Abstract
Aerobic anoxygenic phototrophic (AAP) bacteria are essential for oceanic carbon cycling. However, their architecture and structural adaptations of their photosynthetic systems to ensure adequate light harvesting, electron transport, and oxidative resilience in oxygen-rich environments remain poorly understood. In this study, we present a 2.4-Å cryo-EM structure of the reaction center-light-harvesting 1 (RC–LH1) supercomplex from Dinoroseobacter shibae DFL-12, a marine AAP bacterial symbiont of benthic dinoflagellates. This RC–LH1 supercomplex features a closed LH1 ring comprising 17 αβ-subunits, each containing two spheroidenones per αβ-heterodimer—a previously unreported configuration in phototrophic bacteria. The cytochrome subunit of the RC is truncated to three hemes, in contrast to the four-heme configuration found in anaerobic relatives. The structure also reveals elongated bacteriochlorophyll (BChl) spacing, which may account for its blue-shifted absorption maximum that is optimized for low-light benthic environments. Furthermore, we identify a previously unknown subunit, protein-LRC, which is hypothesized to functionally couple photochemical and respiratory electron transport. Collectively, these specific structural features allow AAP bacteria to balance anoxygenic photosynthesis and protection against oxidative damage, providing a mechanistic framework for them to thrive in oxygenated marine environments. Our study provides insights into the structural and functional variability of bacterial photosynthesis in response to oxygenated marine environments.
Similar content being viewed by others
Introduction
Photosynthesis, the biological process that converts solar energy into chemical energy, is fundamental to life on Earth and drives global biogeochemical cycles1. Photosynthesis can be classified into oxygenic photosynthesis and anoxygenic photosynthesis. Oxygenic photosynthesis, primarily found in cyanobacteria, algae, and plants, releases oxygen through water splitting catalyzed by Photosystem II. In contrast, anoxygenic photosynthesis is distributed across phylogenetically ancient bacterial lineages (e.g., Proteobacteria, Chlorobi, Chloroflexi), which utilize alternative electron donors such as hydrogen sulfide, ferrous iron, or organic compounds for light-driven energy conversion, demonstrating metabolic versatility in adapting to anoxic environments2. Among these organisms, aerobic anoxygenic phototrophic (AAP) bacteria occupy a unique ecological niche by performing anoxygenic photosynthesis in oxygen-rich environments3. AAP bacteria decouple light harvesting from oxygen sensitivity, enabling photosynthesis exclusively under aerobic conditions while utilizing organic substrates for aerobic respiration4.
AAP bacteria make up 1–10% of the total prokaryotes in the surface ocean, and play a crucial role in the ocean’s carbon cycling3,5,6. This ecological prominence underscores their importance in the global carbon cycle and microbial food webs. Despite their wide distribution and significant role in marine ecosystems, the structural and mechanistic bases of their photosynthetic machinery remain poorly understood.
In anaerobic phototrophs, including purple phototrophic bacteria (e.g., Rhodobacter (Rba.) sphaeroides) and green phototrophic bacteria (e.g., Chlorobaculum tepidum), the reaction center-light-harvesting 1 (RC–LH1) supercomplex functions as the central photosynthetic unit, orchestrating photon capture and cyclic electron transport. RC–LH1 supercomplexes exhibit remarkable architectural diversity across species7. Except for the RC–LH1 containing double layered LH1 rings from Gemmatimonas (G.) phototrophica8, RC–LH1 supercomplexes can be mainly divided into three categories: Thermochromatium (Tch.) tepidum9,10,11, Rhodospirillum (Rsp.) rubrum12,13,14, Blastochloris (Blc.) viridis15, Rhodopila (Rpl.) globiformis16, Blastochloris (Blc.) tepida17, Allochromatium (Alc.) tepidum18, Roseospirillum (Rss.) parvum19 exhibits the RC encircled by a closed LH1 ring containing 17/16-subunit αβ-polypeptides, whereas Rhodopseudomonas (Rps.) palustris20,21, Rba. sphaeroides22,23, Rba. blasticus24, Rba. veldkampii25, and Rba. capsulatus26 adopts a C-shaped LH1 array with an opening occupied by protein-W or PufX. Moreover, Rba. sphaeroides23,27,28, Rba. blasticus24, and Rhodobaca (Rbc.) bogoriensis29 can assemble RC–LH1 dimers with an S-shaped LH1 ring architecture. The structural diversity and plasticity of RC–LH1 underlie the evolutionarily conserved mechanisms that enable adaptation to different spectral ranges and the use of light intensity gradients across various light environments.
RC–LH1 structures are crucial for efficient energy and electron transfer in low-oxygen environments. However, AAP bacteria face a specific challenge: their photosynthetic apparatus operates in oxygenated environments where two distinct mechanisms generate reactive oxygen species (ROS). Triplet-excited bacteriochlorophylls (³BChls) produce ROS, whereas electron leakage from the photosynthetic electron transport chain to molecular oxygen exacerbates oxidative stress, collectively reducing photochemical efficiency4.
Here, we report the cryogenic electron microscopy (cryo-EM) structure of the RC–LH1 supercomplex from Dinoroseobacter (D.) shibae DFL-12, a model marine AAP bacteria symbiont of benthic dinoflagellates. The structure reveals that the LH1 ring with elongated BChl distances is optimized for light capture in benthic habitats (short-wavelength light) with varying light intensities. A previously unidentified protein subunit (protein-LRC) is determined to bridge the RC and LH1, potentially integrating photosynthetic and respiratory electron transport. Moreover, the LH1 ring features a narrow quinone transfer channel and contains additional carotenoids, which may serve to mitigate the potential formation of BChl triplet states and quench ROS. Our study provides insights into aerobic anoxygenic photosynthesis and could inform the rational engineering of oxygen-tolerant photosynthetic systems.
Results and discussion
Overall structure of the RC–LH1 supercomplex
RC–LH1 supercomplexes were isolated from D. shibae cells cultivated phototrophically under aerobic conditions through sucrose density gradient ultracentrifugation (Supplementary Fig. 1A). Structural integrity of the purified complexes was confirmed by SDS-PAGE analysis (Supplementary Fig. 1C). The cryo-EM structure of the D. shibae RC–LH1 supercomplex was resolved at 2.49 Å resolution (Supplementary Fig. 2 and Table 1), allowing clear visualization of amino acid side chains (Fig. 1A–C and Supplementary Fig. 3) and enabling robust atomic model construction through iterative refinement (Fig. 1D–F).
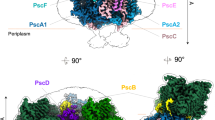
A–C Views of the RC–LH1 density map, colored as in the key at the bottom of the figure. A Side view of the RC–LH1 complex in the membrane plane, showing the height of the RC–LH1 complex. B Top view of the RC–LH1 supercomplex from the cytoplasmic side, showing the diameters of the long axes. C Bottom view of the RC–LH1 supercomplex from the periplasmic side, showing the diameters of the short axes. D–F Structural model of the RC–LH1 complex in three different views corresponding to (A–C).
The RC–LH1 supercomplex exhibits a slightly elliptical architecture in the projection view, with dimensions of 126 Å (short axis), 123 Å (long axis) (Fig. 1B, C), and 133 Å in vertical height (Fig. 1A). To our best knowledge, it represents the largest RC–LH1 structure with a single LH1 ring discovered to date. The RC core is encircled by a closed LH1 ring, which is composed of 17 αβ-heterodimers arranged circumferentially, each binding two bacteriochlorophyll a (BChl a) molecules and two spheroidenone carotenoids (34 total pigments per ring; Fig. 1D). The assignment of spheroidenones was corroborated by characteristic absorption spectra of the purified complexes (Supplementary Fig. 1B) and previous biochemical studies30. Structural modeling of the LH1 subunits revealed a conserved topology: both α- (51 residues) and β-polypeptides (44 residues) possess central transmembrane α-helices, with their N-terminal domains oriented toward the cytoplasmic membrane surface and C-termini extending into the periplasm (Fig. 1D and Supplementary Fig. 3).
The RC is composed of three core subunits (H, L, M) and a cytochrome (Cyt) c subunit that contains three hemes in contrast to the canonical four in anaerobic relatives (Fig. 1D). Structural analysis revealed that the RC contains four BChl a molecules, three bacteriopheophytin (BPhe a) molecules, one spheroidenone molecule, nine lipid molecules, three heme molecules, four ubiquinone-10 molecules, a non-heme iron atom, and an extra chromophore, which could be either a BChl a or a BPhe a (for convenience, modeled as BPhe a in the structural model). Notably, we identified a previously unknown protein subunit, designated protein-LRC (Light-Respiratory Connector) (Fig. 1D–F, Supplementary Figs. 1C and 4, Supplementary Table 1). The structural and functional implications of protein-LRC are discussed in the following sections.
The structure of the D. shibae RC–LH1 supercomplex resembles that of the Blc. viridis RC–LH1 supercomplex (PDB 6ET5)15, both of which feature a closed LH1 ring with 17 αβ-polypeptides surrounding the RC. However, compared to the Blc. viridis counterpart, the D. shibae RC–LH1 supercomplex contains an additional carotenoid in each LH1 αβ-polypeptide, and features a larger LH1 ring diameter. More strikingly, D. shibae RC–LH1 contains a unique tri-heme Cyt c subunit, a subunit protein-LRC, and lacks the γ-apoprotein present in Blc. viridis RC–LH1.
Interestingly, our structure exhibits several notable differences compared to the recent study on the structure of the D. shibae RC–LH1 complex31 (Supplementary Fig. 5). For example, our structure provides a more complete model of protein-LRC (named protein-O in ref. 31), including an additional α-helix on the periplasmic side of RC–LH1 (Supplementary Fig. 5C). Moreover, an additional BPhe/BChl molecule was identified within the RC of our structural model (Supplementary Fig. 5D).
The special protein-LRC
At a 2.49 Å resolution, the cryo-EM density map of the D. shibae RC–LH1 supercomplex revealed the presence of a previously unidentified protein subunit located between the RC and LH1 (Fig. 2A). However, no nucleotide sequences were found within the photosynthetic gene cluster, which encodes the protein that matches the cryo-EM density of this subunit. The sequence of this specific protein was ultimately determined by matching density against the genomic protein sequence. This protein, designated protein-LRC, is hypothesized to be encoded by a gene within the NADH-quinone oxidoreductase gene cluster. In the RC–LH1 density map, only residues from Asp7 to Thr82 of the N-terminal domain of protein-LRC were resolved, adopting a “helix-turn-helix-turn-helix” fold (Fig. 2A). The two α-helices near the N-terminus are transmembrane helices, oriented parallel to the LH1 ring and situated between the RC and LH1-15αβ. The third α-helix extends along one side of the LH1 ring, from LH1-15αβ to LH1-13αβ. The predicted full-length structure of protein-LRC suggests that it consists of an N-terminal domain, a C-terminal domain, and a long, flexible connecting loop (Supplementary Fig. 6). The lack of densities for the C-terminal domain and the connecting loop is likely due to their inherent flexibility or potential degradation/loss during sample preparation.
Protein-LRC forms extensive hydrogen bonds with neighboring LH1 α-subunits and the C-terminus of the RC-L subunit (Fig. 2B and Supplementary Table 2). On the cytoplasmic side, heterodimeric bonds are formed between LRC-Asp62 and LH1-15 α-Arg14, LRC-Val58 and LH1-15 α-Arg15, LRC-Gly60 and LH1-13 α-Arg15, LRC-Arg68 and LH1-14 α-Asp12, as well as LRC-Arg79 and LH1-13 β-Phe8. On the periplasmic side, LRC-Gln38 forms hydrogen bonds with L-Trp266 and L-Glu269. Additionally, the residues LRC-Cys10 and LRC-Cys59 stabilize the spatial arrangement of the two transmembrane helices of protein-LRC by forming disulfide bonds.
Comparison of protein-LRC and PufY/protein-Y/protein-U identified in Rba. sphaeroides RC–LH1 revealed that, despite the similar positions (yet not identical) of the transmembrane helices in the N-terminal region of protein-LRC and PufY/protein-Y/protein-U within their respective RC–LH1 complexes, the orientation of these transmembrane helices and the overall structures of protein-LRC differ substantially from those of and PufY/protein-Y/protein-U (Supplementary Fig. 7)22,23,32. Specifically, protein-LRC features a distinctive “helix–turn–helix–turn–helix” motif in its N-terminal region, whereas PufY/protein-Y/protein-U lack the additional helical and peripheral features observed in protein-LRC. Additionally, protein-LRC possesses a long loop region and a C-terminal domain, which are absent in PufY/protein-Y/protein-U.
Bioinformatic and structural analysis revealed that the C-terminus of protein-LRC exhibits high sequence and structural similarity to the C-terminal domain of Clade A NADH-quinone oxidoreductase subunit E (Supplementary Figs. 8 and 9). In certain photosynthetic bacteria, both Clade A and Clade E NADH-quinone oxidoreductases coexist, whereas in others, only one type is present. The main distinction between them lies in the presence of an additional C-terminal domain of NADH-quinone oxidoreductase subunit E found in Clade A, which is not found in Clade E. These two NADH-quinone oxidoreductases are responsible for distinct adaptive responses to various environmental conditions33. Rba. sphaeroides contains both types of NADH-quinone oxidoreductases and regulates their expression depending on growth conditions, whereas Rba. capsulatus only contains the Clade A type but can cleave the extra C-terminal domain of subunit E under specific growth conditions to adapt to its environment34,35. These findings suggest that the C-terminal domain of subunit E plays a crucial role in modulating the function of NADH-quinone oxidoreductase, implying that protein-LRC is likely involved in regulating NADH-quinone oxidoreductase activity. Nonetheless, the functional role of protein-LRC remains unclear due to limited experimental evidence. Further research is required to evaluate if it plays a role in the coupling of photochemical and respiratory electron transport and modulating the QA Em in the RC.
The special cytochrome c subunit
There are two types of RCs in anoxygenic phototrophic bacteria: one features a tightly bound Cyt c subunit on the periplasmic side that donates electrons to the photo-oxidized RC core, whereas the other lacks this Cyt c subunit and accepts electrons directly from water-soluble carriers36. Structural studies of RCs in several photosynthetic bacteria have shown that the Cyt c subunit typically contains four c-type hemes aligned along its long axis10,15. In contrast, the Cyt c subunit in D. shibae contains only three hemes—specifically, Hemes 2, 3, and 4, with Met and His residues serving as the sixth axial ligands for the heme irons—while the heme near the periplasmic side (Heme 1) is absent (Fig. 3A, B). This observation is consistent with bioinformatic analysis, which reveals that the first heme-binding motif (Cys-Xaa-Xaa-Cys-His) is missing in the sequence of the Cyt c subunit from D. shibae (Fig. 3A and Supplementary Fig. 10).
The loss of Heme 1 results in the absence of the traditional Cyt c2 binding site, suggesting the electron transfer pathway of this unique Cyt c subunit differs from that of the typical 4-heme Cyt c subunit. Recent AlphaFold3 predictions proposed that electron transfer from Cyt c2 to the RC occurs via the RC-proximal heme (Heme 4) in subunit C31. However, if this is the case, the actual functions of the other hemes (Heme 1-3) remain mysterious. Further studies, e.g., incorporating molecular genetics approaches, are required to decipher the electron transfer pathways and their connection to environmental adaptation.
In addition to the unique heme arrangement, the N-terminus of the Cyt c subunit also exhibits distinctive features. It is attached to the LH1 ring and forms hydrogen bonds with LH1-1αβ. Heterodimeric bonds are formed between C-Lys12 and LH1-1 α-Arg14, as well as between C-Asp16 and LH1-1 α-Arg15 (Fig. 4C; Supplementary Table 3). This creates an additional interaction beyond the typical RC–LH1 ring interactions, and likely provides a suitable target for the assembly of the RC–LH1 ring. The assembly of D. shibae RC–LH1 is further discussed below.
Interactions within the LH1 ring and between RC and LH1
The interactions within the LH1 ring and between RC and LH1 of the D. shibae RC–LH1 supercomplex closely resemble those observed in other RC–LH1 supercomplexes. The structural integrity of the LH1 ring is maintained through an extensive hydrogen-bonding network, primarily localized within the N-terminal domains of the αβ-heterodimers (Fig. 4A, B and Supplementary Table 4). Intra-heterodimer stabilization is facilitated by specific hydrogen bonds, including those between α-Tyr5 and β-Asp13, α-Leu9 and β-Phe8, as well as by interactions of α-Trp8 with both β-His20 and β-Thr9. Notably, the transmembrane α-helical regions of LH1 subunits do not participate in direct hydrogen bonding. Inter-heterodimer connectivity is established on the cytoplasmic face through hydrogen bonds linking α(n)-Lys6 of one heterodimer to β(n-1)-Glu18 of the adjacent subunit (Fig. 4B).
The interactions between RC and LH1 are mainly concentrated in three regions on the cytoplasmic side (Fig. 4C and Supplementary Table 3). C-Asp16, L-Trp26, and H-Gly53 interact with Arg15 residues of neighboring LH1 α-subunits. In addition to these hydrogen bond positions, H-Ala50 and C-Lys12 interact with Arg14 residues of neighboring LH1 α-subunits. Together, these interfacial interactions anchor the RC core within the LH1 antenna complex while preserving structural flexibility that is essential for quinone transport.
Assembly of RC–LH1
During our structural analysis of D. shibae RC–LH1, in addition to the density map for a complete RC–LH1 supercomplex, 3D classification also revealed two high-resolution density maps corresponding to D. shibae RC–LH1 supercomplexes with incomplete LH1 rings, termed RC–LH1-A and RC–LH1-B (Fig. 5A). Their structures were determined at resolutions of 2.44 Å and 2.75 Å, respectively (Fig. 5B, Supplementary Fig. 2 and Table 1).
The RC structures in RC–LH1-A and RC–LH1-B are identical to those in the complete D. shibae RC–LH1. However, unlike the complete D. shibae RC–LH1 with 17 LH1 αβ-subunits, RC–LH1-A contains only 10 αβ-subunits, whereas RC–LH1-B has 14. Moreover, due to the absence of LH1-13 αβ and LH1-14 αβ, protein-LRC is also missing in RC–LH1-A, as the critical interactions required for its stabilization are no longer present.
The occurrence of RC–LH1-A and RC–LH1-B is likely due to the instability of αβ-subunit binding within the LH1 ring during the purification process. Interestingly, in both RC–LH1-A and RC–LH1-B, the LH1 ring always begins to lose subunits from LH1-17 αβ. This suggests that the LH1-1 αβ side has greater structural stability when interacting with the RC. As mentioned earlier, the N-terminal region of the Cyt c subunit provides additional interactions between LH1-1 αβ and RC. This indicates that the N-terminal region of the Cyt c subunit may function similarly to PufX in Rba. sphaeroides, serving as an anchoring site for the binding of the first LH1 αβ subunit23.
These findings allow us to propose an assembly pathway for the D. shibae RC–LH1 supercomplex. Its assembly begins with the formation of the RC core, which is composed of the H, L, M, and Cyt c subunits (Fig. 5C). Next, free LH1 αβ-subunits bind to the RC with the assistance of the N-terminal region of the Cyt c subunit. The assembly of the LH1 complex is initiated by the incorporation of the first αβ-subunit, followed by the sequential binding of additional αβ-subunits around the RC, ultimately forming a closed ring. When the LH1 ring reaches the LH1-14 αβ position, protein-LRC begins to bind. (Fig. 5C). The initiation assembly site and directional propagation for D. shibae RC–LH1 appear to be consistent with previously proposed assembly models for PufX-containing RC–LH1 complexes22,23,25. The precise mechanisms of this assembly and whether the proposed assembly process can be applied to other RC–LH1 supercomplexes similar to D. shibae RC–LH1 remain to be investigated.
Arrangement of pigments and its adaption to aerobic habitats with varying light intensity
In the LH1 ring, the pigments form a tightly stacked array, systematically organized into two distinct groups: a ring consisting of 34 closely coupled BChls and another ring comprising 34 carotenoids (Fig. 6A). The orientations between BChls in D. shibae are similar to those observed in other photosynthetic bacteria7. Due to the elliptical shape of the LH1 ring, the distance between BChls fluctuates, varying between 9.97 Å and 8.31 Å (Supplementary Fig. 11). Both the Mg-Mg distances between BChls within individual LH1 αβ-heterodimers (9.65 Å on average) and between BChls of adjacent LH1 αβ-subunits (8.76 Å on average) (Supplementary Fig. 11) are within 10 Å, ensuring efficient exciton coupling and energy resonance within LH1. We compared the BChl ring of the RC–LH1 supercomplexes of D. shibae and Blc. viridis (PDB 6ET5)15, both of which contain 17 αβ-heterodimers. Our analysis showed that D. shibae RC–LH1 has a significantly larger radius and an increased average Mg-Mg distance (Supplementary Fig. 12). The intra-subunit (8.8 Å) or inter-subunit (8.5 Å) Mg-Mg distances of BChl b in Blc. viridis RC–LH1 supercomplex are relatively smaller, while D. shibae RC–LH1 has a larger average intra-subunit Mg-Mg distance of 9.65 Å, which exceeds those typically found in other RC–LH1 structures, such as those of Tch. tepidum (8.88 Å)9, and Rps. palustris (9.53 Å)20. The Mg-Mg distance within the BChl pairs reflects the degree of electronic coupling, with shorter distances indicating red shifts in the absorption spectrum and larger distances suggesting blue shifts15. The enhanced Mg-Mg distance may account for the blue-shift in the absorption peak (866.5 nm) of BChls in LH1 from D. shibae (Supplementary Fig. 1B). Moreover, D. shibae RC–LH1 possesses BChl a, instead of BChl b that is present in Blc. viridis RC–LH1 (Supplementary Fig. 13), and does not contain the γ-apoprotein that constrains the free movement of the LH1 ring and stabilizes the BChl b pairs15. These structural variations are presumed to contribute to the distinct absorption spectra of the two RC–LH1 supercomplexes, with a blue shift in D. shibae and red shift in Blc. viridis15.
A Pigments and cofactors viewed from the cytoplasmic side. B Side view of the arrangement of pigments and cofactors in the membrane plane. C Arrangement of pigments, quinones, and lipids associated with RC. The extra chromophore is marked in the figure. Although it cannot be definitively identified as either BChl or BPhe, for convenience it was modeled as BPhe in the structural model. D Maximum and minimum Mg-Mg distances between the BChls in the LH1 ring and the special pair BChls.
It should be noted that the absorption spectra we measured represent a mixture of RC–LH1 complexes containing both complete and incomplete LH1 rings. However, given that the average Mg–Mg distances are nearly identical in both forms (Supplementary Figs. 14–16 and Supplementary Table 5), and that the absorption spectra from different purification batches (possessing varying ratios of complete and incomplete LH1 rings) remained consistently comparable (Supplementary Fig. 16B), we infer that any spectral differences between open and closed LH1 rings are negligible. Consequently, the absorption spectrum of the fully closed RC–LH1 supercomplex is expected to be resemble that of the mixture.
Moreover, the increased radius of the BChl ring results in an increase in the distances between the BChls in the LH1 ring and the special pair of BChls in the RC, ranging from 52.6 to 40.1 Å (Fig. 6D). The minimal distances between LH1 and RC in D. shibae RC-LH1 is 40.1 Å, greater than those of several representative RC-LH1 complexes from Blc. viridis (38.7 Å)15, Rba. sphaeroides (38.3 Å)22,23, and Rba. capsulatus (39.0 Å)26. This increased distance may reduce the efficiency of excitation energy transfer (EET), potentially leading to the accumulation of 3BChl under high light conditions. This could result in the production and accumulation of ROS during aerobic photosynthesis. Compared to Blc. viridis, D. shibae thrives in oxygen-rich environments and features a larger LH1 ring with a greater diameter, making it more susceptible to severe oxidative stress37. Consequently, specific protective mechanisms are necessary to mitigate oxidative damage. A notable structural difference is that each LH1 subunit in D. shibae RC–LH1 contains two carotenoid molecules, whereas Blc. viridis has only one (Supplementary Fig. 13). The primary carotenoid in D. shibae is spheroidenone, which is produced through the oxidation of spheroidene and OH-spheroidene via spheroidene monooxygenase (CrtA) under aerobic conditions. Typically, only one carotenoid is located between the αβ-polypeptides of the closed LH1 ring structure, facilitating the formation of channels for quinone trafficking across the LH1 αβ-polypeptides. However, D. shibae RC–LH1 incorporates two carotenoids per LH1 αβ-polypeptide within the closed LH1 ring. In addition to increasing pigment levels to enhance light harvesting under low-light conditions, the higher carotenoid concentration may help suppress the formation of BChl triplet states and quench ROS. The increased inter-BChl distances attenuate excitonic coupling in the LH1 BChls, leading to a more uniform excitation distribution across the antenna system. This structural modification consequently necessitates additional carotenoids for effective triplet-state quenching. This provides a plausible explanation for the observed doubling of carotenoids in the LH1 complex. This dual function may be particularly important under high-light conditions, where the probability of triplet state formation and ROS generation increases. Collectively, these adaptations may enhance oxidative stress protection and overall light capture efficiency.
As we described above, the cofactors in the RC included four BChls a, three BPhes a, one spheroidenone, nine lipids, four ubiquinone-10 molecules, three hemes, one iron atom, and an extra chromophore, which could be either a BChl a or a BPhe a (Fig. 6C). The composition and arrangement of pigments in the D. shibae RC are consistent with those of most purple phototrophic bacteria, whereas the major differences lie in the presence of an additional BPhe/BChl in the RC–LH1 supercomplex and the absence of a heme molecule. The limited resolution of the cryo-EM density map precludes an unambiguous determination of magnesium ion occupancy in this additional chromophore, preventing its definitive assignment as either BChl a or BPhe a (Fig. 6C). In the Rba. veldkampii RC-LH1 complex, an extra chromophore density occupies the corresponding position and was assigned as BPhe a. (Supplementary Fig. 17)25.This special BPhe/BChl in D. shibae RC–LH1 is located between the RC-H, RC-L subunits, and LH1-1αβ, where it stabilizes the spatial structure by interacting with the main chain oxygen of L-Ile221 and is positioned closer to the BPhe a on the non-electron-transmitting side (Fig. 6C). The exact role of the extra BPhe/BChl remains to be examined.
Pathways of quinone/quinol exchange
The special pair of BChls acts as the primary electron donor and is excited to P880*; this excited state then transfers an electron to BChl a and BPhe a, and subsequently to QA. QB accepts electrons from QA. Once quinone receives two electrons, it binds two protons from the cytoplasm and escapes from the RC to the neighboring Cyt bc1. In RC–LH1s with open LH rings, quinones/quinols are assumed to primarily shuttle through the gap in the LH1 ring, such as Rba. sphaeroides23, Rba. veldkampii25, and Rba. capsulatus26. In contrast, in RC–LH1s with closed LH rings forming a palisade around the RC, quinones/quinols are proposed to diffuse through relatively small channels between two adjacent LH1 αβ subunits.
Four UQ-10 molecules were identified in the density map of D. shibae RC–LH1 (Figs. 6C and 7A). Two UQ-10 molecules function as the primary (QA) and secondary (QB) quinone acceptors (Fig. 7A). The head of QA is hydrogen bonded to His220 and Ala261 residues of RC-M, and the head of QB is hydrogen bonded to RC-L residues His191, Ser224, Ile225, and Gly226 (Supplementary Fig. 18 and Supplementary Table 6). Two additional putative UQ-10 molecules (Q3 and Q4) are located in the gap between LH1 and the RC and are mainly surrounded by nonpolar residues of RC-L, RC-M, and protein-LRC. Q3 shares the same orientation as QB and is in close proximity to the isoprenoid tail of QB (Fig. 7A), suggesting that Q3 is in an appropriate position for the exchange of QB after double reduction and protonation.
Owing to the presence of additional carotenoid molecules in the LH1 ring, the gaps between most LH1 αβ subunits appear limited in size to enable efficient quinone passage. A narrow pore exists between LH1-1αβ and LH1-17αβ (Fig. 7B). This pore formation is facilitated by the binding of the N-terminal domain of the Cyt c subunit, which induces conformational changes in the N-terminal regions of LH1-17α and effects the binding conformation of BChl a (Supplementary Figs. 19 and 20). This pore is relatively small (10.2 Å in diameter) compared to that in Blc. viridis RC–LH1 (13.4 Å in diameter) (Supplementary Fig. 21). This may serve as a viable channel for quinone exchange.
Conclusions
The cryo-EM structure of the D. shibae RC–LH1 supercomplex unveiled key structural innovations driving its adaptation to aerobic anoxygenic photosynthesis. The cytochrome subunit of D. shibae RC–LH1 is integrated with three hemes instead of four hemes as found in anaerobic relatives. The LH1 ring exhibits elongated BChl spacing, a configuration likely optimized for efficient light harvesting in the fluctuating light conditions of benthic ecosystems. The tightly closed LH1 architecture is enriched with carotenoids and features a constricted quinone channel. More strikingly, a previously unidentified bridging subunit (protein-LRC) physically connects the RC and LH1, offering a potential molecular conduit for coordinating photosynthetic electron transport with respiratory pathways. These structural features provide the mechanistic basis of oxygen-tolerant photosynthesis in AAP bacteria and highlight their physiological adaptations in oxygen-dominated environments.
Methods
Growth condition of D. shibae
Wild-type Dinoroseobacter shibae DFL-12 (DSM-16493) was obtained from DSMZ. D. shibae cells were grown phototrophically under aerobic conditions in liquid 2216e medium at 30 °C in glass bottles under a light intensity of 25 μmol photons s−1m−2 (Bellight 70 W halogen bulbs). The bacterial culture period ranges from five to seven days, during which the color gradually transitions to red. Bacteria reaching the logarithmic growth phase will be utilized for subsequent experiments.
Purification of RC − LH1 complexes
Cells were harvested by centrifugation at 5000 × g for 10 min at 4 °C, washed three times with Tris-HCl (pH 8.0), and resuspended in 20 mM HEPES (pH 8.0). The cells were disrupted by passage through a French press three times at 16,000 psi. Cell debris was removed by centrifugation at 20,000 × g for 30 min. Membranes were collected by centrifuging the resulting supernatant at 125,000 × g for 90 min and solubilized by the addition of β-DDM (n-dodecyl β-D-maltoside) to a final concentration of 3% (w/v) for 30 min to 60 min in the dark at 4 °C with gentle stirring. Unsolubilized proteins were removed by centrifugation at 21,000 × g for 30 min. The supernatant was then applied onto 10–25% (w/v) continuous sucrose gradients made with working buffer containing 0.01% (w/v) β-DDM. Gradients were centrifuged at 230,000 × g for 18 h. The RC–LH1 complexes in the sucrose gradient solution were collected, and the purity of RC–LH1 complexes was characterized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and absorption spectra (Supplementary Fig. 1).
Absorption spectra
Following purification, the RC-LH1 complexes were successfully isolated and collected from the sucrose density gradients. To characterize their spectral properties, the absorbance of the purified samples was subsequently measured at room temperature. The measurement was performed across a broad wavelength spectrum ranging from 300 to 900 nm using a Libra S22 spectrophotometer (Biochrom, United Kingdom). Data points were collected at 1-nm intervals to ensure a high-resolution absorption spectrum for detailed analysis.
Cryo-EM data collection
A 4 μL aliquot of RC–LH1 sample was applied to a freshly glow-discharged holey carbon grid (Quantifoil Au R2/1, 200 mesh) with a continuous carbon support. The grid was blotted for 2 s at 100% humidity and 10 °C, with a force level of 0, and immediately plunge-frozen into liquid ethane cooled by liquid nitrogen using a Vitrobot Mark IV (Thermo Fisher, USA). The grids were then loaded into a 300 kV Titan Krios G3i microscope (Thermo Fisher, USA) equipped with a Falcon 4 direct electron detector (Gatan, USA) for data acquisition. A total of 5962 movie stacks were automatically recorded using EPU (Thermo Fisher, USA)38 at a total dose of 40 e− Å−2 per stack, with a defocus range of −0.8 to −1.8 μm and a pixel size of 0.97 Å.
Data processing
Data processing was conducted using cryoSPARC (v4.4.1)39. Patch motion correction and contrast transfer function (CTF) correction were performed, and micrographs with a resolution worse than 6 Å were discarded. From the remaining 5854 micrographs, 1,011,021 particles were picked using cryoSPARC’s Blob picking algorithm. The particles were extracted and subjected to three rounds of 2D classification, yielding 271,106 particles for further analysis. These particles were used for Ab initio reconstruction and 3D classification into four classes. Three “best-looking” classes were selected for non-uniform refinement in cryoSPARC (v4.4.1), using the ‘optimize per-particle defocus’ option, yielded final maps with resolutions of 2.49 Å for the D. shibae RC–LH1 supercomplex, 2.44 Å for the RC–LH1-A supercomplex, and 2.75 Å for the RC–LH1-B supercomplex. Resolution estimates were based on the gold-standard fourier shell correlation at 0.143.
Model building and refinement
The structure of the RC–LH1 supercomplex of Blc. viridis (DSM-133) (PDB ID: 6ET5) was initially docked into the cryo-EM map of the RC–LH1 complex using UCSF Chimera (v1.17)40. The model was manually revised and refined based on cryo-EM density using Coot (v 0.9.4)41, followed by real-space refinement using Phenix (v1.20.1)42. Figures were generated using UCSF Chimera (v1.17) and ChimeraX (v1.16)40,41,42,43.
Statistics and reproducibility
All purification experiments of the D. shibae RC–LH1 supercomplex were independently repeated at least three times, and the biochemical properties were consistently observed, demonstrating the robustness of the experimental procedures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The cryo-EM density map of D. shibae RC–LH1 supercomplex, RC–LH1-A supercomplex, and RC–LH1-B supercomplex were deposited in the Electron Microscopy Data Bank (EMDB, www.ebi.ac.uk/pdbe/emdb/) under the accession code EMDB-63379, EMDB-63381, and EMDB-63382. Atomic coordinates of D. shibae RC–LH1supercomplex, RC–LH1-A supercomple,x and RC–LH1-B supercomplex were deposited in the Protein Data Bank (PDB, www.rcsb.org) with the accession code 9LTS, 9LTU, and 9LTV. The uncropped and unedited gel image corresponding to Supplementary Fig. 1C is provided in Supplementary Fig. 22. Source data underlying Supplementary Figs. 1B and 16B are available in Supplementary Data 1 and Supplementary Data 2, respectively. All other data are available from the corresponding author on reasonable request.
References
Field, C. B., Behrenfeld, M. J., Randerson, J. T. & Falkowski, P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240 (1998).
Blankenship, R. E. Early evolution of photosynthesis. Plant Physiol. 154, 434–438 (2010).
Kolber, Z. S. et al. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science 292, 2492–2495 (2001).
Yurkov, V. V. & Beatty, J. T. Aerobic anoxygenic phototrophic bacteria. Microbiol. Mol. Biol. Rev. 62, 695–724 (1998).
Jiao, N. et al. Distinct distribution pattern of abundance and diversity of aerobic anoxygenic phototrophic bacteria in the global ocean. Environ. Microbiol. 9, 3091–3099 (2007).
Koblížek, M. Ecology of aerobic anoxygenic phototrophs in aquatic environments. FEMS Microbiol. Rev. 39, 854–870 (2015).
Liu, L. N., Bracun, L. & Li, M. Structural diversity and modularity of photosynthetic RC-LH1 complexes. Trends Microbiol. 32, 38–52 (2024).
Qian, P. et al. 2.4-Å structure of the double-ring Gemmatimonas phototrophica photosystem. Sci. Adv. 8, eabk3139 (2022).
Kawakami, T. et al. Crystal structure of a photosynthetic LH1-RC in complex with its electron donor HiPIP. Nat. Commun. 12, 1104 (2021).
Niwa, S. et al. Structure of the LH1-RC complex from Thermochromatium tepidum at 3.0 Å. Nature 508, 228–232 (2014).
Yu, L. J., Suga, M., Wang-Otomo, Z. Y. & Shen, J. R. Structure of photosynthetic LH1-RC supercomplex at 1.9 Å resolution. Nature 556, 209–213 (2018).
Qian, P. et al. Cryo-EM structure of the Rhodospirillum rubrum RC-LH1 complex at 2.5 Å. Biochem. J. 478, 3253–3263 (2021).
Christianson, B. et al. Characterization of the structure and function of the photosynthetic RC–LH1 core supercomplex from Rhodospirillum rubrum. Physiol. Plant. 17, e70275 (2025).
Tani, K. et al. Cryo-EM structure of the photosynthetic LH1-RC complex from Rhodospirillum rubrum. Biochemistry 60, 2483–2491 (2021).
Qian, P., Siebert, C. A., Wang, P., Canniffe, D. P. & Hunter, C. N. Cryo-EM structure of the Blastochloris viridis LH1-RC complex at 2.9 Å. Nature 556, 203–208 (2018).
Tani, K. et al. An LH1-RC photocomplex from an extremophilic phototroph provides insight into origins of two photosynthesis proteins. Commun. Biol. 5, 1197 (2022).
Kimura, Y. et al. The thermal-stable LH1-RC complex of a hot spring purple bacterium powers photosynthesis with extremely low-energy near-infrared light. Biochemistry 64, 170–179 (2025).
Tani, K. et al. A Ca2+-binding motif underlies the unusual properties of certain photosynthetic bacterial core light-harvesting complexes. J. Biol. Chem. 298, 101967 (2022).
Wang, X. P. et al. Insights into the divergence of the photosynthetic LH1 complex obtained from structural analysis of the unusual photocomplexes of Roseospirillum parvum. Commun. Biol. 7, 1658 (2024).
Roszak, A. W. et al. Crystal structure of the RC-LH1 core complex from Rhodopseudomonas palustris. Science 302, 1969–1972 (2003).
Swainsbury, D. J. K. et al. Structures of Rhodopseudomonas palustris RC-LH1 complexes with open or closed quinone channels. Sci. Adv. 7, eabe2631 (2021).
Qian, P. et al. Cryo-EM structure of the monomeric Rhodobacter sphaeroides RC-LH1 core complex at 2.5 Å. Biochem. J. 478, 3775–3790 (2021).
Cao, P. et al. Structural basis for the assembly and quinone transport mechanisms of the dimeric photosynthetic RC-LH1 supercomplex. Nat. Commun. 13, 1977 (2022).
Wang, P. et al. Architectures of photosynthetic RC-LH1 supercomplexes from Rhodobacter blasticus. Sci. Adv. 10, eadp6678 (2024).
Bracun, L. et al. Cryo-EM structure of the photosynthetic RC-LH1-PufX supercomplex at 2.8-Å resolution. Sci. Adv. 7, eabf8864 (2021).
Bracun, L., Yamagata, A., Christianson, B. M., Shirouzu, M. & Liu, L. N. Cryo-EM structure of a monomeric RC-LH1-PufX supercomplex with high-carotenoid content from Rhodobacter capsulatus. Structure 31, 318–328.e313 (2023).
Qian, P. et al. Three-dimensional structure of the Rhodobacter sphaeroides RC-LH1-PufX complex: dimerization and quinone channels promoted by PufX. Biochemistry 52, 7575–7585 (2013).
Tani, K. et al. Asymmetric structure of the native Rhodobacter sphaeroides dimeric LH1-RC complex. Nat. Commun. 13, 1904 (2022).
Semchonok D. A. et al. Cryo-EM structure of the Rhodobaca bogoriensis RC-LH1-PufX dimeric complex at 2.9 Å. bioRxiv, 481955 (2022).
Biebl, H. et al. Dinoroseobacter shibae gen. nov., sp. nov., a new aerobic phototrophic bacterium isolated from dinoflagellates. Int. J. Syst. Evolut. Microbiol. 55, 1089–1096 (2005).
Wang, W. et al. Cryo-EM analysis of a tri-heme cytochrome-associated RC-LH1 complex from the marine photoheterotrophic bacterium Dinoroseobacter Shibae. Adv. Sci. 12, e2413456 (2025).
Tani, K. et al. A previously unrecognized membrane protein in the Rhodobacter sphaeroides LH1-RC photocomplex. Nat. Commun. 12, 6300 (2021).
Spero, M. A., Aylward, F. O., Currie, C. R. & Donohue, T. J. Phylogenomic analysis and predicted physiological role of the proton-translocating NADH:quinone oxidoreductase (complex I) across bacteria. mBio 6, e00389–00315 (2015).
Spero, M. A. et al. Different functions of phylogenetically distinct bacterial complex I isozymes. J. Bacteriol. 198, 1268–1280 (2016).
Herter, S. M., Kortlüke, C. M. & Drews, G. Complex I of Rhodobacter capsulatus and its role in reverted electron transport. Arch. Microbiol. 169, 98–105 (1998).
Masuda, S., Yoshida, M., Nagashima, K. V., Shimada, K. & Matsuura, K. A new cytochrome subunit bound to the photosynthetic reaction center in the purple bacterium, Rhodovulum sulfidophilum. J. Biol. Chem. 274, 10795–10801 (1999).
Piwosz, K., Kaftan, D., Dean, J., Šetlík, J. & Koblížek, M. Nonlinear effect of irradiance on photoheterotrophic activity and growth of the aerobic anoxygenic phototrophic bacterium Dinoroseobacter shibae. Environ. Microbiol. 20, 724–733 (2018).
Thompson, R. F., Iadanza, M. G., Hesketh, E. L., Rawson, S. & Ranson, N. A. Collection, pre-processing and on-the-fly analysis of data for high-resolution, single-particle cryo-electron microscopy. Nat. Protoc. 14, 100–118 (2019).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 66, 213–221 (2010).
Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci. 32, e4792 (2023).
Acknowledgements
We thank Qiu-Yao Jiang (Shandong First Medical University, China) for cryo-EM data collection. We thank Xiao-Ju Li (Shandong University, China) for their assistance in TEM. This work was supported by the National Key R&D Program of China (2023YFA0914600, 2021YFA0909600, 2024YFC2816000), the National Natural Science Foundation of China (32330001, 32070109, 32170127, 32370136), Shandong Province Science Fund for Excellent Young Scholars (ZR2025QB18), Program of Shandong for Taishan Scholars (tspd20240806, tsqn202408064), the SKLMT Frontiers and Challenges Project (SKLMTFCP-2023-06), the University of Liverpool-Chinese Scholarship Council PhD studentship to Y.-Y.Z.
Author information
Authors and Affiliations
Contributions
Y.-Z.Z., L.-N.L., and P.W. conceived the study. P.W. and L.-N.L. designed the experiments. P.W., Z.-K.L., Y.-Y.Z., J.-X.L., K.L., J.-L.L. performed the experiments. Z.-K.L., P.W., Y.-Y.Z., and J.-L.L. purified the protein samples and conducted optical spectral analysis. P.W., J.-X.L., K.L. collected cryo-EM data and processed the cryo-EM data. P.W. and Z.-K.L. assembled and generated the structural model and performed structural analysis. P.W., Z.-K.L., X.-L.C., Y.-Z.Z., and L.-N.L. wrote the manuscript. All the authors contributed to the discussion and improvement of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Seiji Akimoto and the other anonymous reviewer(s) for their contribution to the peer review of this work. Primary handling editors: Janesh Kumar and David Favero.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, ZK., Li, JX., Zhang, YY. et al. Structural basis for aerobic anoxygenic photosynthesis in the reaction center–light-harvesting 1 (RC–LH1) supercomplex of Dinoroseobacter shibae. Commun Biol 8, 1565 (2025). https://doi.org/10.1038/s42003-025-08935-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08935-7