Abstract
Phospholipase Cε (PLCε) cleaves phosphatidylinositol lipids to increase intracellular Ca2+ and activate protein kinase C (PKC) in response to stimulation of cell surface receptors. PLCε is activated via direct binding of small GTPases at the cytoplasmic leaflets of cellular membranes. In the cardiovascular system, the RhoA GTPase regulates PLCε to initiate a pathway that protects against ischemia/reperfusion injuries, but the underlying molecular mechanism is not known. We present here the cryo-electron microscopy (cryo-EM) reconstruction of RhoA bound to PLCε, showing that the G protein binds a unique insertion within the PLCε EF hands. Deletion of or mutations to this PLCε insertion decrease RhoA-dependent activation without impacting its regulation by other G proteins. Together, our data support a model wherein RhoA binding to PLCε allosterically activates the lipase and increases its interactions with the membrane, resulting in maximum activity and cardiomyocyte survival.
Similar content being viewed by others
Introduction
Mammalian phospholipase C (PLC) enzymes are translocated and activated at the cytoplasmic leaflet of membranes in response to diverse stimuli. All PLCs cleave phosphatidylinositol-4,5-bisphosphate (PIP2) at the plasma membrane, producing inositol-1,4,5-triphosphate (IP3) and diacylglycerol (DAG). IP3 stimulates intracellular Ca2+ release, which, together with DAG, activates protein kinase Cs (PKCs)1. PLCε also cleaves phosphatidylinositol-4-phosphate (PI4P) at the perinuclear membrane, where local increases in DAG activate PKCs and protein kinase D2,3.
PLCε is activated downstream of G protein-coupled receptors and receptor tyrosine kinases (RTKs) through binding of the Rap1A, RhoA, and Ras GTPases, as well as the Gβγ heterodimer. Activation likely proceeds through simultaneous membrane localization and activation, with the G protein dictating the location of activation2,3,4. In the cardiovascular system, PLCε activation has been best studied in response to stimulation of Gs- and G12/13-coupled receptors. Stimulation of the β-adrenergic receptors leads to activation of the Rap1A GTPase, which in turn activates PLCε at the perinuclear membrane. Increased PI4P hydrolysis activates PKC- and PKD-dependent pathways that maximize Ca2+-induced Ca2+ release and contractility5,6. However, sustained activation leads to upregulation of genes that promote cardiac hypertrophy7,8,9,10,11. Gβγ-dependent activation of PLCε, downstream of the endothelin-1 receptor, results in a similar pathological response10,11,12. Intriguingly, PLCε has been reported to have a cardioprotective role in response to ischemia/reperfusion injuries. Stimulation of G12/13-coupled receptors, such as the sphingosine-1-phosphate receptor (S1PR), activates RhoA2,3. In this pathway, RhoA activates PLCε at the plasma membrane, where PIP2 hydrolysis increases intracellular Ca2+ and PKC activity. The latter activates a PKD-dependent pathway that protects the mitochondria from oxidative stress, a major cause of acute cardiomyocyte cell death under ischemic conditions13,14,15,16,17.
The ability of PLCε to hydrolyze substrates at different intracellular sites is due to its subfamily-specific regulatory domains and insertions. Like other PLCs, PLCε contains a pleckstrin homology (PH) domain, four EF hand repeats (EF1-4), the catalytic TIM barrel, and a C2 domain (Fig. 1a)3,18. The core is flanked by an N-terminal region and a CDC25 domain that is a guanine nucleotide exchange factor (GEF) for the Rap1A GTPase. At its C-terminus are two Ras association (RA) domains, RA1 and RA2. RA1 stabilizes the catalytic core, while RA2 binds Rap1A and Ras GTPases3,19,20. Finally, the TIM barrel contains two insertions: the X–Y linker and the Y-box. As in PLCβ and PLCδ, the PLCε X–Y linker occludes the active site and must be displaced via an interfacial activation mechanism to allow substrate binding3,21,22. The Y-box is a ~70 amino acid insertion unique to the PLCε subfamily23, but whether it has a role in basal activity is less clear.
a Domain architecture of R. norvegicus PLCε, with domain boundaries shown above. Variants used in this study are shown below, open boxes indicate internal deletions. b (Left) Basal and RhoAG14V-stimulated activities of PLCε and variants retaining EF1/2 and/or the Y-box. At least three independent experiments from independent transfections were carried out for each variant, and data are shown as the average of triplicate measurements ± SD. Data was analyzed using an unpaired, one-tailed t-test with Welch’s correction comparing the basal and RhoA-stimulated activities of each variant. ****p < 0.0001, for PH-C **p < 0.0053, EF-C **p < 0.0034, PLCεΔY **p < 0.0075, EF-CΔY *p < 0.012. (Right) The change in maximal activity ± SD was calculated by subtracting RhoAG14V-stimulated activity from the basal activity of each variant. Data were analyzed using a one-way ANOVA and Kruskal–Wallis test comparing each variant to PLCε, followed by a Dunn’s multiple comparisons test. For EF3-C **p < 0.0053, PLCεΔY **p < 0.0030, and EF-C-ΔY *p > 0.0159. Representative Western blots are shown below, with empty pCMV vector (EV) and β-actin used as loading controls. Differences in expression were not found to be statistically significant but may still contribute to variation in activities. PLCε variants express a C-terminal FLAG tag and are detected with an anti-FLAG antibody, while RhoA contains an N-terminal HA tag and is detected with an anti-HA antibody.
RhoA is the most robust activator of PLCε, increasing its activity ~5-10-fold over basal in cell-based assays3,24,25. Initial studies demonstrated that only the active form the GTPase directly interacted with the lipase to increase activity. Efforts to map its binding site relied on a series of N- and C-terminally truncated PLCε variants that narrowed its binding site to a region between the EF hands and C2 domains (Fig. 1a). Given that only PLCε contains the Y-box, it emerged as a possible binding site for the GTPase, especially because its deletion eliminated RhoA-dependent activation. However, N-terminally truncated PLCε variants, with or without a Y-box, were shown to pull down the active GTPase to similar extents23,26. Although the Y-box may be required for RhoA-mediated activation, it is clearly not the binding site.
In this work, we define the minimal structural requirements for RhoA-dependent activation of PLCε. Prenylated RhoA·GTP is required for maximum activation, but a soluble mutant also stimulates the lipase, demonstrating the mechanism likely involves both membrane localization and allosteric components. Informed by newly annotated PLCε domain boundaries, we show PLCε variants retaining the PH domain and EF hands 1/2 (EF1/2) are robustly activated by the GTPase, whereas variants that lack the Y-box, the N-terminus, CDC25, PH domain, and/or EF1/2 hands have decreased basal and RhoA-stimulated activities. Our cryo-electron microscopy (cryo-EM) reconstruction of a RhoA·GTP–PLCε complex reveals an integral role of the EF hands in the mechanism, with RhoA·GTP binding to a PLCε subfamily-specific insertion in this domain, ~60 Å away from the active site and Y-box in the TIM barrel. Mutation or deletion of the PLCε EF hand insertion compromises RhoA-dependent activation in cells. Comparison of the RhoA·GTP–PLCε reconstruction to other PLCε structures shows that RhoA binding induces conformational changes within the EF hands that likely contribute to allosteric activation.
Results
RhoA-dependent activation of PLCε requires EF hands ½
Previous studies investigating RhoA-dependent activation of PLCε used variants truncated at the N-terminus, removing all or parts of the CDC25 domain, PH domain, and EF1/2, based on sequence conservation, or the C-terminal RA domains14,27,28. Because we recently established domain boundaries for the PH and EF hand domains18, we used this approach to test the contributions of the N-terminal regions and Y-box in basal and RhoA-dependent activation in cells. In this assay, cells are metabolically labeled with [3H]-myoinositol, which is incorporated into their lipid head groups. Transfected PLCε species cleave the [3H]-labeled phosphatidylinositol phospholipids, producing DAG and [3H]-inositol species ([3H]-IPx), the latter of which are quantified by scintillation counting29,30. RhoAG14V increases WT PLCε activity ~5-fold over basal, consistent with previous reports (Fig. 1a, b)23,31. PLCε PH-C, which lacks the N-terminal 836 residues, is similarly activated by RhoAG14V. PLCε EF-C, which lacks the N-terminus and PH domain, is also significantly activated by RhoAG14V, but its maximum activity is ~4-fold lower than that of RhoA-activated PLCε. PLCε EF3-C, which is further truncated to remove EF1/2, is unresponsive to the GTPase (Fig. 1a, b). We also reassessed the role of the Y-box in activation in the context of PLCε and the EF-C variant. Deletion of this element significantly decreased basal and eliminated RhoA stimulation (Fig. 1b), indicating the Y-box is required for lipase activity in general, and not specifically for activation by RhoA.
Maximum RhoA-dependent activity of PLCε variants decreased progressively as its N-terminus was truncated. To confirm the role of the EF hands, particularly EF1/2, in RhoA activation of the holoenzyme, we generated a series of chimeras between PLCε and PLCβ3, which is not regulated by RhoA (Fig. 2a). In these chimeras, either the entire EF1-4 module of PLCε was replaced with that of PLCβ3 (PLCε/β3 EF), or only the EF1/2 (PLCε/β3 EF1/2) or EF3/4 (PLCε/β3 EF3/4) module was replaced (Fig. 2a). The chimeras expressed similarly and were properly folded and functional (Supplementary Fig. 1). However, only PLCε/β3 EF1/2 showed significantly activation by RhoAG14V, with a ~3-fold increase over basal activity (Fig. 2b).
a Schematic of the PLCε/β3 EF hand chimeras. b Basal and RhoAG14V-stimulated activities of PLCε, PLCβ3, and PLCε/β3 chimeras. (Left) Only PLCε variants that retain the EF1/2 hands are responsive to RhoA-dependent activation. At least three independent experiments from independent transfections were performed for each variant, and data are shown as the average of triplicate measurements ± SD. Data were analyzed using an unpaired, one-tailed t-test with Welch’s correction comparing the basal and RhoA-stimulated activities of each variant. ****p < 0.0001, **p < 0.0067, *p < 0.0167. (Right) Changes in maximal activity of each variant were analyzed using a one-way ANOVA and Kruskal–Wallis test comparing each variant to PLCε, followed by a Dunn’s multiple comparisons test. ***p < 0.0485, *p < 0.0198. Representative Western blots are shown below, with empty pCMV vector (EV) and β-actin used as loading controls. PLCε variants express a C-terminal FLAG tag and are detected with an anti-FLAG antibody, and RhoA contains an N-terminal HA tag for detection using an anti-HA antibody.
Maximum activity requires prenylated RhoA
For PLC enzymes regulated by G proteins, maximum lipase activation requires the G protein activators to be prenylated and/or acylated3,32. We compared the ability of wild-type, prenylated RhoA·GTPγS and soluble forms of RhoA·GTPγS and RhoAG14V·GTP to directly activate purified PLCε PH-C using a modified version of the commercially available IP-One assay. Briefly, phosphatidylinositol (PI) is incorporated into liposomes, and the activity of the lipase produces DAG, which remains in the liposome, and free inositol phosphate (IP1)33,34. Wild-type (prenylated) RhoA·GTPγS and soluble RhoAG14V·GTP significantly increased lipase activity ~7 and ~5-fold over basal, respectively. Soluble RhoA·GTPγS increased lipase activity ~3-fold over basal (Supplementary Fig. 2). The fact that soluble RhoA variants partially activate PLCε PH-C confirms that membrane localization mediated by RhoA alone is insufficient to achieve full activation of the lipase by RhoA.
RhoA·GTP binds to the PLCε E2α′ helix of the EF hands
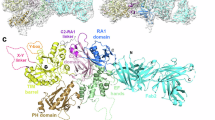
Cryo-EM single particle analysis (SPA) was used to determine the structure of RhoA·GTP bound to PLCε PH-C. This variant is activated by RhoA (Fig. 1a, b) and is the largest variant that has been purified in sufficient quantities for biophysical studies18,30,35. PLCε PH-C and prenylated RhoA·GTP were incubated in a 3:1 molar ratio before being applied to grids. From an initial data set of 1,329,298 particles, two distinct populations containing 184,875 and 106,370 particles were identified. Because the first population was larger and the resulting volume showed more structural features, it was selected for further processing and used to generate an initial 3.5 Å map. A structure of PLCε PH-C (PDB ID 9B13, in review18) was fit into the map, revealing unmodeled density around the EF hands. Given the importance of the EF hands in RhoA-dependent activation (Figs. 1 and 2), the crystal structure of RhoA·GMPPNP (PDB ID 1S1C)36 was placed in the density such that its switch regions, which had the strongest density, were adjacent to the EF hands. The resulting RhoA–PLCε PH-C model was then used as a template for additional rounds of particle picking and refinement, resulting in a final map at 3.3 Å resolution (209,463 particles, Supplementary Figs. 3–5, Table 1).
In the RhoA·GTP–PLCε PH-C reconstruction, clear density is observed for the PH-RA1 domains (Fig. 3). No density is observed for the RA2 domain, despite its presence in the protein, because it is flexibly connected to the rest of the lipase19,30. The architecture of PLCε is similar to that of the lipase bound to an Fab fragment, with an r.m.s.d. of 0.87 Å for 786 Cα atoms (out of 841 resolved residues, PDB ID 9B13)18. The PH domain and EF1/2 pack adjacent to the PLCε catalytic core, which includes the EF3/4-RA1 domains35. The active site remains blocked by the C-terminus of the X–Y linker. However, the rest of the X–Y linker (residues 1525–1631) and the Y-box (residues 1662–1730) were not resolved, consistent with these regions being highly dynamic in solution.
a The 3.3 Å cryo-EM map of the RhoA·GTP–PLCε PH-C complex and b fitted with the ribbon diagram of the RhoA·GTP–PLCε PH-C complex. The domains in PLCε are colored as in Fig. 1a, and RhoA·GTP is shown in light blue. c Ribbon diagram of the RhoA·GTP–PLCε PH-C colored as in (a). The PLCε active site Ca2+ is shown as a black sphere, the E2α′ helix and adjacent loop that bind RhoA are shown in firebrick. Disordered regions, including the X–Y linker (hot pink) and Y-box (orange), are shown as dashed lines. RhoA·GTP is shown in light blue, Mg2+ as a black sphere, and GTP in light blue sticks. The N- and C-termini of each protein are labeled. d Density (gray mesh) for the E2α′ helix (shown in red) and e loop connecting the helix to the EF3/4 subdomain.
RhoA·GTP binds exclusively to the EF hands via its switch I and switch II, consistent with other RhoA–effector enzyme complexes (Fig. 3, Supplementary Fig. 6)36,37,38. Switch I and II of RhoA·GTP could be resolved in the density (Supplementary Fig. 7) and bind to the PLCε E2α′ helix (residues 1273–1287), located in a subfamily-specific insertion within EF3/418,39. The E2α′ helix is followed by an extended loop (residues 1288–1302) that reenters the EF3/4 module. Residues 1288–1296 are ordered and poised to interact with residues on the β3 strand of the GTPase (Figs. 3d and 4; Supplementary Fig. 7). Overall, the interaction is largely hydrophobic, burying ~1800 Å2 surface area. Within the PLCε E2α′ helix, residues Asn1275, Ile1279, Ala1282, Ile1283, and Ala1286 interact with Phe39 in switch I and Leu69 and Leu72 in switch II of RhoA·GTP (Figs. 3d and 4). PLCε Ile1295, on the loop following E2α′, also interacts with RhoA Phe39. Finally, PLCε Arg1049 and Trp1051, located on the loop connecting EF1 and EF2, interact with Leu72 and Pro75 at the C-terminus of the RhoA·GTP switch II helix (Figs. 3d and 4). The orientation of RhoA·GTP and PLCε places the prenylated C-tail of the GTPase in the same plane as the PLCε PH domain and the active site in the TIM barrel, which would allow these elements to simultaneously engage the membrane as expected during activation18,35.
a Schematic showing the boundaries for internal deletions in the PLCε EF3/4 module. b (Left) Basal and RhoAG14V-stimulated activities of PLCε variants lacking the E2α′ helix (Δ1275–1289), the loop connecting it to the F3α helix (Δ1287–1298) in the EF3/4 module, or both (Δ1275–1298). Deletion of any of these regions largely eliminates RhoA-dependent activation. At least three independent experiments from independent transfections were performed for each variant. Data shown represents the average of triplicate measurements ± SD, and analyzed using unpaired, one-tailed t-test with Welch’s correction to compare the basal and RhoA-stimulated activities of each variant. ****p < 0.0001, ***p < 0.0003, **p < 0.0080, *p < 0.0112. (Left) The change in maximal activity ± SD was calculated by subtracting the RhoA-stimulated activity from the basal activity of each variant. Data were analyzed using a one-way ANOVA and Kruskal–Wallis test comparing each variant to PLCε, followed by a Dunn’s multiple comparisons test. For Δ1275–1289, **p < 0.0041, for Δ1287–1298, *p < 0.0134, and for Δ1275–1298, **p < 0.0097. c The PLCε E2α′ helix (red) binds to the switch regions of RhoA (light blue). Additional contacts with RhoA are made by residues in the EF1/2 module and the loop linking E2α′ to the F3α helix. Labeled residues were subjected to site-directed mutagenesis, and their impact on RhoA-dependent activation was quantified. d (Left) Mutations in the G protein–PLCε interface decrease RhoA-dependent activation. At least three independent experiments from independent transfections were carried out for each variant. Data shown represents the average of triplicate measurements ± SD, and was analyzed using unpaired, one-tailed t-test with Welch’s correction to compare the basal and RhoA-stimulated activities of each variant. ****p < 0.0001, ***p < 0.0003, **p < 0.0080, *p < 0.0112. (Right) Mutation of PLCε Trp1051 in EF1/2, residues Ala1282, Ile1283, and Ala1286 in E2α′, and Ile1295 in the E2α′-F3α loop significantly decreases maximum RhoA-dependent activation. Data was analyzed using a one-way ANOVA and Kruskal–Wallis test comparing each variant to PLCε, followed by a Dunn’s multiple comparisons test. Representative Western blots are shown below, with empty pCMV vector (EV) and β-actin used as loading controls. Differences in expression were not found to be statistically significant but may still contribute to variation in activities. PLCε variants express a C-terminal FLAG tag and are detected with an anti-FLAG antibody, while RhoA contains an N-terminal HA tag and is detected using an anti-HA antibody.
There are differences in the intramolecular interactions between the RhoA complex and the PLCε EF3-RA1 and Fab–PLCε PH-C structures18,30. Residue-residue (RR) distance analysis40 was used to identify conformational changes across the three structures in an unbiased manner (Supplementary Figs. 8 and 9). Within each individual structure, the PH and TIM barrel domains move as a single unit, as do EF3/4 and the C2 domain. In contrast, the RA1 domain is conformationally distinct in each structure, potentially due to the influence of crystal packing interactions in the EF3-RA1 structure or Fab binding in the Fab–PLCε PH-C structure (Supplementary Figs. 8 and 9)18,30. Binding of RhoA·GTP to the EF hands induces a shift in the position of EF3/4, moving it ~2 Å closer to the TIM barrel domain relative to its position in the other structures. This conformational change may reflect an allosteric component of RhoA-mediated activation (Supplementary Fig. 10).
The PLCε E2α′ helix is required for RhoA·GTP-dependent activation
We first tested whether the PLCε E2α′ helix and flanking loop are needed for RhoA-dependent activation (Fig. 4a, b, Supplementary Fig. 7). Deletion of the disordered region preceding the E2α′ helix (PLCε Δ1226–1270) did not eliminate RhoA-dependent activation but decreased maximum activity (Fig. 4a, b). Deletions of the E2α′ helix (Δ1275–1289), the flanking loop (Δ1287–1298), or both (Δ1275–1298) eliminated RhoA-dependent activation. The E2α′ helix is only required for activation by RhoA, as PLCε and PLCε Δ1275–1289 had the same fold activation when co-transfected with two other well-established G protein activators, Rap1AQ63E and the Gβγ heterodimer (Supplementary Fig. 11)20,41. The E2α′ helix is also sufficient to confer sensitivity to RhoA-mediated activation. Replacement of the PLCβ3 E2α-F2α helices in the EF hands (residues 183–221) with the corresponding region of PLCε (F2α-E2α′, PLCε residues 1196–1284), or insertion of E2α′ between PLCβ3 EF1/2 and EF3/4 subdomains (PLCβ3 residues 221–222), increased activity upon cotransfection with RhoAG14V (Supplementary Fig. 12).
Residues in PLCε that interact with RhoA·GTP in the structure were also mutated to test their role in activation (Fig. 4c, Supplementary Fig. 7). PLCε Arg1049, Trp1051, Phe1187 and Val1189 are positioned to interact with the C-terminus of the switch II helix in RhoA (Fig. 4c). PLCε R1049A, in the EF1 module, and F1187E and V1189E, in the loop immediately preceding the E2α′ helix, had ~2-fold lower maximum activity. PLCε W1051E resulted in a significant ~5-fold reduction in its maximum activity, confirming the importance of this residue in packing against switch II (Fig. 4c). PLCε Ile1279, Ala1282, Ile1283, and Ala1286 are located on the surface of the E2α′ helix and interact with Phe39, Leu69, and Leu72 in the switch regions of RhoA·GTP (Fig. 4c). PLCε I1279E had a ~2-fold decrease in its maximum activity, whereas A1282K, I1283E, and A1286K significantly decreased maximum activity, with the I1283E mutation eliminating the response to the G protein (Fig. 4d). PLCε Ile1295E, located on the loop connecting E2α′ back to EF3, also caused a significant ~4-fold decrease in maximum activity relative to wild-type (Fig. 4d). Together, PLCε Trp1051, the E2α′ helix, and Ile1295 make the required interactions with the RhoA in order for activation to occur.
Discussion
In this study, we provide the first molecular insights into the mechanism by which RhoA activates PLCε. Using cell-based assays, we show that the EF hands are critical for RhoA-dependent activation, whereas the Y-box, previously thought to be involved in the process, is needed for general lipase function. The role of the Y-box is not yet known, but because these variants are detected by Western blot at the correct molecular weight, the lack of activity seems unlikely to reflect a folding defect. To define the regions of the EF hands necessary for activation, we generated chimeras wherein the PLCε EF hands were replaced, in whole or in part, with those of PLCβ3. However, only a chimera containing the PLCβ3 EF1/2 subdomain could be activated by RhoA in cells (Figs. 1 and 2). We then used cryo-EM to determine a reconstruction of the RhoA·GTP–PLCε PH-C complex (Fig. 3). Although the quality of the overall RhoA·GTP density was weak, the overall shape of the density was consistent, and the switch regions could be resolved, allowing us to fit RhoA in the map. Site-directed mutagenesis and cell-based assays support the model for the complex. RhoA·GTP binds to the E2α′ helix in the PLCε EF hands, making additional contacts with residues in EF1 and the loop connecting E2α′ to the EF3/4 module. Deletion or mutation of the E2α′ helix, EF1/2, or the loop connecting E2α′ and EF3/4 all but eliminate RhoA-dependent activation (Fig. 4) while having little impact on regulation by other G proteins (Supplementary Fig. 11). These findings strongly indicate that E2α′ is the binding site for RhoA.
PLCε must interact with the inner leaflet of the plasma membrane to hydrolyze its PIP2 substrate. In addition to the PIP2 binding site in the active site of the TIM barrel, the CDC25 and PH domains may also contribute to membrane association, and together could define a common membrane engagement surface18. In the reconstruction of the RhoA·GTP–PLCε PH-C complex, the C-terminal thirteen residues of the GTPase, which are disordered in prior atomic structures, allow the prenylated C-tail of RhoA to reach the same plane of this potential membrane surface. Thus, the architecture captured in our reconstruction is compatible with a functional complex at a membrane. Although membrane localization is an important component of the RhoA activation mechanism, we showed this alone is insufficient for maximum activation, as soluble, active RhoA proteins stimulate PLCε PH-C to a submaximal threshold in liposome-based assays (Supplementary Fig. 2). RR distance plots comparing the structures of PLCε PH-C18 and PLCε EF3-RA130 show that the EF3/4 module is closer to the TIM barrel domain when RhoA·GTP is bound (Supplementary Fig. 8). This may promote rearrangements within the catalytic domain that, when the complex is at the membrane, facilitate displacement of the autoinhibitory X–Y linker and allow substrate binding (Fig. 5). This is supported by mutations in EF3/4 and the TIM barrel that decrease maximum RhoA-dependent activation ~3-fold (Supplementary Fig. 10). The role of the Y-box however remains undetermined but is clearly functionally important given that its deletion impairs both basal and G protein-stimulated activities (Fig. 1a, b).
Under basal conditions, PLCε is autoinhibited by the X–Y linker in the cytoplasm. RhoA is activated downstream of G12/13-coupled receptors by a RhoA guanine nucleotide exchange factor (RhoGEF). The activated G protein binds the lipase via the E2α′ helix, which, together with the PLCε CDC25, PH, and TIM barrel domains, may define a common membrane interaction surface. RhoA binding induces conformational changes within the lipase, moving the EF hands closer to the TIM barrel domain, stabilizing the EF1/2-EF3/4 interface and the CDC25-PH domain module closer to the TIM barrel. These long-range conformational changes may facilitate displacement of the X–Y linker from the active site, exposing the active site once at the membrane. The RhoA·GTP–PLCε PH-C reconstruction is shown in the boxed inset. The prenylated C-tail of RhoA is shown as a dashed line, and together with the TIM barrel and PH domain, may form a shared membrane interaction surface. The ends of the PLCε X–Y linker are indicated by hot pink asterisks.
Whether other G protein activators of PLCε regulate lipase activity through similar mechanisms has yet to be determined. It seems that membrane localization will always be a key component because the autoinhibitory X–Y linker would need to be displaced via interfacial activation at the membrane3,21, and all known activators of PLCε are prenylated G proteins3. At a minimum, regulation by Gβγ and Rap1A occurs independently of the PLCε E2α′ helix because its deletion did not alter the fold change in activity by these G proteins. The Gβγ binding site has not been determined, but the CDC25 and RA2 domains are both required for maximum activation by this G protein41. It is possible that two Gβγ molecules bind to the lipase, promoting translocation to the membrane, a scenario that has been observed in other Gβγ–effector enzyme complexes, such as the Gβγ–PI3K42 and Gβγ–PLCβ343 complexes. In the case of Rap1A, the GTPase binds the C-terminal RA2 domain of PLCε, but activation requires the PH and EF1/2 domains, indicating that there are allosteric components. Indeed, small-angle X-ray scattering showed that binding of constitutively active Rap1A to PLCε PH-C induced long-range conformational changes, stabilizing the lipase in an extended state20. This could potentially be explained either by the Rap1A-RA2 module interacting with the PH and/or EF1/2 domains, or by the latter two domains constituting a second Rap1A binding site3.
In the future, studies that integrate the role of the membrane in PLCε function are needed for a complete understanding of regulation. PLCε translocates to the cytoplasmic leaflets of the plasma and perinuclear membranes, hydrolyzing PIP2 or PI4P, respectively2,3. These membranes differ in their composition and biophysical properties, and whether and how these factors impact interfacial activation, membrane engagement, and/or G protein activation remain to be explored.
Methods
Cloning of PLCε and variants
A pCMV vector encoding R. norvegicus PLCε with a C-terminal FLAG tag (gift from A.V. Smrcka, U. Michigan) was used as a template to generate PLCε PH-C (residues 837–2282), EF-C (residues 1038–2282), and EF3-C (residues 1284–2282). The Y-box (residues 1667–1728) was deleted in PLCε and EF-C by Q5-site-directed mutagenesis (New England BioLabs, Inc.). In-Fusion cloning (Takara Bio USA, Inc.) was used to generate PLCε and PLCβ3 (in a pCI-neo vector, Promega) chimeras. For PLCε/β3 EF, PLCε residues 1038–1355 were replaced with PLCβ3 152–304, PLCε/β3 EF1/2 replaced PLCε residues 1038–1284 with residues 152–216 from PLCβ3, and PLCε/β3 EF3/4 replaced PLCε residues 1035–1355 with PLCβ3 residues 217–304. Internal deletions in the PLCε EF hands (Δ1275–1289, Δ1287–1298, and Δ1275–1298) and point mutants were generated with In-Fusion cloning (Takara Bio USA, Inc.). PLCε PH-C was subcloned into pFastBac HTA (ThermoFisher) for protein expression and purification41. All constructs were sequenced over the coding region. Primer sequences are shown in Table 2.
Expression and purification of PLCε variants
R. norvegicus PLCε PH-C was expressed in baculovirus-infected Spodoptera frugiperda (Sf9) (RRID: CVCL_0549) cells at an MOI of ~1 for 48 h and harvested by centrifugation. The pellets were resuspended in lysis buffer (20 mM HEPES, pH 8.0, 50 mM NaCl, 10 mM β-mercaptoethanol (β-Me), 0.1 mM EDTA, 0.1 mM EGTA, and EDTA-free protease inhibitor tablets (Roche) at one-third strength), homogenized and lysed by dounce, and centrifuged at 100,000 × g for 1 h. The supernatant was filtered and diluted to a final volume of 320 mL with 20 mM imidazole, 300 mM NaCl, and 10 mM β-Me prior to loading on a 5 mL HisTrap column (Cytiva) pre-equilibrated with binding buffer (20 mM HEPES, pH 8.0, 300 mM NaCl, 10 mM β-Me, 0.1 mM EDTA, 0.1 mM EGTA, and 20 mM imidazole). The column was washed with 8 column volumes (CVs) of binding buffer and eluted with a 0–500 mM imidazole gradient. Fractions containing the protein were concentrated to ~1 mL and exchanged into low salt buffer (20 mM HEPES, pH 8.0, 50 mM NaCl, 0.1 mM EDTA, 0.1 mM EGTA, and 2 mM DTT) before loading on a 1 mL MonoQ column (Cytiva) pre-equilibrated with the low salt buffer. The protein was eluted with a 0–500 mM NaCl gradient. Fractions containing the protein were pooled, concentrated to ~1 mL, and then applied to tandem Superdex 200 Increase 10/300 GL columns (Cytiva) equilibrated with 20 mM HEPES pH 8.0, 200 mM NaCl, 0.1 mM EDTA, 0.1 mM EGTA, and 2 mM DTT. Fractions containing the final, purified protein were identified by SDS-PAGE, concentrated to 4–5 mg/mL, flash frozen in liquid nitrogen, and stored at −80 °C.
Cloning of RhoA variants
The cDNA encoding wild-type human RhoA (residues 1–194, gift from J.J.G. Tesmer, Purdue U.) was subcloned into pcDNA 3.1 and an HA-tag installed at the N-terminus. Soluble RhoA was generated by subcloning the human cDNA into a pMALc2H10T vector (gift from J. J. G. Tesmer, Purdue). The G14V mutation was introduced using Q5-site-directed mutagenesis (New England BioLabs, Inc.). RhoA and RhoAG14V were also subcloned into pFastBac HTA for expression and purification. All constructs were sequenced over the coding region. Primer sequences are shown in Table 2.
Expression and purification of RhoA variants
RhoA and RhoAG14V were expressed in baculovirus-infected High5 cells, cultured in Lonza Insect-XPRESS media (Fisher Scientific) at an MOI of ~1 and harvested after 48 h. Pellets were resuspended in lysis buffer (20 mM HEPES, pH 8.0, 150 mM NaCl, 0.1 mM EDTA, 10 mM β-Me, 10% glycerol, 20 mM GDP, 1 mM leupeptin and lima bean (LL) protease, 1 mM phenylmethanesulfonylfluoride (PMSF), and 5 mM MgCl2). Cells were lysed by four freeze-thaw cycles and centrifuged at 100,000 × g for 1 h. The membrane pellet containing RhoA or RhoAG14V was resuspended in solubilization buffer (20 mM HEPES, pH 8.0, 150 mM NaCl, 10 mM β-Me, 10% glycerol, 20 mM GDP, 1 mM LL, 1 mM PMSF, and 5 mM MgCl2) and sodium cholate added to a final concentration of 1% (w/v). The slurry was stirred for 1 h at 4 °C to solubilize the membrane fraction, then centrifuged at 100,000 × g for 45 min. The supernatant was diluted 5-fold with load dilution buffer (solubilization buffer supplemented with 1% (w/v) sodium cholate) and loaded on an Ni-NTA affinity column (Roche cOmplete Ni-NTA resin) equilibrated with 10 CVs load dilution buffer. The column was washed with 10 CV of wash 1 buffer (20 mM HEPES pH 8.0, 150 mM NaCl, 10 mM β-Me, 10% glycerol, 20 mM GDP, 1 mM MgCl2, and 1% (w/v) sodium cholate) and 20 CVs of wash 2 buffer (20 mM HEPES pH 8.0, 300 mM NaCl, 10 mM β-Me, 10% glycerol, 20 mM GDP, 1 mM MgCl2, 10 mM CHAPS, and 20 mM imidazole). RhoA or RhoAG14V was eluted in 10 CVs of elution buffer (20 mM HEPES pH 8.0, 150 mM NaCl, 10 mM β-Me, 10% glycerol, 20 mM GDP, 1 mM MgCl2, 10 mM CHAPS, and 250 mM imidazole), and concentrated to ~1 mL. The protein was loaded on a Superdex 75 Increase 10/300 GL (Cytiva) column equilibrated with 20 mM HEPES pH 8.0, 150 mM NaCl, 1 mM DTT, 40 mM GDP, 1 mM MgCl2, and 6 mM CHAPS. Fractions containing the purified protein were identified by SDS-PAGE, flash frozen in liquid nitrogen, and stored at −80 °C.
For soluble RhoA and RhoAG14V, E. coli BL21(DE3) was transformed and grown to an OD600 of 0.4–0.6. Expression was induced by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 16–18 h at 18 °C. Cells were harvested by centrifugation and flash frozen in liquid nitrogen. Proteins were purified as described above, with some modifications. Sodium cholate and CHAPS were omitted from all buffers. After the pellets were resuspended in lysis buffer, 1 mg/mL lysozyme (Fisher BioReagents) was added and incubated on ice for 30 min. To ensure complete lysis, samples were then sonicated on ice for 20 cycles (15 s pulse, 45 s recovery), then centrifuged at 100,000 × g for 1 h. After Ni-NTA affinity chromatography, the protein concentration was measured using a Bradford assay and incubated with 8% (w/w) TEV protease overnight at 4 °C in dialysis buffer (20 mM HEPES, pH 8.0, 150 mM NaCl, 10 mM β-Me, 10% glycerol, 40 mM GDP, and 5 mM MgCl2). The next day, the Ni-NTA column was washed with 20 CV of dialysis buffer, and the dialysate was passed five times over the resin. The TEV-cleaved, soluble RhoA or RhoAG14V was concentrated to ~1 mL, and purified on tandem Superdex 75 Increase 10/300 GL (Cytiva) columns.
Purified RhoA proteins were activated using nucleotide exchange. Briefly, the proteins were incubated with a 10-fold molar excess of GTP or GTPγS and a 4-fold molar excess of EDTA for 1.5 h on ice. The reaction was quenched by the addition of a 10-fold molar excess of MgCl2 and incubated for 30 min on ice23. The proteins were flash frozen in liquid nitrogen and stored at −80 °C.
Rap1AQ63E and Avi-Gβγ cloning
The cDNA encoding human Rap1A (residues 1–184) was subcloned into pcDNA 3.1 with an N-terminal HA tag. The Q63E mutation was introduced using the Q5-Site-Directed Mutagenesis Kit (New England BioLabs Inc). Human Avi-tagged Gβ1 and Gγ2 were subcloned into a pCI-neo vector (gift from A.V. Smrcka, U. Michigan). Constructs were sequenced using whole-plasmid sequencing (Plasmidsaurus).
[3H]-IPx accumulation assay
COS-7 cells (gift from A.V. Smrcka, U. Michigan, (RRID: CVCL_0224)) were seeded at a density of 100,000 cells/well in 12-well plates in Dulbecco’s Modified Eagle’s Medium (Corning) supplemented with 10% fetal bovine serum (FBS, BioTechne), 1% Glutamax (Gibco), and 1% penicillin-streptomycin (Corning) and incubated for 24 h. The cells were co-transfected with 750 ng empty pCMV, 750 ng PLCε variant DNA alone or with 375 ng of RhoAG14V, 375 ng of Rap1AQ63E, or 375 ng Avi-Gβ1 and 375 ng Gγ2 DNA. After 24 h, the cells were washed with serum- and inositol-free Ham’s F-10 media (Invitrogen) and incubated for 16–18 h in Ham’s F-10 media supplemented with 1.5 mCi/well myo[2-3H(N)] inositol (Revvity). Ten millimoles of LiCl was added to each well and incubated for 1 h to inhibit inositol phosphatases. The media was aspirated, and cells were washed once with ice-cold PBS. Cells were lysed on ice by the addition of 1 mL ice-cold 50 mM formic acid. Lysates containing [3H]-labeled inositol phosphates were loaded onto pre-equilibrated Dowex AGX8 anion exchange columns (BioRad), washed twice with 50 mM formic acid, once with 100 mM formic acid, eluted with 1.2 M ammonium formate and 0.1 M formic acid into scintillation vials. Total [3H]-IPx was quantified by scintillation counting (Uniscint BD scintillation cocktail, National Diagnostics)29,30. All experiments were performed at least three times in triplicate from independent transfections.
Immunoblotting
Cells were plated, transfected, and incubated in Ham’s F-10 media 24 h post-transfection, replicating the conditions used for the [3H]-IPx accumulation assays. After 48 h, cells were washed once with cold PBS and scraped into 100 μL of 1X SDS loading dye (100 mM Tris-HCl, pH 6.8, 6% w/v sucrose, 2% w/v SDS, 5% v/v β-Me, and 0.02% bromophenol blue) and incubated at 90 °C for 10 min prior to loading on a 10% SDS-PAGE gel. Samples were transferred to a PVDF membrane overnight in Towbin buffer (25 mM Tris, 192 mM glycine, 20% (v/v) methanol) at 4 °C. The membrane was blocked with 5% bovine serum albumin (BSA) in 1X Tris-buffered saline supplemented with 0.1% Tween-20 (TBST) for 1 h, washed three times with 1X TBST, and incubated overnight at 4 °C with an anti-FLAG rabbit antibody (Cell Signaling Technology Cat# 14793, RRID:AB_2572291), anti-HA rabbit or mouse antibody (Cell Signaling Technology Cat# 2367, RRID:AB_10691311), and anti-actin mouse antibody (Cell Signaling Technology Cat# 3700, RRID:AB_2242334) at 1:1000 dilutions. Endogenous and transfected Avi-Gβγ were detected using an anti-GNB1 rabbit antibody (Invitrogen). The next day, the blot was washed three times in 1X TBST and incubated with goat anti-mouse (Sigma-Aldrich Cat# 12-349, RRID:AB_390192) or anti-rabbit (Cell Signaling Technology Cat# 7074, RRID:AB_2099233) secondary antibody conjugated with HRP (Sigma-Aldrich) at a 1:10,000 dilution at 27 °C for 1 h. The blot was washed three times in 1X PBS, and the West Pico ECL substrate was added (ThermoFisher Scientific)30. Blots were imaged using a GeneGnome, and densitometry analysis was performed in ImageJ24.
Liposome-based activity assays
100 μM of hen egg white phosphatidylethanolamine (PE) and 250 μM of soybean phosphatidylinositol (PI, Avanti) were mixed, dried under nitrogen, and stored at −20 °C. Lipids were resuspended by bath sonication in buffer containing 50 mM HEPES, pH 7.4, 80 mM KCl, 2 mM EGTA, and 1 mM DTT. Purified PLCε PH-C was diluted to a final amount of 2–5 ng in assay buffer (100 mM HEPES pH 7.4, 160 mM KCl, 6 mM EGTA, and 1 mM DTT), 3 mg/mL BSA, and 3 mM DTT. RhoA buffer (20 mM HEPES pH 8, 150 mM NaCl, 1 mM DTT, 40 mM GTP, 1 mM MgCl2, and 6 mM CHAPS) alone or containing a final concentration of 3 mM RhoA variant was then added. All samples were transferred to 30 °C and incubated with 10 μL liposomes for 2 min before the reaction was initiated by the addition of 5 μL free Ca2+ solution (1X assay buffer, 1 mM DTT, and 18 mM CaCl2). Reactions were incubated for 15 min at 30 °C, then quenched with 5 μL Ca2+ chelating solution (1X assay buffer, 1 mM DTT, and 210 mM EGTA). Negative controls lacked free Ca2+18. All assays were performed at least three times in triplicate using proteins purified from at least two independent preparations.
Statistics and reproducibility
All cell-based assays were performed in technical triplicates at least three times from independent transfections. Assays using purified proteins were carried out in technical duplicates or triplicates at least three times using protein from independent preparations. Assay data is shown as the average of technical triplicates ± SD. Data were analyzed using an unpaired, one-tailed t-test with Welch’s correction comparing the basal and RhoA-stimulated activities of each variant. The change in maximal activity ± SD was calculated by subtracting Rthe hoAG14V-stimulated activity from the basal activity of each variant. Data was analyzed using a one-way ANOVA and Kruskal–Wallis test comparing each variant to PLCε, followed by a Dunn’s multiple comparisons test.
Cryo-EM sample preparation
PLCε PH-C at 0.6 mg/mL was incubated with wild-type RhoA•GTP in a 1:3 molar ratio in 20 mM HEPES pH 8.0, 150 mM NaCl, 2 mM DTT, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM MgCl2, 40 mM GTP, 0.5 mM CaCl2, and incubated on ice for 1 h. The reaction was supplemented with CHAPS (Millipore Sigma) to a final concentration of 2.5 mM, and 3.5 μL of the reaction was applied to a glow-discharged Quantifoil 1.2/1.3 300 mesh holey copper grid. Grids were blotted at blot force 2 for 3 s, at 4.2 °C with 100% humidity, and plunge frozen in liquid ethane using a Vitrobot Mark IV (ThermoFisher Scientific).
Cryo-EM data acquisition
Grids containing RhoA•GTP–PLCε PH-C were imaged on a Titan Krios G4 (ThermoFisher Scientific) electron microscope equipped with a post-GIF K3 Summit Direct Electron Detector (Gatan, Inc.) and Gatan quantum GIF energy filter. 6378 movies were collected using 300 kV at a magnification of 81,000× (pixel size of 0.527 Å) and defocus range of 0.6–2.0 μm using EPU44. Each movie stack recorded 40 frames, for a total dose of 57.8 electrons/Å2, and a total exposure time of 3.21 s per stack.
Cryo-EM data processing
The cryo-EM workflow and validation for the RhoA•GTP–PLCε PH-C complex are shown in Supplementary Figs. 4 and 5 and Table 1. Patch motion correction and contrast transfer function (CTF) were calculated using cryoSPARC45. From a total of 6378 exposures, 1,329,298 particles were accepted. As a first step, ~400 particles were manually selected, generating twenty 2D classes for template picking, and fifty 2D class averages were generated. Nine classes (291,245 particles) were selected for an ab initio reconstruction that generated two initial models, Volumes A1 and B2. Volume A1 was larger and contained more particles, and so was used as the input for a non-uniform refinement to generate Volume A2 (3.52 Å, 184,875 particles). An AlphaFold2 model of PLCε PH-C, as well as the PLCε PH-C reconstruction from a Fab-bound complex (PDB ID 9B1318), could be fit in the density of Volume A2. Additionally, unmodeled density was observed adjacent to the EF hands, and heterogeneous refinement was carried out using the particles in Volumes A2 and B1, as well as 291,245 particles identified in the ab initio reconstruction, to improve the resolution of this region. The resulting Volume A3 (194,157 particles) was subjected to non-uniform refinement against the same volume class, yielding a 3.44 Å map. Rigid-body fitting the PLCε PH-C structure into Volume 4A revealed stronger density adjacent to the EF hands. The crystal structure of RhoA (PDB ID 1S1C36) was fit into this density with its switch regions poised to interact with the E2α′ helix. This model was then used to generate twenty 2D volume classes in EMAN, which were used as templates for particle picking from the initial micrographs. 5,828,020 particles were extracted, with 1,047,728 particles accepted after inspection and used to generate fifty 2D classes. The best ten 2D classes (209,103 particles) were selected and used to generate two volumes, C1 and D1. Volume C1 was unique compared to the initial volumes (Volumes A1 and B1), while Volume D1 was very similar to Volume A1. To maximize the particle number for the final reconstruction, the 114,829 particles in Volume C1 were used in a non-uniform refinement, generating Volume C2 at 6.67 Å. These 209,103 particles were combined with the original 291,245 particles, and after removal of duplicates, the remaining 419,418 particles were used in heterogeneous refinement against volumes A2, B1, and C2. The resulting Volume A5 (209,463 particles) was subjected to non-uniform refinement, resulting in the final 3.32 Å map.
Model building, refinement, and validation
An AlphaFold2 model of PLCε PH-C and the crystal structure of RhoA·GMPPNP (PDB ID: 1S1C28) were rigid-body fit into the cryo-EM density using COOT46,47. Alternating rounds of manual model building in COOT and refinement in PHENIX46 were carried out, guided by the DAQ collaboratory48. Stereochemistry of the final model was evaluated using MolProbity and CaBLAM in PHENIX46,49. Coordinates for the RhoA•GTP–PLCε PH-C reconstruction, volume map, half maps A and B, refinement mask, and FSC curve for Volume A were deposited in the EMDB and PDB as accession numbers EMD-43927 and 9AX5, respectively. Raw micrograph data were also deposited in EMPIAR-12069.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Annotated, uncropped, and unedited Western blots for the main text and supplementary figures are shown in Supplementary Figs. 13 and 14. Cryo-EM maps and coordinates were deposited in the EMDB and PDB as accession numbers EMD-43927 and 9AX5, respectively. Raw micrograph data were also deposited in EMPIAR-12069. Vectors encoding PLCε PH-C, EF-C, and EF3-C are available from Addgene (IDs: 244967, 244968, 244969, respectively). All source data shown in the main text and supplementary information are annotated and available in the Purdue University Research Repository (purr.purdue.edu) under doi:10.4231/ZMJZ-PM07.
References
Kadamur, G. & Ross, E. M. Mammalian phospholipase C. Annu. Rev. Physiol. 75, 127–154 (2013).
Smrcka, A. V., Brown, J. H. & Holz, G. G. Role of phospholipase Cε in physiological phosphoinositide signaling networks. Cell Signal. 24, 1333–1343 (2012).
Muralidharan, K., Van Camp, M. M. & Lyon, A. M. Structure and regulation of phospholipase Cbeta and epsilon at the membrane. Chem. Phys. Lipids 235, 105050 (2021).
Smrcka, A. V. Regulation of phosphatidylinositol-specific phospholipase C at the nuclear envelope in cardiac myocytes. J. Cardiovasc. Pharmacol. 65, 203–210 (2015).
Oestreich, E. A. et al. Epac and phospholipase Cε regulate Ca2+ release in the heart by activation of protein kinase Cε and calcium-calmodulin kinase II. J. Biol. Chem. 284, 1514–1522 (2009).
Oestreich, E. A. et al. Epac-mediated activation of phospholipase Cε plays a critical role in beta-adrenergic receptor-dependent enhancement of Ca2+ mobilization in cardiac myocytes. J. Biol. Chem. 282, 5488–5495 (2007).
Wang, H. et al. Phospholipase C ε modulates β-adrenergic receptor-dependent cardiac contraction and inhibits cardiac hypertrophy. Circ. Res. 97, 1305–1313 (2005).
Zhang, L., Malik, S., Kelley, G. G., Kapiloff, M. S. & Smrcka, A. V. Phospholipase C ε scaffolds to muscle-specific A kinase anchoring protein (mAKAPβ) and integrates multiple hypertrophic stimuli in cardiac myocytes. J. Biol. Chem. 286, 23012–23021 (2011).
Zhang, L. et al. Phospholipase cepsilon hydrolyzes perinuclear phosphatidylinositol 4-phosphate to regulate cardiac hypertrophy. Cell 153, 216–227 (2013).
Nash, C. A., Brown, L. M., Malik, S., Cheng, X. & Smrcka, A. V. Compartmentalized cyclic nucleotides have opposing effects on regulation of hypertrophic phospholipase Cepsilon signaling in cardiac myocytes. J. Mol. Cell. Cardiol. 121, 51–59 (2018).
Wei, W. & Smrcka, A. V. Internalized β2-adrenergic receptors oppose PLC-dependent hypertrophic signaling. Circ. Res. 135, e24–e38 (2024).
Malik, S. et al. G protein betagamma subunits regulate cardiomyocyte hypertrophy through a perinuclear Golgi phosphatidylinositol 4-phosphate hydrolysis pathway. Mol. Biol. Cell 26, 1188–1198 (2015).
Means, C. K. & Brown, J. H. Sphingosine-1-phosphate receptor signalling in the heart. Cardiovasc. Res. 82, 193–200 (2009).
Xiang, S. Y. et al. PLCepsilon, PKD1, and SSH1L transduce RhoA signaling to protect mitochondria from oxidative stress in the heart. Sci. Signal. 6, ra108 (2013).
Yung, B. S. et al. Selective coupling of the S1P 3 receptor subtype to S1P-mediated RhoA activation and cardioprotection. J. Mol. Cell. Cardiol. 103, 1–10 (2017).
Xiang, S. Y. et al. RhoA protects the mouse heart against ischemia/reperfusion injury. J. Clin. Investig. 121, 3269–3276 (2011).
Brand, C. S., Tan, V. P., Brown, J. H. & Miyamoto, S. RhoA regulates Drp1 mediated mitochondrial fission through ROCK to protect cardiomyocytes. Cell Signal. 50, 48–57 (2018).
Samassekou, K. M. et al. Cryo-EM Structure of Phospholipase Cε Defines N-terminal Domains and their Roles in Activity. Preprint at bioRxiv 2024.09.11.612521 https://doi.org/10.1101/2024.09.11.612521 (2024).
Bunney, T. D. et al. Structural and mechanistic insights into ras association domains of phospholipase C. Mol. Cell 21, 495–507 (2006).
Sieng, M. et al. Functional and structural characterization of allosteric activation of phospholipase Cepsilon by Rap1A. J. Biol. Chem. 295, 16562–16571 (2020).
Hicks, S. N. et al. General and versatile autoinhibition of PLC isozymes. Mol. Cell 31, 383–394 (2008).
Lyon, A. M., Begley, J. A., Manett, T. D. & Tesmer, J. J. Molecular Mechanisms of Phospholipase C beta3 Autoinhibition. Structure 22, 1844–1854 (2014).
Seifert, J. P. et al. RhoA activates purified phospholipase C-ε by a guanine nucleotide-dependent mechanism. J. Biol. Chem. 279, 47992–47997 (2004).
Kelley, G. G., Kaproth-Joslin, K. A., Reks, S. E., Smrcka, A. V. & Wojcikiewicz, R. J. G-protein-coupled receptor agonists activate endogenous phospholipase Cε and phospholipase Cβ3 in a temporally distinct manner. J. Biol. Chem. 281, 2639–2648 (2006).
Kelley, G. G., Reks, S. E. & Smrcka, A. V. Hormonal regulation of phospholipase Cε through distinct and overlapping pathways involving G12 and Ras family G-proteins. Biochem. J. 378, 129–139 (2004).
Seifert, J. P., Zhou, Y., Hicks, S. N., Sondek, J. & Harden, T. K. Dual activation of phospholipase C-ε by Rho and Ras GTPases. J. Biol. Chem. 283, 29690–29698 (2008).
Dusaban, S. S. et al. Phospholipase C{varepsilon} links G protein-coupled receptor activation to inflammatory astrocytic responses. Proc. Natl. Acad. Sci. USA 110, 3609–3614 (2013).
Wing, M. R., Snyder, J. T., Sondek, J. & Harden, T. K. Direct activation of phospholipase C-ε by Rho. J. Biol. Chem. 278, 41253–41258 (2003).
de Rubio, R. G. et al. Phosphatidylinositol 4-phosphate is a major source of GPCR-stimulated phosphoinositide production. Sci. Signal. 11, eaan1210 (2018).
Rugema, N. Y. et al. Structure of phospholipase Cepsilon reveals an integrated RA1 domain and previously unidentified regulatory elements. Commun. Biol. 3, 445 (2020).
Citro, S. et al. Phospholipase Cε is a nexus for Rho and Rap-mediated G protein-coupled receptor-induced astrocyte proliferation. Proc. Natl. Acad. Sci. USA 104, 15543–15548 (2007).
Lyon, A. M., Dutta, S., Boguth, C. A., Skiniotis, G. & Tesmer, J. J. Full-length Galpha(q)-phospholipase C-beta3 structure reveals interfaces of the C-terminal coiled-coil domain. Nat. Struct. Mol. Biol. 20, 355–362 (2013).
Esquina, C. M. et al. Intramolecular electrostatic interactions contribute to phospholipase Cbeta3 autoinhibition. Cell Signal. 62, 109349 (2019).
Hudson, B., Jessup, R. E., Prahalad, K. K. & Lyon, A. M. Galphaq and the phospholipase Cbeta3 X-Y linker regulate adsorption and activity on compressed lipid monolayers. Biochemistry 58, 3454–3467 (2019).
Garland-Kuntz, E. E. et al. Direct observation of conformational dynamics of the PH domain in phospholipases C and beta may contribute to subfamily-specific roles in regulation. J. Biol. Chem. 293, 17477–17490 (2018).
Dvorsky, R., Blumenstein, L., Vetter, I. R. & Ahmadian, M. R. Structural Insights into the Interaction of ROCKI with the Switch Regions of RhoA. J. Biol. Chem. 279, 7098–7104 (2004).
Chen, Z. et al. Activated RhoA binds to the pleckstrin homology (PH) Domain of PDZ-RhoGEF, a potential site for autoregulation. J. Biol. Chem. 285, 21070–21081 (2010).
Kristelly, R., Gao, G. & Tesmer, J. J. G. Structural determinants of RhoA binding and nucleotide exchange in leukemia-associated rho guanine-nucleotide exchange factor. J. Biol. Chem. 279, 47352–47362 (2004).
Essen, L. O., Perisic, O., Cheung, R., Katan, M. & Williams, R. L. Crystal structure of a mammalian phosphoinositide-specific phospholipase C δ. Nature 380, 595–602 (1996).
Chen, J. E., Huang, C. C. & Ferrin, T. E. RRDistMaps: a UCSF Chimera tool for viewing and comparing protein distance maps. Bioinformatics 31, 1484–1486 (2015).
Madukwe, J. C., Garland-Kuntz, E. E., Lyon, A. M. & Smrcka, A. V. G protein betagamma subunits directly interact with and activate phospholipase Cepsilon. J. Biol. Chem. 293, 6387–6397 (2018).
Chen, C.-L. et al. Molecular basis for Gβγ-mediated activation of phosphoinositide 3-kinase γ. Nat. Struct. Mol. Biol. 31, 1198–1207 (2024).
Falzone, M. E. & MacKinnon, R. Gβγ activates PIP2 hydrolysis by recruiting and orienting PLCβ on the membrane surface. Proc. Natl. Acad. Sci. USA 120, e2301121120 (2023).
Thompson, R. F., Iadanza, M. G., Hesketh, E. L., Rawson, S. & Ranson, N. A. Collection, pre-processing and on-the-fly analysis of data for high-resolution, single-particle cryo-electron microscopy. Nat. Protoc. 14, 100–118 (2018).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. Sect. D Struct. Biol. 75, 861–877 (2019).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Casañal, A., Lohkamp, B. & Emsley, P. Current developments in Coot for macromolecular model building of electron cryo-microscopy and crystallographic data. Protein Sci. 29, 1055–1064 (2020).
Terashi, G., Wang, X., Maddhuri Venkata Subramaniya, S. R., Tesmer, J. J. G. & Kihara, D. Residue-wise local quality estimation for protein models from cryo-EM maps. Nat. Methods 19, 1116–1125 (2022).
Prisant, M. G., Williams, C. J., Chen, V. B., Richardson, J. S. & Richardson, D. C. New tools in MolProbity validation: CaBLAM for CryoEM backbone, UnDowser to rethink “waters,” and NGL Viewer to recapture online 3D graphics. Protein Sci. 29, 315–329 (2019).
Acknowledgements
We thank Dr. A.V. Smrcka (U. Michigan) for DNA encoding R. norvegicus PLCε, human Avi-tagged Gβ and Gγ, pCI-neo vector, and COS-7 cells, and Dr. J.J.G. Tesmer for DNA encoding human RhoA and the pMALc2H10T vector. We are grateful to Dr. Thomas Klose, Dr. Frank Vago, and Steve Wilson for their guidance in cryo-EM data collection and computational support. This work is supported by NIH 1R01HL141076-01 (A.M.L.), and support from the Purdue Institute for Cancer Research Shared Resource Award to the Purdue Cryo-EM Facility is gratefully acknowledged (P30CA023168). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
V.O. and A.M.L. designed the experimental approach. V.O., K.S., K.M., E.E.G., and A.M.L. cloned, expressed, and purified PLCε variants. V.O., K.S., K.M., E.E.G.-K., and W.C.H. performed and analyzed activity assays, and V.O. and B.D. carried out Western blots. V.O. prepared cryo-EM samples, and with K.S., E.E.G., and I.J.F., collected cryo-EM data. V.O. and A.M.L. modeled the structure. V.O. and A.M.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Henrik Dohlman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Dario Ummarino and Xiaohui “Frank” Zhang. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ohri, V., Samassekou, K., Muralidharan, K. et al. RhoA allosterically activates phospholipase Cε via its EF hands. Commun Biol 8, 1368 (2025). https://doi.org/10.1038/s42003-025-08742-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08742-0