. Using this method, we ultimately yielded nine chromosome-scale scaffolds with no apparent large-scale misassemblies. The final Hi-C contact map was visualized with Juicebox [v1.11]91 to identify and correct potential misassemblies.
In addition, the complete Wolbachia genome (1,236,102 bp) was assembled using the Flye assembler and polished as part of the aforementioned process. The Wolbachia genome quality was evaluated using BUSCO scores with the rickettsiales_odb10 database. We annotated the Wolbachia genome with Prokka [v1.1.1]29. We assembled and annotated the mitochondrial genome (199,511 bp) and using the MitoHiFi [v3.2]92.
Identification of telomeres and centromeres
We employed quarTeT93 to predict both telomeres and centromeres. Telomeres were predicted using quarTeT TeloExplorer module with the parameters: ‘-c other -m 100’. Centromeres were predicted using quarTeT CentroMiner module with the parameters: ‘-n 100 -m 200 -s 0.8 -d 10 -e 0.00001 -g 50000 -i 100000 --trf 2 7 7 80 10 50 -r 3 --TE EDTA.TEanno.gff3’. Additionally, centromFind, with default parameters, was used to further validate centromere positions. These tools utilize complementary approaches for centromere prediction. CentroMiner identifies centromere candidates based on genome data and transposable element (TE) annotation, focusing on tandem repeats and surrounding TE regions. In contrast, CentromFind uses Hi-C matrices to detect centromeres by identifying regions with low interaction frequencies, which are indicative of centromeric repetitive sequences. The integrated results from the quarTeT CentroMiner module and centromFind. When both tools identified centromeres, their intersection was taken. If only one tool produced predictions, those results were directly used.
Annotation of repetitive sequences
De novo repeat library prediction and homology-based comparisons were used for the annotation of repetitive sequences. We used RepeatModeler [v2.0.2] (https://github.com/Dfam-consortium/RepeatModeler) with default parameters to build the de novo repeat library. LTR_FINDER [v1.70]94 and LTR_retriever [v2.9.0]95, both with default parameters, were applied to identify long terminal repeat (LTR) sequences in the genome. Annotation and masking were performed using RepeatMasker [v4.1.5]96, following the pipeline (https://darencard.net/blog/2022-07-09-genome-repeat-annotation/). Specifically, the genome was annotated and masked in a stepwise manner using RepeatMasker: simple repeats, were annotated first, followed by curated repeat elements from existing databases. Next, known species-specific repeats identified from the sixth iteration of RepeatModeler were annotated, and finally, unknown species-specific repeats from the same cycle were incorporated.
Genome annotation
The genomic region containing repetitive sequences was masked and utilized for subsequent analyses. We employed three complementary strategies to predict protein-coding genes: homology-based prediction, transcriptome-based prediction, and ab initio gene prediction. For the transcriptome-based approach, we first mapped clean transcriptome sequences to the assembled genome using HISAT2 [v2.1.0]97. Transcript structures were then reconstructed with StringTie [v2.2.1]98, and coding regions were identified using TransDecoder [v5.5.0] (https://github.com/TransDecoder/TransDecoder). To refine these predictions, we used BLAST and HMMER to compare TransDecoder’s predicted coding sequences with known protein databases. The results from these homology searches were integrated into the transcriptome-based gene predictions to improve accuracy. For homology-based annotation, we used Miniprot [v0.13]99 to align homologous protein sequences (Arthropoda) to the genome, enabling the inference of unknown gene structures. This approach allowed us to predict gene functions, domains, and other characteristics by comparing the target genome with known homologous protein sequences. For ab initio gene prediction, we utilized Braker3 [v3.0.6]100 and Augustus [v3.5.0]101 with default parameters to perform gene prediction on the repeat-masked genome assembly. During the Braker3 run, we incorporated protein homology information from OrthoDB of arthropoda to enhance prediction accuracy. Finally, we integrated gene models from the different prediction methods using EvidenceModeler [v2.1.0]102 to produce a consolidated set of gene predictions. Following gene prediction, we performed functional annotation by matching predicted genes against various databases. Specifically, we used BLASTP [v2.12.0] with a threshold of 1e-5 to compare predicted genes against the SwissProt protein database. Additionally, we employed the eggNOG-mapper web server (http://eggnog-mapper.embl.de/) for functional annotation, including Gene Ontology (GO) terms, KEGG pathway annotations, and Pfam domain annotations.
Preliminary analysis of resequencing data to identify Wolbachia Infection using HAYSTAC
Wolbachia, like most endosymbionts, cannot be cultured independently16. Consequently, even when enrichment protocols are employed, host DNA is typically co-sequenced with Wolbachia in the same experiment. This presents challenges related to sequencing coverage and potential contamination, as the host genome is often two to three orders of magnitude larger than the Wolbachia genome. Therefore, implementing a bioinformatics pipeline is crucial for detecting Wolbachia within host resequencing datasets. Here, we screened 89 Analcellicampa resequncing data using HAYSTAC103 with default parameters to detect the Wolbachia genome. Before examination, a custom database of complete Wolbachia genomes was compiled (Supplementary Data 14), including 24 complete genomes from Wolbachia parasitizing Hymenoptera and other hosts, obtained from the NCBI RefSeq database, along with the Wolbachia genome assembled in this study. For samples with a higher number of confidently assigned Wolbachia reads ( > 3000). We further ensured the results’ validity by using IGV104 to checking a detailed of the coverage and distribution of reads on Wolbachia genome. Both approaches provide robust support for confirming Wolbachia infection.
Mapping and variant calling
We first performed quality control on raw sequencing reads using fastp90 for trimming and filtering. High-quality paired-end resequencing reads were then mapped to AD genome with BWA “BWA-MEM” algorithm105 using the default parameters. Post-alignment processing included sorting, marking duplicates, indel realignment, and base quality recalibration, which were carried out using Picard (http://picard.sourceforge.net, last accessed January 5, 2021) and Genome Analysis Toolkit (GATK)106 software. Genomic variants for each individual were then identified using the HaplotypeCaller module and the GVCF model in GATK. followed by merging all GVCF files. SNPs were called with the HaplotypeCaller module. We then kept SNP sites if they passed the recommended hard filtering thresholds (QD > 2, FS < 60, MQ > 40, MQRankSum > −12.5, and ReadPosRankSum > 15) as used previously107. Additional filtering was applied, retaining SNPs with a minor allele frequency (MAF) > 0.01 and SNP call rates > 80%. The resulting high-quality variants were used for downstream analyses. The transition-to-transversion (Ts/Tv) ratio was calculated via VCFtools108 to validate the quality of our SNPs set. Called genotypes were phased using SHAPEIT4109. SnpEff (v5.0)61 was used for annotating and predicting the effects of identified SNPs. We used SIFT62 based on the AD reference genome to predict the functional impact of amino acid substitutions; scores < 0.05 indicate potentially deleterious changes.
Phylogenetic and population genetic analyses
A maximum-likelihood tree was constructed using iqtree program (version: 1.6.12)110 in ASC model sites with optimal substitution model determined by the software and with 1000 bootstrap replicates. Principal Component Analysis (PCA) was conducted using the “SNPRelate” package in R111 based on whole-genome SNPs dataset. ADMIXTURE [v1.3.0]36 was used to quantify the genome wide admixtures among all samples. Admixture was run for each possible genetic clusters (K = 2 to 9) with 10 replicates for each K. We ran CLUMPAK112 on ADMIXTURE outputs to generate consensus solutions for each K and finally identified the optimal K cluster supported by the data based on cross validation (CV) errors. Population splitting and admixture analyses were further carried out using TreeMix program113. This method requires unlinked markers, so we used SNPrelate to prune out markers with high linkage (r2 ≥ 0.2 within 100 Kb). We selected an appropriate number of migration edges based on the decay in variation explanation improvements with successive numbers. BITE114 was used to do 100 bootstrap replicates, and to visualize the consensus trees with bootstrap values and migration edges when doing TreeMix analyses. Identical scores (ISs) were calculated to evaluate the similarities of the sequenced genomes to the AD reference genome according to the following formula:
where Si is the number of alleles identical to the AD reference allele at a given SNP site i, n is the total number of SNPs within a 50-kb window and n′ is the total number of missing SNPs within a 50-kb windows.
Demographic history and selection analyses
We used PSMC38 to estimate variation in Ne over historical time. For diploid females, consensus sequences were generated using the ‘mpileup’ command from the SAMtools (http://samtools.sourceforge.net/) package, applying the recommended PSMC settings (-C 50, -O, -D 2*reads_depth, -d 1/3*reads_depth). Input files were created using the ‘fq2psmcfa’ tool. For each sample, the PSMC algorithm was run for 30 iterations. We estimated piecewise constant ancestral Ne over 90 time intervals, following the methodology of Wallberg et al115. For haploid males, we applied the established “pseudo-diploid” method by analyzing sequence divergence between pairs of males to infer demographic parameters. This approach is appropriate for detecting how shared population history manifests differently across genomic lineages with distinct inheritance patterns. To account for variance in Ne, we performed 100 bootstrap replications for representative samples from each subspecies. These steps ensured robust validation of historical population size fluctuations.
Since PSMC analyzes a single individual per population, we also employed SMC + + (v1.15.2)41 to infer population size dynamics using multiple individuals per population. To ensure robust mapping, uniquely mappable regions were identified using the SNPable toolkit (http://lh3lh3.users.sourceforge.net/snpable.shtml) with settings -k = 35 and r = 0.9. When analyzing population size dynamics separately for males and females, we focused on populations with more than one individual for each sex (AD, AX, and AM). As all individuals had a sequencing coverage exceeding 20-fold, there was no need for sample exclusion due to low coverage. Input files for SMC + + were prepared following the recommended pipeline from the SMC + + GitHub repository (https://github.com/popgenmethods/smcpp). All analyses were conducted using default parameters to ensure consistency across datasets.
Finally, we used momi237, which fits models to the site-frequency spectrum, to further assess the underlying population history based on six populations (AD, AW, AX, AM, AA, and AE) and Outgroup, assuming a mutation rate of 5.27 × 10-9 per site per generation115 and a generation time of 1 year. Given that our study primarily focuses on changes in Ne across species, we tested two models: one assuming constant population size and another allowing for variable population size over the past 30,000 years, reflecting potential effects of the LGM and corresponding to the Ne declines observed during this period in PSMC analyses (Fig. 2h and Supplementary Fig. 8). The best-fitting model was selected based on the Akaike Information Criterion (AIC) value. Then, we performed 20 independent runs with different starting parameters and kept the model with the biggest log-likelihood value for the optimal model. To ensure robust inference of population parameters, we conducted 50 bootstraps, providing confidence intervals for the estimated values.
Nucleotide diversity (π) and dxy values were calculated for six species in and between Analcellicampa genus using pixy [v1.2.7]116 with a 10-kb window and a 5-kb sliding window. The average π or dxy was calculated using the formula: sum(window count_diffs)/sum(window comparisons). To detect genomic regions under selection in AD, we calculated the log2(π-AD/π-AX). We alsp used updated XP-CLR117 to compare the allele frequency distributions for the AD and AX populations to detect selective sweeps based on phased genotypes that were inferred by SHAPEIT4. The command line was xpclr --input input.vcf --samplesA AD_list --samplesB AX_list –chr chrN –phased --size 10000 --step 5000 -maxsnps 600 –out outputFile. The parameters used included overlapping sliding windows of 10 kb with a 5 kb step and a maximum of 600 SNPs per window. Genomic regions falling within the top 5% of XP-CLR values and log2(π ratio) values across the whole genome were identified as candidate selective sweeps. We carried out Gene Ontology (GO) enrichment analysis on the PSGs within candidate selective sweep regions using the enrichGO function from clusterProfiler [v3.14.3]118 and the org.Dm.eg.db database.
Statistics and reproducibility
Detailed explanations of the statistical methods used in this study can be found in the relevant sections of the Methods. For the selection scan, 30 AD individuals and 26 AX individuals were used. When performing enrichment analysis using clusterProfiler v3.14.3, the Benjamini-Hochberg method is used to adjust the p-values. For figures, each data point corresponds to an individual animal.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The genome assembly has been deposited in the figshare and is available at: https://figshare.com/articles/dataset/sawfly5_3_fa/29650295. All DNA-seq and RNA-seq data are deposited in GSA database under Bioproject ID PRJCA043463. The data can be accessed via https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA043463. The numerical source data behind the graphs are listed in Supplementary Data S1-14.
Code availability
The analysis scripts used for conducting the data analyses in this study are publicly available on Zenodo (https://doi.org/10.5281/zenodo.16403754119).
References
Moreira, L. A. et al. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139, 1268–1278 (2009).
Klepzig, K., Adams, A., Handelsman, J. & Raffa, K. Symbioses: a key driver of insect physiological processes, ecological interactions, evolutionary diversification, and impacts on humans. Environ. Entomol. 38, 67–77 (2009).
Porter, S. S. & Sachs, J. L. Agriculture and the disruption of plant–microbial symbiosis. Trends Ecol. Evol. 35, 426–439 (2020).
Brucker, R. M. & Bordenstein, S. R. Speciation by symbiosis. Trends Ecol. Evol. 27, 443–451 (2012).
Weeks, A. R., Reynolds, K. T. & Hoffmann, A. A. Wolbachia dynamics and host effects: what has (and has not) been demonstrated?. Trends Ecol. Evol. 17, 257–262 (2002).
Hilgenboecker, K., Hammerstein, P., Schlattmann, P., Telschow, A. & Werren, J. H. How many species are infected with Wolbachia?–a statistical analysis of current data. FEMS Microbiol. Lett. 281, 215–220 (2008).
LePage, D. & Bordenstein, S. R. Wolbachia: can we save lives with a great pandemic?. Trends Parasitol. 29, 385–393 (2013).
Cordaux, R., Bouchon, D. & Grève, P. The impact of endosymbionts on the evolution of host sex-determination mechanisms. Trends Genet 27, 332–341 (2011).
Gong, J. T. et al. Wolbachia-based strategies for control of agricultural pests. Curr. Opin. Insect Sci. 57, 101039 (2023).
Hughes, G. L. & Rasgon, J. L. Transinfection: a method to investigate Wolbachia–host interactions and control arthropod-borne disease. Insect Mol. Biol. 23, 141–151 (2014).
Sanaei, E., Charlat, S. & Engelstädter, J. Wolbachia host shifts: routes, mechanisms, constraints and evolutionary consequences. Biol. Rev. Camb. Philos. Soc. 96, 433–453 (2021).
Henry, L. P., Bruijning, M., Forsberg, S. K. & Ayroles, J. F. The microbiome extends host evolutionary potential. Nat. Commun. 12, 5141 (2021).
Werren, J. H., Baldo, L. & Clark, M. E. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6, 741–751 (2008).
Clark, M. et al. Wolbachia modification of sperm does not always require residence within developing sperm. Heredity 101, 420–428 (2008).
Niu, G. -y et al. A new species of Analcellicampa Wei (Hymenoptera: Tenthredinidae), feeding in fruit of the cherry Cerasus pseudocerasus× Cerasus avium. Sci. Silvae. Sin. 57, 113–120 (2021).
Scholz, M. et al. Large scale genome reconstructions illuminate Wolbachia evolution. Nat. Commun. 11, 5235 (2020).
Gerth, M., Gansauge, M. T., Weigert, A. & Bleidorn, C. Phylogenomic analyses uncover origin and spread of the Wolbachia pandemic. Nat. Commun. 5, 5117 (2014).
Ranallo-Benavidez, T. R., Jaron, K. S. & Schatz, M. C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 11, 1432 (2020).
Oeyen, J. P. et al. Sawfly genomes reveal evolutionary acquisitions that fostered the mega-radiation of parasitoid and eusocial Hymenoptera. Genome Biol. Evol. 12, 1099–1188 (2020).
Vurture, G. W. et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33, 2202–2204 (2017).
Kolmogorov, M., Yuan, J., Lin, Y. & Pevzner, P. A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546 (2019).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Bankevich, A., Bzikadze, A. V., Kolmogorov, M., Antipov, D. & Pevzner, P. A. Multiplex de Bruijn graphs enable genome assembly from long, high-fidelity reads. Nat. Biotechnol. 40, 1075–1081 (2022).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Vicari, M. R., Bruschi, D. P., Cabral-de-Mello, D. C. & Nogaroto, V. Telomere organization and the interstitial telomeric sites involvement in insects and vertebrates chromosome evolution. Genet. Mol. Biol. 45, e20220071 (2022).
Shen, F., Xu, S., Shen, Q., Bi, C. & Lysak, M. A. The allotetraploid horseradish genome provides insights into subgenome diversification and formation of critical traits. Nat. Commun. 14, 4102 (2023).
Jayakodi, M. et al. The giant diploid faba genome unlocks variation in a global protein crop. Nature 615, 652–659 (2023).
Melters, D. P. et al. Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14, 1–20 (2013).
Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014).
Jacobs, J. et al. Complete de novo assembly of Wolbachia endosymbiont of Drosophila willistoni using long-read genome sequencing. Sci. Rep. 14, 17770 (2024).
Grant, J. R. et al. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res 51, W484–W492 (2023).
Klasson, L. et al. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol. Biol. Evol. 25, 1877–1887 (2008).
Neupane, S., Bonilla, S. I., Manalo, A. M. & Pelz-Stelinski, K. S. Complete de novo assembly of Wolbachia endosymbiont of Diaphorina citri Kuwayama (Hemiptera: Liviidae) using long-read genome sequencing. Sci. Rep. 12, 125 (2022).
Zhang, Z. et al. Complete De Novo Assembly of Wolbachia Endosymbiont of Frankliniella intonsa. Int. J. Mol. Sci. 24, 13245 (2023).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491 (2011).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19, 1655–1664 (2009).
Kamm, J., Terhorst, J., Durbin, R. & Song, Y. S. Efficiently inferring the demographic history of many populations with allele count data. J. Am. Stat. Assoc. 115, 1472–1487 (2020).
Li, H. & Durbin, R. Inference of human population history from individual whole-genome sequences. Nature 475, 493–496 (2011).
Haber, M. et al. Chad genetic diversity reveals an African history marked by multiple Holocene Eurasian migrations. Am. J. Hum. Genet. 99, 1316–1324 (2016).
Wang, M.-S. et al. Ancient hybridization with an unknown population facilitated high-altitude adaptation of canids. Mol. Biol. Evol. 37, 2616–2629 (2020).
Choi, J. Y., Bubnell, J. E. & Aquadro, C. F. Population genomics of infectious and integrated Wolbachia pipientis genomes in Drosophila ananassae. Genome Bio. l Evol. 7, 2362–2382 (2015).
Kriesner, P., Hoffmann, A. A., Lee, S. F., Turelli, M. & Weeks, A. R. Rapid sequential spread of two Wolbachia variants in Drosophila simulans. PLoS Pathog. 9, e1003607 (2013).
Stahlhut, J. K. et al. The mushroom habitat as an ecological arena for global exchange of Wolbachia. Mol. Ecol. 19, 1940–1952 (2010).
Terhorst, J., Kamm, J. A. & Song, Y. S. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 49, 303–309 (2017).
Strunov, A., Lerch, S., Blanckenhorn, W. U., Miller, W. J. & Kapun, M. Complex effects of environment and Wolbachia infections on the life history of Drosophila melanogaster hosts. J. Evol. Biol. 35, 788–802 (2022).
Verma, S., Ramani, R., Sachan, A. & Chandra, R. The role of Wolbachia and the environment on sex determination of the Indian lac insect, Kerria lacca (Coccoidea: Tachardiidae). J. Asia Pac. Entomol. 26, 102019 (2023).
Fujimoto, Y. et al. Homeoproteins Six1 and Six4 regulate male sex determination and mouse gonadal development. Dev. Cell. 26, 416–430 (2013).
Kong, R. & Somers, W. G. dRTEL1 is essential for the maintenance of Drosophila male germline stem cells. PLoS Genet 17, e1009834 (2021).
Anderson, M. A. et al. Asunder is a critical regulator of dynein–dynactin localization during Drosophila spermatogenesis. Mol. Biol. Cell. 20, 2709–2721 (2009).
Noda, T. et al. Sperm membrane proteins DCST1 and DCST2 are required for sperm-egg interaction in mice and fish. Commun. Biol. 5, 332 (2022).
Linnemannstöns, K. et al. The PTK7-related transmembrane proteins off-track and off-track 2 are co-receptors for Drosophila Wnt2 required for male fertility. PLoS Genet 10, e1004443 (2014).
Whitten, S. J. & Miller, M. A. The role of gap junctions in Caenorhabditis elegans oocyte maturation and fertilization. Dev. Biol. 301, 432–446 (2007).
Carvalho, A. B., Lazzaro, B. P. & Clark, A. G. Y chromosomal fertility factors kl-2 and kl-3 of Drosophila melanogaster encode dynein heavy chain polypeptides. Proc. Natl Acad. Sci. Usa. 97, 13239–13244 (2000).
Ma, B. et al. Serotonin signaling modulates sexual receptivity of virgin female. Drosoph. Neurosci. Bull. 38, 1277–1291 (2022).
Grosjean, Y., Grillet, M., Augustin, H., Ferveur, J.-F. & Featherstone, D. E. A glial amino-acid transporter controls synapse strength and courtship in. Drosoph. Nat. Neurosci. 11, 54–61 (2008).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012).
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009).
Pai, L. M., Barcelo, G. & Schüpbach, T. D-cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell 103, 51–61 (2000).
Kiger, A. A. et al. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science 294, 2542–2545 (2001).
Jaenike, J. Spontaneous emergence of a new Wolbachia phenotype. Evolution 61, 2244–2252 (2007).
Hurst, G. D. & Frost, C. L. Reproductive parasitism: maternally inherited symbionts in a biparental world. Cold Spring Harb. Perspect. Biol. 7, a017699 (2015).
Saubin, M. et al. Neutral genetic structuring of pathogen populations during rapid adaptation. J. Hered. 116, 62–77 (2025).
Wiwatanaratanabutr, I. & Grandjean, F. Impacts of temperature and crowding on sex ratio, fecundity and Wolbachia infection intensity in the copepod, Mesocyclops thermocyclopoides. J. Invertebr. Pathol. 141, 18–23 (2016).
Gora, N. V. et al. Climate factors and Wolbachia infection frequencies in natural populations of Drosophila melanogaster. Cytol. Genet. 54, 189–198 (2020).
Normark, B. B. The evolution of alternative genetic systems in insects. Annu. Rev. Entomol. 48, 397–423 (2003).
Burt, A. & Trivers, R. Genes in conflict: the biology of selfish genetic elements. Harvard University Press, (2006).
Charlat, S., Hurst, G. D. & Merçot, H. Evolutionary consequences of Wolbachia infections. Trends Genet. 19, 217–223 (2003).
Nielsen, R. Molecular signatures of natural selection. Annu. Rev. Genet. 39, 197–218 (2005).
Kraaijeveld, K. Male genes with nowhere to hide: sexual conflict in haplodiploids. Anim. Biol. 59, 403–415 (2009).
Kraaijeveld, K. et al. Does a parthenogenesis-inducing Wolbachia induce vestigial cytoplasmic incompatibility?. Naturwissenschaften 98, 175–180 (2011).
Kremer, N. et al. A new case of Wolbachia dependence in the genus Asobara: evidence for parthenogenesis induction in Asobara japonica. Heredity 103, 248–256 (2009).
Reynolds, L. A. et al. Suppression of Wolbachia-mediated male-killing in the butterfly Hypolimnas bolina involves a single genomic region. PeerJ 7, e7677 (2019).
Raychoudhury, R., Baldo, L., Oliveira, D. C. & Werren, J. H. Modes of acquisition of Wolbachia: horizontal transfer, hybrid introgression, and codivergence in the Nasonia species complex. Evolution 63, 165–183 (2009).
Baldo, L. et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 72, 7098–7110 (2006).
Porter, J. & Sullivan, W. The cellular lives of Wolbachia. Nat. Rev. Microbiol 21, 750–766 (2023).
Zabalou, S. et al. Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. Proc. Natl Acad. Sci. Usa. 101, 15042–15045 (2004).
Hoffmann, A. A. et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476, 454–457 (2011).
Liu, C. et al. Most invasive species largely conserve their climatic niche. Proc. Natl Acad. Sci. Usa. 117, 23643–23651 (2020).
McCulloch, G. A. & Waters, J. M. Rapid adaptation in a fast-changing world: Emerging insights from insect genomics. Glob. Chang. Biol. 29, 943–954 (2023).
Zhu, Y. X. et al. Wolbachia modify host cell metabolite profiles in response to short-term temperature stress. Environ. Microbiol. Rep. 16, e70013 (2024).
Crossley, M. S. et al. No net insect abundance and diversity declines across US Long Term Ecological Research sites. Nat. Ecol. Evol. 4, 1368–1376 (2020).
Meicai, W. Two new genera of Hoplocampinae from China with a key to known genera of the subfamily in the world (Hymenoptera: Nematidae). J. Cent. South. Coll. 18, 12–18 (1998).
Kokot, M., Długosz, M. & Deorowicz, S. KMC 3: counting and manipulating k-mer statistics. Bioinformatics 33, 2759–2761 (2017).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
De Coster, W., D’hert, S., Schultz, D. T., Cruts, M. & Van Broeckhoven, C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669 (2018).
Huang, N. & Li, H. compleasm: a faster and more accurate reimplementation of BUSCO. Bioinformatics 39, btad595 (2023).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010).
Zhou, C., McCarthy, S. A. & Durbin, R. YaHS: yet another Hi-C scaffolding tool. Bioinformatics 39, btac808 (2023).
Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27, 722–736 (2017).
Hu, J., Fan, J., Sun, Z. & Liu, S. NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics 36, 2253–2255 (2020).
Durand, N. C. et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016).
Uliano-Silva, M. et al. MitoHiFi: a python pipeline for mitochondrial genome assembly from PacBio high fidelity reads. BMC Bioinforma. 24, 288 (2023).
Lin, Y. et al. quarTeT: a telomere-to-telomere toolkit for gap-free genome assembly and centromeric repeat identification. Hortic. Res. 10, uhad127 (2023).
Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 35, W265–W268 (2007).
Ou, S. & Jiang, N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176, 1410–1422 (2018).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinforma. 25, 10 (2009). 11-14.10. 14.
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Li, H. Protein-to-genome alignment with miniprot. Bioinformatics 39, btad014 (2023).
Hoff, K. J., Lomsadze, A., Borodovsky, M. & Stanke, M. Whole-genome annotation with BRAKER. Methods Mol. Biol. 1962, 65–95 (2019).
Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res 34, W435–W439 (2006).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9, 1–22 (2008).
Dimopoulos, E. A. et al. HAYSTAC: A Bayesian framework for robust and rapid species identification in high-throughput sequencing data. PLoS Comput. Biol. 18, e1010493 (2022).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform 14, 178–192 (2013).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303 (2010).
Zhang, M., Yang, Q., Ai, H. & Huang, L. Revisiting the evolutionary history of pigs via de novo mutation rate estimation in a three-generation pedigree. Genom. Proteom. Bioinf. 20, 1040–1052 (2022).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Delaneau, O., Zagury, J. F., Robinson, M. R., Marchini, J. L. & Dermitzakis, E. T. Accurate, scalable and integrative haplotype estimation. Nat. Commun. 10, 5436 (2019).
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Zheng, X. et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328 (2012).
Kopelman, N. M., Mayzel, J., Jakobsson, M., Rosenberg, N. A. & Mayrose, I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 15, 1179–1191 (2015).
Pickrell, J. K. & Pritchard, J. K. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet 8, e1002967 (2012).
Milanesi, M. et al. BITE: an R package for biodiversity analyses. BioRxiv, 181610 (2017).
Wallberg, A. et al. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat. Genet. 46, 1081–1088 (2014).
Korunes, K. L. & Samuk, K. pixy: Unbiased estimation of nucleotide diversity and divergence in the presence of missing data. Mol. Ecol. Resour. 21, 1359–1368 (2021).
Chen, H., Patterson, N. & Reich, D. Population differentiation as a test for selective sweeps. Genome Res 20, 393–402 (2010).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics: a J. Integr. Biol. 16, 284–287 (2012).
Zhang, M. Scripts for the article: ‘Telomere-to-telomere genome assembly uncovers Wolbachia-driven recurrent male bottleneck effect and selection in a sawfly’. (Zenodo, https://doi.org/10.5281/zenodo.16403754 (2025).
Acknowledgements
This work was financially by grants from National Natural Science Foundation of China (Grant No. 32370500), Jiangxi Provincial Natural Science Foundation (Grant No. 20232BAB215017), and Science and Technology Research Project of Jiangxi Provincial Department of Education (Grant No. GJJ2200344).
Author information
Authors and Affiliations
Contributions
M.W. designed research; G.N., B.T. and D.W. contributed samples; M.Z. performed research; M.Z., R.Z., G.M. and J.C. analyzed data; M.Z. and M.W. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics
All procedures involving animals were based on the care and use guidelines of experimental animals established by the Ministry of Science and Technology of China. We have complied with all relevant ethical regulations for animal use.
Peer review
Peer review information
Communications Biology thanks Ken Kraaijeveld and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Aylin Bircan, Johannes Stortz. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, M., Zhai, R., Niu, G. et al. Telomere-to-telomere genome assembly uncovers Wolbachia-driven recurrent male bottleneck effect and selection in a sawfly. Commun Biol 8, 1211 (2025). https://doi.org/10.1038/s42003-025-08629-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08629-0