Abstract
Contagious bovine pleuropneumonia (CBPP), caused by Mycoplasma mycoides subsp. mycoides (Mmm), is a devastating cattle disease with high morbidity and mortality. Although mass vaccination or stamping-out strategy has controlled CBPP in many countries, it remains a major threat to cattle productivity in sub-Saharan Africa. An attenuated vaccine derived from serial passages of the virulent Ben-1 strain has eradicated CBPP in China, but the underlying immunoprotective mechanisms are unclear. While interferon-inducible GTPases are known to play a crucial role in host defense, IRG-47, an interferon-gamma (IFN-γ)-inducible GTPase, was previously thought to have limited anti-infective activity. Here, we demonstrate that IRG-47 is critical for defending against Mmm and is specifically upregulated by the Ben vaccine strain. Screening potential protective proteins in this vaccine strain revealed that Ben_0691 is a key factor responsible for boosting IRG-47 to combat Mmm infection. Mechanistically, Ben_0691 binds directly to the host cell ATP synthase subunit alpha (ATP5A1) and stabilizes ATP5A1 by reducing its autophagic-degradation. This stabilization promotes IRG-47 expression, which disrupts Mmm’s early cell-associativity and provides resistance to Mmm infection. Notably, Ben_0691 N-terminal amino acids 66–72 are required for its interaction with ATP5A1, the induction of IRG-47 and anti-Mmm action. The AMPK-mTOR-IFN-γ pathway is further characterized as an essential signaling for ATP5A1 to facilitate IRG-47 expression. Together, our study reveals a previously unrecognized role of IRG-47 in defending against Mmm infection, and the protective mechanism of Ben_0691 as an IRG-47 activator. These findings provide new insights into vaccine-based protection strategies and may inform future CBPP control strategies.
Similar content being viewed by others
Introduction
Contagious bovine pleuropneumonia (CBPP) is a severe respiratory infectious disease affecting Bovidae, caused by Mycoplasma mycoides subsp. mycoides (Mmm)1. As a member of the genus Mycoplasma, a large group of the smallest self-replicating bacteria lacking cell walls, Mmm infection trigger substantial pathologic damage in the respiratory tracts and lungs of cattle2,3. When CBPP first appears in a herd, the mortality rate may exceed 50%, and can reach up to 90% during an outbreak, severely affecting animal welfare and livestock production4,5. CBPP is one of the six priority diseases officially recognized by the World Organization for Animal Health (WOAH, www.woah.org), and it is the only non-viral one. It was first reported in Europe in the 16th century, with global spread beginning in the 19th century6. Many countries have successfully eradicated the disease through culling or post-immunization eradication strategies, however CBPP remains endemic in sub-Saharan Africa. Previous estimates suggest that CBPP causes direct and indirect losses of approximately €44.8 million annually in these regions, and this figure is likely to rise, posing a serious threat to livestock production2,7,8. Additionally, insufficient surveillance and reporting have left the prevalence of CBPP in several Asian countries unknown2.
CBPP spread widely prevalent in China during the early 20th century, and led to significant financial losses. Since the 1960s, an attenuated vaccine developed by serial passages of the highly virulent isolate Ben-1 in heterologous animals was implemented, leading to the effective control and eradication of domestic CBPP4. This vaccine achieves protection rates of 95–100% over 2 years, outperforming other mycoplasma vaccines and most commercial ruminant vaccines. Nonetheless, its production process (intraperitoneal inoculation of sheep) is no longer acceptable under modern animal welfare standards9. The currently available attenuated vaccines T1/SR and T1/44, prepared from Tanzanian T1 strains, also face limitations in terms of a short duration of protection and severe adverse reactions2. Consequently, there is an urgent need for new or improved CBPP vaccines to prevent the disease outbreaks or re-epidemics. A comprehensive understanding of host-vaccine interactions and vaccine-induced protective immune responses are a prerequisite for the development of effective next-generation vaccines.
Interferon-gamma (IFN-γ) has been proposed as a key element required for protecting against CBPP, as its level was inversely correlated with disease severity. Although IFN-γ is rarely induced in naturally infected cattle, it is significantly upregulated in infected but recovered cattle10. The protective functions of IFN-γ are multiple and complex. One well-characterized mechanism is the transcriptional activation of over 1200 interferon-stimulated genes (ISGs), whose products promote a robust immune response against microbes11. Among these ISGs, several interferon-inducible GTPase families stand out, including the 65 kDa guanylate-binding proteins (GBPs) and the 47 kDa immunity-related GTPases (IRGs). Although the IRG system is complex and largely absent in most mammals, its orthologs have been identified in at least 12 species, including cows, pigs and humans12. Its only known function has been characterized in mice, where IRGs plays a critical role in attacking the vacuolar membranes of intravacuolar parasites13. Many IRGs are key mediators of resistance against pathogen infections in mice. For example, IRGM1 (LRG-47) is essential for resistance to Listeria monocytogenes, Mycobacterium tuberculosis, Mycobacterium avium and Toxoplasma gondii infections14,15,16,17; IRGM2 (GTPI) provides resistance against Toxoplasma gondii and Chlamydia psittaci18,19; IRGM3 (IGTP) defends against Leishmania major and Chlamydia trachomatis20,21; IRGA6 (IIGP1) is resistant to Chlamydia trachomatis and rabies viruses; while IRGB10 provides resistance to Chlamydia trachomatis and Francisella novicida22,23,24,25,26. Whether and what IFN-γ-inducible proteins are involved in protecting the host against CBPP is currently unknown.
In this study, we reveal the role of IRG protein IRG-47 in combating Mmm infection. We further identify Ben_0691 as a potential Mmm-protective protein, and elucidate the mechanism by which Ben_0691 utilizes IRG-47 for anti-Mmm infection. We hope that this study will provide a theoretical basis for improving/developing vaccines against Mmm, and may offer new avenues for developing innovative approaches and novel drugs to combat Mmm infection.
Results
Interferon-gamma-inducible protein IRG-47 enhances host cell resistance to Mmm infection
Given our previous observation that host IRG-47 expression correlates with vaccination outcomes against Mmm, and considering the critical role of the immune-related GTPase (IRG) family in host defense against pathogen infections21, we hypothesized that IRG-47 may contribute to host cell resistance against Mmm infection. To test this, we first constructed three small interfering RNAs (siRNAs) targeting bovine IRG-47 and a Myc-tagged IRG-47 eukaryotic expression plasmid. Embryonic bovine lung epithelial (EBL) cells are widely used as an in vitro model system for the study of bovine respiratory pathogen-host interactions27. It was demonstrated that the three specific siRNAs (siIRG-47-1, -2 or -3) transfection significantly decreased IRG-47 levels compared to control siRNA (siNC) transfection (Fig. 1A), while IRG-47-Myc plasmid transfection promoted IRG-47 expression in EBL cells compared to control plasmid pCAGGS-Myc transfection (Fig. 1B). CCK-8 assays confirmed that IRG-47 knockdown or overexpression did not affect EBL cell viability (Fig. 1C and Supplementary Table 1). We subsequently transfected EBL cells with siIRG-47-1 to knockdown IRG-47 or with IRG-47-Myc plasmid to overexpress IRG-47, followed by exposure to highly virulent Mmm strains (Ben-1 or Mu-1) at a multiplicity of infection (MOI) of 50 for 12 h. TaqMan qPCR detection of Mmm genomic DNA was used to assess whether the IRG-47 level affected Mmm infection to EBL cells (total numbers of Mmm adhered and entered into the cell). We observed that IRG-47 knockdown significantly increased the number of Mmm infecting EBL cells, whereas IRG-47 overexpression dramatically reduced the number of Mmm infecting EBL cells, compared to control cells co-transfected with siNC and pCAGGs-Myc (Fig. 1D, E). These results were further confirmed by an indirect immunofluorescence assay (IFA) targeting Mmm. As shown in Fig. 1F, G, green fluorescence (Mmm signal) was enhanced in IRG-47 knockdown cells and diminished in IRG-47 overexpression cells, compared with siNC and pCAGGs-Myc co-transfected EBL cells. According to these findings, IRG-47 level is strongly associated with host cell anti-Mmm infection.
A EBL cells were transfected with siRNAs (siIRG-47-1, -2 and -3) for 48 h and harvested for Western blotting analysis to verify the efficiency of IRG-47 knockdown. siNC represents the control siRNA. B EBL cells were transfected with 1 μg IRG-47-Myc or vector (pCAGGs-Myc), and cell lysates were analyzed for IRG-47 and β-actin levels by Western blotting. C CCK-8 assays showed that siRNA (siIRG-47-1) and IRG-47-Myc transfection had no effect on the cell viability of EBL cells. D–G EBL cells were transfected with siNC or siIRG-47-1 and pCAGGS-Myc or IRG-47-Myc, and then incubated with Mmm (Ben-1 and Mu-1) at an MOI of 50 for 12 h. These cells were harvested for TaqMan qPCR to analyze the amount of Mmm infected to EBL cells (D, E), or cells were subjected for Indirect immunofluorescence assay (IFA) using mouse anti-Mmm monoclonal antibody (green) to detect Mmm infection level to EBL cells. DAPI was used to stain cellular nuclei (blue) (F, G). The data are represented the means ± SDs and were normalized to the corresponding values in control cells. Significance was assessed by one-way ANOVA with Dunnett’s multiple comparison tests relative to the control (C, n = 6 independent experiments) or with Tukey’s multiple comparison test (D, E, n = 3 independent experiments). **p < 0.01; ***p < 0.001; ****p < 0.0001.
Ben_0691 upregulates IRG-47 to resist Mmm infection of host cells
Based on in vivo challenge experiments conducted in cattle in the 1980s, the Mmm Ben series vaccine strain Ben-181 (Ben-181-vacc) conferred high protection against Mmm infection, whereas the strain Ben-468 (Ben-468-lowprot) provided only markedly reduced protection. We next evaluated the ability of these two strains in regulating host cell IRG-47. EBL cells were infected with Ben-181-vacc or Ben-468-lowprot at an MOI of 50. As expected, Ben-181-vacc upregulated host cell IRG-47 expression, whereas Ben-468-lowprot downregulated host cell IRG-47 expression compared to the mock (Fig. 2A), suggesting that IRG-47 upregulation may be linked to the Ben vaccine’s anti-infection immunoprotection. Given that Ben-468-lowprot lacks 35 genes compared to Ben-181-vacc28 and fails to induce host cell IRG-47, we hypothesized that one or more of these missing genes could upregulate IRG-47 expression. To test this point, the 35 genes were bioinformatically analyzed and functionally predicted, and several candidates were chosen to detect their actions on the IRG-47 protein expression. Eukaryotic plasmids of these candidate genes with a Flag or GFP tag were constructed to transfect EBL cells. Among them, only Ben_0691 increased IRG-47 protein level in EBL cells and was thus selected for subsequent studies (Fig. 2B, C and Supplementary Fig. 1A). In addition, we purified MBP-tagged Ben_0691 protein (Supplementary Fig. 1B), and incubated EBL cells with different concentrations (0.25, 0.5, or 1 μg/ml) of rBen_0691 for 12 and 24 h. Western blotting assay revealed that rBen_0691 incubation increased IRG-47 levels in EBL cells compared to the MBP-tag protein incubation (Fig. 2D, E), further confirming that Ben_0691 upregulates host cell IRG-47 protein levels. Unfortunately, we were unable to directly demonstrate Ben_0691-induced IRG-47 expression in vivo due to the lack of accurate and efficient mycoplasma genome editing tools29. We therefore purified a specific antibody against Ben_0691 from immunized rabbit serum, and used it to preincubate Mmm Ben-181-vacc for blocking its surface Ben_0691. Compared to the anti-IgG antibody treated Ben-181-vacc infection, this antibody-treated Ben-181-vacc showed an attenuated effect on the upregulation of IRG-47, confirming the role of Ben_0691 on Mmm cells surface in inducing host cell IRG-47 (Fig. 2F).
A EBL cells were incubated with Ben-181-vacc or Ben-468-lowprot at an MOI of 50 for 24 h, and then collected to assess IRG-47 protein levels by Western blotting analysis. The promotion of IRG-47 expression by Ben_0691 was confirmed by Western blotting of the IRG-47 levels in EBL cells transfected with Ben_0691-Flag/GFP or vector plasmid (B, C); or in EBL cells treated with rBen_0691 or MBP protein for 12 and 24 h (D, E). The Western blot images of β-actin, GFP and IRG-47 in panel C were also used in Fig. 4C. F Ben_0691 blockage attenuated the effect of Ben-181-vacc upregulation of host cell IRG-47. Mmm Ben-181-vacc was incubated with the purified antibodies against Ben_0691 (10, 20, and 50 μg/ml) or isotype IgG for 30 min and then added to EBL cells. The IRG-47 level in EBL cells was determined by Western blotting. G–J EBL cells were transfected with 0.5 μg Ben_0691-Flag/GFP or vector followed by incubation with Mmm (Ben-1 and Mu-1) at an MOI of 50 for 12 h. The cells were harvested for TaqMan qPCR to analyze the amount of Mmm infected to EBL cells (G, H), or cells were subjected for IFA using mouse anti-Mmm monoclonal antibody (green) to detect Mmm infection level to EBL cells. DAPI was used to stain cellular nuclei (blue) (I, J). All data are presented as the means ± SDs of the results from three independent experiments, and were normalized to the corresponding values in control cells. Significance was assessed by a two-tailed Student’s t test. ****p < 0.0001.
We further investigated the effect of Ben_0691 on Mmm infection. EBL cells were transfected with Ben_0691-GFP or pEGFP-C1, and then incubated with Mmm Ben-1 or Mu-1 at an MOI of 50 for 12 h. TaqMan qPCR assays showed a significant reduction in the number of Mmm infection in EBL cells transfected with Ben_0691-GFP plasmid, compared to the cells transfected with the control plasmid pEGFP-C1, suggesting that Ben_0691 overexpressing decreases Mmm infection to EBL cells (Fig. 2G, H). These findings were further verified by IFA analysis of Mmm signals on EBL cells transfected with Ben_0691-Flag or pCAGGS-Flag (Fig. 2I, J).
Overall, these results indicate that we successfully identified Mmm Ben_0691 from Ben vaccine candidate protective antigens, which upregulates IRG-47 protein level and thereby reduces Mmm infection in EBL cells.
Ben_0691-induced IRG-47 upregulation reduces early cell-associated Mmm
We next investigated whether Ben_0691 promotes host cell resistance against Mmm at the onset of infection. To this end, EBL cells were transfected with Ben_0691-GFP or pEGFP-C1, and the total number of Mmm associated with these cells was monitored during the early infection phases (30 min, 1 h, 2 h, 3 h, and 6 h post-infection). TaqMan qPCR analysis revealed a significant reduction of cell-associated Mmm in overexpressing Ben_0691 cells compared to those overexpressing GFP (Fig. 3A). This finding was corroborated by a decrease in the Mmm green fluorescent signal detected via IFA (Fig. 3B). Notably, this reduction was already evident at 30 min post-infection, a time point at which mycoplasma replication and cell entry are negligible30,31,32,33, implying that Ben_0691 reduces early pool of cell-associated Mmm, likely by disturbing bacterial adherence.
A, B EBL cells were transfected with Ben_0691-Flag/GFP or vector, followed by incubation with Mmm Ben-1 for 30 min, 1 h, 2 h, 3 h, and 6 h. The cells were harvested for TaqMan qPCR to determin the number of Mmm adhering to EBL cells (A), or cells were subjected for IFA using mouse anti-Mmm monoclonal antibody (green) to detect cell-associated Mmm level. DAPI was used to stain cellular nuclei (blue). The data were normalized to the corresponding values in control cells (B). C The positional changes of IRG-47 (Red) were observed by laser confocal microscopy in EBL cells transfected with vector or Ben_0691-GFP. Cell nuclei were stained with DAPI (blue). Scale bar 5 μm. D, E The number of Mmm adhering to vector- or IRG-47-Myc-transfected EBL cells was measured TaqMan qPCR or IFA. Data are presented as the means ± SDs from five independent experiments, and significance was assessed by a two-tailed Student’s t test. *p < 0.05; ***p < 0.001; ****p < 0.0001.
IRG-47 is a protein localized on the endoplasmic reticulum (ER) membrane34. To determine whether Ben_0691 interferes with Mmm adherence through altering IRG-47 localization, we utilized laser confocal microscopy to analyze IRG-47 subcellular distribution in EBL cells transfected with Ben_0691-GFP or pEGFP-C1. As illustrated by the red fluorescence in Fig. 3C, Ben_0691 did not affect IRG-47 localization, suggesting that its disturbance of Mmm adherence may be related to upregulation of IRG-47 protein levels rather than relocalization. To confirm this, we assessed the number of cell-associated Mmm in EBL cells overexpressing IRG-47. It was observed that increased IRG-47 expression significantly reduced cell-associated Mmm levels compared to control plasmid-transfected cells (Fig. 3D, E).
Altogether, these results suggest that Ben_0691 decreases early cell-associated Mmm by upregulating IRG-47 expression, thereby enhancing host cell resistance to Mmm infection.
ATP5A1 is essential for Ben_0691 to upregulate host cell IRG-47
To gain a deeper understanding of how Ben_0691 upregulates host cell IRG-47, we performed pull-down assays to identify the key host molecules involved. Magnetic beads bound to rBen_0691 or MBP protein were incubated with EBL cell lysate, and the bound host proteins were eluted, separated by SDS-PAGE and then silver-stained (Fig. 4A). Specific protein bands pulled down by rBen_0691 were analyzed by mass spectrometry (MS). Thirty-three host proteins were identified, of which the MS data showed 11 peptide matches to ATP synthase subunit alpha (ATP5A1), with approximately 28% sequence coverage (Fig. 4B).
A Screening for the host proteins interacting with Ben_0691 by the MBP pull-down assay. 100 μg MBP or rBen_0691 protein was conjugated to anti-MBP magnetic beads, followed by incubation with EBL whole cell lysates for 4 h. The protein complexes binding to the beads were eluted, separated by SDS-PAGE and silver-stained. B The amino acid sequence of bovine ATP5A1 is shown and matching peptides detected in MS analysis was highlighted in red. C The enhancement of ATP5A1 and IRG-47 expression by Ben_0691 were confirmed by Western blotting of the ATP5A1, IRG-47, and β-actin levels in EBL cells transfected with vector or Ben_0691-GFP. D Transfection of the ATP5A1-Myc plasmid in EBL cells enhanced IRG-47 expression. E EBL cells were transfected with Ben_0691-GFP or vector for 12 h, and then transfected with siNC or siATP5A1-1 for 48 h. Cell lysates were collected to assay the levels of IRG-47, ATP5A1, and β-actin by Western blotting. The Western blot images of β-actin, GFP, IRG-47, and ATP5A1 in (E) were also used in Fig. 8F. F EBL cells were transfected with vector or Ben_0691-Flag for 48 h, and harvested for qPCR to detect ATP5A1 mRNA level. The data are represented the means ± SDs from three independent experiments, and were normalized to the corresponding values in control cells. Significance was assessed by one-way ANOVA with Dunnett’s multiple comparison tests relative to the control. *p < 0.05. G EBL cells expressing Ben_0691-GFP or GFP (pEGFP-C1) were treated with the protein synthesis inhibitor cycloheximide (CHX, 100 μg/ml) for 12 h. ATP5A1 and β-actin levels were determined by Western blotting analysis. Densitometry analysis of ATP5A1 levels relative to actin (ATP5A1/actin) normalized to no CHX treatment. H EBL cells expressing Ben_0691-GFP or GFP were incubated with the autophagy inhibitor 3-MA (100 μM, 12 h), and then subjected to analyze ATP5A1 and β-actin levels by Western blotting.
ATP5A1 is the alpha subunit of the soluble F1 catalytic core of mitochondrial ATP synthase that catalyzes ATP synthesis, and is essential for normal mitochondrial function and metabolism35. To assess whether ATP5A1 mediates the effect of Ben_0691 on IRG-47 upregulation, we transfected EBL cells with the Ben_0691-GFP plasmid and detected an enhanced ATP5A1 expression (Fig. 4C). ATP5A1 overexpression by ATP5A1-Myc transfection also led to elevated IRG-47 expression (Fig. 4D). These data indicate that Ben_0691 interacts with ATP5A1 and promotes its expression, which in turn upregulates IRG-47 protein level. Moreover, we constructed three specific siRNAs (siATP5A1-1, 2 and 3) targeting bovine ATP5A1, and all three siRNAs significantly reduced ATP5A1 expression in EBL cells (Supplementary Fig. 2A). CCK-8 detection showed that ATP5A1 knockdown or overexpressing did not impaired cell viability (Supplementary Fig. 2B). We observed that IRG-47 protein levels were significantly increased in Ben_0691-overexpressing EBL cells (siNC + Ben_0691-GFP) compared to control cells (siNC + pEGFP-C1). However, using siATP5A1-1 to knockdown ATP5A1 in Ben_0691-overexpressing cells (siATP5A1-1 + Ben_0691-GFP), the upregulation of IRG-47 was almost eliminated (Fig. 4E), suggesting that ATP5A1 is crucial for Ben_0691 to upregulate host cell IRG-47.
Ben_0691 stabilizes the ATP5A1 protein
To investigate the mechanism by which Ben_0691 increases ATP5A1 protein levels, we first examined whether it regulates ATP5A1 mRNA transcription. As shown in Fig. 4F, ATP5A1 mRNA levels remained unchanged in EBL cells transfected with Ben_0691-Flag, indicating that the increase in ATP5A1 protein levels is not due to transcriptional upregulation. We hence speculated whether Ben_0691 affects ATP5A1 protein stability through its interaction with ATP5A1. We subsequently monitored ATP5A1 protein decay in EBL cells transfected with the control vector pEGFP-C1 or Ben_0691-GFP, upon the addition of the protein synthesis inhibitor cycloheximide (CHX, 100 μg/ml). The data revealed that ATP5A1 levels decreased by 29% in the presence of Ben_0691, whereas a reduction of 63% occurred in its absence, suggesting that Ben_0691 stabilizes the ATP5A1 protein by preventing its degradation (Fig. 4G).
The ubiquitin-proteasome system and autophagic-lysosomal system are the two major intracellular protein degradation pathways36. To investigate which pathway is involved in Ben_0691-mediated ATP5A1 upregulation, we treated EBL cells expressing Ben_0691-GFP or pEGFP-C1 with the proteasome inhibitor MG132 or the autophagy inhibitor 3-MA. Western blotting analysis showed that ATP5A1 levels increased in GFP-expressing cells treated with 3-MA compared to untreated control cells, indicating that ATP5A1 is degraded through the autophagic-lysosomal pathway. In contrast, 3-MA treatment had no effect on ATP5A1 levels in Ben_0691-GFP-expressing cells, demonstrating that Ben_0691 itself inhibits ATP5A1 autophagic-degradation, rendering 3-MA ineffective (Fig. 4H). Additionally, MG132 treatment did not alter ATP5A1 levels in either GFP- or Ben_0691-GFP-expressing cells (Supplementary Fig. 2C). These results suggest that Ben_0691 stabilizes ATP5A1 by reducing its autophagic degradation, thereby enhancing its protein levels.
Ben_0691 directly interacts with host ATP5A1
To further confirm the specific interaction of Ben_0691 with host ATP5A1, we performed a coimmunoprecipitation (Co-IP) analysis. HeLa cells were co-transfected with Ben_0691-GFP and ATP5A1-Myc together or alone, and reciprocal interactions were observed using either magnetic beads conjugated with anti-GFP or anti-Myc antibodies to conduct IP (Fig. 5A, B). Confocal microscopy was also employed and showed the colocalization of Ben_0691 with ATP5A1 in the cytoplasm of HeLa cells (Fig. 5C). These data indicate that Mmm Ben_0691 interacts with host ATP5A1.
A, B Ben_0691-GFP coimmunoprecipitated with ATP5A1-Myc. Hela cells were transfected with Ben_0691-GFP or ATP5A1-Myc, or the control plasmids pEGFP-C1 or pCAGGS-Myc for 36 h. Cell lysates were collected for Co-IP with beads conjugated with anti-Myc antibody (A) and GFP antibody (B). C Colocalization of the Ben_0691 and ATP5A1 proteins. D EBL cells were incubated with Mmm Ben-18-vacc, and the interaction of endogenous ATP5A1 and Ben_0691 protein in the cell lysates was detected by coimmunoprecipitation with antibody against ATP5A1 proteins rather than IgG and protein A-conjugated beads. E EBL cells were incubated with anti-ATP5A1 or isotype IgG antibody followed by incubation with an Alexa Fluor 488-labeled goat anti-rabbit secondary antibody. Flow cytometry analysis of 10,000 cells from each sample to examine ATP5A1 expression on the EBL cells surface. F Far-Western blotting analysis showed the direct binding of ATP5A1 with the Ben_0691 protein. The proteins rBen_0691 and MBP (negative control) were transferred onto nitrocellulose (NC) membranes. Left: The NC membrane was incubated with anti-MBP antibodies; Middle: The NC membrane was first incubated with the ATP5A1-His protein and then with anti-ATP5A1 antibodies; Right: The NC membrane was incubated with the His-tag protein and then with anti-His antibodies.
In addition, we asked whether endogenous ATP5A1 interacts with Ben_0691, we incubated EBL cells with Ben-181-vacc (MOI = 50, 12 h) and conducted a Co-IP assay with these cell lysates using magnetic beads conjugated with antibodies to IgG or ATP5A1 protein. As shown in Fig. 5D, endogenous ATP5A1 was found to interact with Ben_0691 protein during Ben-181-vacc incubation of EBL cells.
ATP5A1 is primarily localized in mitochondria, but it has also been reported to localized on many cell membranes37. We then used flow cytometry to detect whether ATP5A1 is expressed on the EBL cell surface. As illustrated in Fig. 5E, ATP5A1 was indeed detected on the surface of EBL cells. Based on these findings, we hypothesized that the interaction between Ben_0691 and ATP5A1 is likely to be physically direct. To verify this, we performed a Far-Western blotting analysis on rBen_0691-immobilized membranes using commercially available His-tagged recombinant human ATP5A1 protein. The direct interaction was confirmed by observing the ATP5A1 band at the rBen_0691 position (Fig. 5F).
Collectively, these findings suggest that the ATP5A1 protein is an interacting partner of the Mmm Ben_0691 protein.
Ben_0691 N-terminal 66 to 72 aa region is responsible for the interaction with the host ATP5A1 C-terminal domain
To identify the key amino acids (aa) region in Ben_0691 responsible for its interaction with ATP5A1, we constructed truncated Ben_0691 plasmids with a GFP tag at their N-terminus: Ben_0691-P1-GFP (aa 1–52), Ben_0691-P2-GFP (aa 53–103) and Ben_0691-P3-GFP (aa 104–154). Each construct was co-transfected with ATP5A1-Myc into HeLa cells, and cell lysates were analyzed by Co-IP using magnetic beads conjugated with anti-Myc antibodies. All three truncated Ben_0691 constructs were well expressed (Fig. 6A), and only Ben_0691-P2-GFP was immunoprecipitated with ATP5A1 protein. A reverse Co-IP analysis using magnetic beads conjugated anti-GFP antibodies also confirmed that ATP5A1 only interacted with the Ben_0691-P2-GFP protein (Fig. 6B).
A The fragment of Ben_0691 aa 53–103 interacted with the ATP5A1 protein. Upper: Schematic of the Ben_0691 truncates tagged with GFP and locations of the aa residues are noted. Lower: Hela cells were co-transfected with ATP5A1-Myc and truncated Ben_0691 constructs, and the cell lysates were collected for Co-IP assays using magnetic beads conjugated with anti-Myc antibody. B Hela cells were treated as described in (A) and harvested for Co-IP assays using magnetic beads conjugated with anti-GFP antibody. C Ben_0691-GFP plasmids mutated aa 63–72 did not immunoprecipitate with ATP5A1. Upper: Schematic of the Ben_0691 mutation tagged with GFP and locations of the aa residues are noted. Lower: Hela cells were co-transfected with ATP5A1-Myc and five Ben_0691-P2-GFP mutants. Cell lysates were collected for Co-IP assay using magnetic beads conjugated with anti-Myc antibody. D Ben_0691 plasmid mutated aa 66–68 or 69–72 did not immunoprecipitate with the ATP5A1. Hela cells were co-transfected with ATP5A1-Myc and the four Ben_0691-P2-GFP mutation constructs, and then collected to perform a Co-IP assay. E The fragment of ATP5A1 aa 422–553 interacted with the Ben_0691 protein. Upper: Schematic of the ATP5A1 truncates tagged with Myc and locations of the aa residues are noted. Lower: Hela cells were co-transfected with Ben_0691-GFP and truncated ATP5A1 constructs, and the cell lysates were collected for Co-IP assays using magnetic beads conjugated with anti-Myc antibody.
To further map the specific interaction site, we mutated 10 or 11 consecutive amino acids of Ben_0691-GFP-P2 to alanine, respectively. Five mutant constructions were generated: Ben_0691-P2-GFP M1 (aa 53–62), M2 (aa 63–72), M3 (aa 73–82), M4 (aa 83–92) and M5 (aa 93–103), and each was subjected to Co-IP with ATP5A1. It was shown that only M2 mutant abolished the interaction with ATP5A1 (Fig. 6C), suggesting that the aa 63–72 region of Ben_0691 is responsible for the interaction with ATP5A1. Within this critical region aa 63 to 72, we further introduced alanine substitutions of 3 or 4 consecutive amino acids to construct three mutant plasmids: Ben_0691-P2-GFP M2 (aa 63–65), Ben_0691-P2-GFP M2 (aa 66–68), and Ben_0691-P2-GFP M2 (aa 69–72). Co-IP assays showed that mutations in the aa 66–68 or 69–72 regions disrupted the interaction (Fig. 6D), suggesting that amino acids at positions 66 to 72 in the Ben_0691 N-terminal region are critical for its interaction with host ATP5A1.
The bovine ATP5A1 protein consists of 553 aa residues, including an N-terminal domain (aa 69–135), a nucleotide-binding domain (NBD, aa 195–415) and a C-terminal domain (aa 422–547)38. We also investigated the key structural domain in ATP5A1 necessary for the interaction with Ben_0691. We constructed four ATP5A1 truncates, and found that the D3 and D4 truncates interacted with Ben_0691, whereas the D1 and D2 truncates did not (Fig. 6E), suggesting that the C-terminal domain (aa 422–553) of ATP5A1 is indispensable for its interaction with Ben_0691.
Together, these findings demonstrate that the Ben_0691 N-terminal aa 66–72 region targeting interacts with the aa 422–553 region of ATP5A1.
Interaction with ATP5A1 is required for Ben_0691 to upregulate IRG-47 and thereby exert anti-infective effects
We further analyzed whether Ben_0691 upregulates host IRG-47 by interacting with ATP5A1. Transfection of the Ben_0691-GFP plasmid was determined to promote both ATP5A1 and IRG-47 expressions (Fig. 7A). Subsequently, we constructed a mutant plasmid Ben_0691-M(66–68)-GFP, in which the three key amino acid sites 66 to 68 responsible for interacting with ATP5A1 were mutated to alanine. It was observed that this mutant neither increased the expression of ATP5A1 nor IRG-47, compared to the cells transfected with pEGFP-C1 (Fig. 7A), indicating that the loss of interaction with ATP5A1 abolishes Ben_0691-mediated IRG-47 upregulation. In addition, TaqMan qPCR and IFA assays revealed that Ben_0691-GFP/Flag transfection significantly reduced Mmm infection in EBL cells, compared to the control plasmid transfection. However, the Mmm infection level in EBL cells transfected with the mutant Ben_0691-GFP/Flag plasmid was comparable to those in control plasmid-transfected cells (Fig. 7B–E). These results demonstrate that the interaction with host ATP5A1 is necessary for Ben_0691 to upregulate IRG-47 and thus exert anti-Mmm action.
A Ben_0691 did not upregulate host cell ATP5A1 and IRG-47 levels when it lost its binding to ATP5A1. B–E EBL cells were transfected with pEGFP-C1, Ben_0691-M(66–68)-GFP or Ben_0691-GFP, followed by exposure to Mmm (Ben-1 and Mu-1) at an MOI of 50 for 12 h. The cells were harvested for TaqMan qPCR to analyze the amount of Mmm infected to EBL cells (B, C), or cells were collected for IFA using mouse anti-Mmm monoclonal antibody (green) to detect Mmm infection level to EBL cells (D, E). The data represented the means ± SDs from three independent experiments and were normalized to the corresponding values in control cells. Significance was assessed by one-way ANOVA with Tukey’s multiple comparison test. ***p < 0.001.
Ben_0691 upregulates host cell IRG-47 through the ATP5A1-AMPK-mTOR-IFN-γ pathway
Previous studies have shown that ATP5A1 affects the adenosine monophosphate-activated protein kinase (AMPK)-mTOR signaling pathway39, which may regulate the IRG-47 expression. To test this possibility, we transfected EBL cells with siATP5A1-1 to knockdown ATP5A1, or with ATP5A1-Myc to overexpress it. ATP5A1 knockdown enhances AMPK phosphorylation (P-AMPK/AMPK) and cascaded to reduce mTOR phosphorylation level (P-mTOR/mTOR). Conversely, ATP5A1 overexpression decreased AMPK phosphorylation while increasing mTOR phosphorylation in EBL cells. These data suggest that ATP5A1 activates the AMPK-mTOR signaling pathway and upregulates IRG-47 expression (Fig. 8A). We next incubated EBL cells with the AMPK activator Acadesine (AICAR) or the mTOR inhibitor Rapamycin (RAPA) for 12 h to interfere with the activation of the AMPK-mTOR signaling pathway (Fig. 8B). CCK-8 detection confirmed that the selected concentrations of AICAR and RAPA did not affect EBL cell viability (Fig. 8C, D). As shown in Fig. 8E, F, ATP5A1 overexpression no longer upregulated IRG-47 protein levels in AICAR- or RAPA-treated EBL cells, implying that ATP5A1 promotes IRG-47 protein expression by activating the AMPK-mTOR signaling pathway.
A EBL cells were transfected with pCAGGS-Myc or ATP5A1-Myc for 12 h, and then transfected with siNC or siATP5A1-1 for 48 h. Cell lysates were collected to analyze the levels of P-AMPK, AMPK, P-mTOR, mTOR, IRG-47, ATP5A1 and β-actin by Western blotting. B EBL cells were incubated with AICAR (100 μM) or RAPA (1 μg/ml) for 12 h, and harvested for Western blotting to analyze the phosphorylation levels of AMPK (P-AMPK/AMPK,) or mTOR (P-mTOR/mTOR). C, D The selected concentrations of AICAR or RAPA had no effect on cell viability in EBL. E, F EBL cells were transfected with pCAGGS-Myc or ATP5A1-Myc for 36 h, and then incubated with AICAR or RAPA for 12 h. Cell lysates were collected for Western blotting to assay the levels of P-AMPK, AMPK, P-mTOR, mTOR, IRG-47, ATP5A1, and β-actin. G EBL cells were transfected with pEGFP-C1 or Ben_0691-GFP for 12 h, and then transfected with siNC or siATP5A1-1 for 48 h. Cell lysates were subjected for Western blotting of the levels of P-AMPK, AMPK, P-mTOR, mTOR, IRG-47, ATP5A1 and β-actin. H EBL cells were transfected with pCAGGS-Myc or ATP5A1-Myc, followed by incubation with or without AICAR or RAPA. The cells were harvested for qPCR to detect IFN-γ mRNA level. All data are presented as the means ± SDs and were normalized to the corresponding values in control cells. Significance was assessed by a two-tailed Student’s t test (C, D, n = 6 independent experiments) or by one-way ANOVA with Tukey’s multiple comparison test (H, n = 3 independent experiments). *p < 0.05.
We further validated this above finding in EBL cells overexpressing Ben_0691. We found that increased Ben_0691 level significantly promoted ATP5A1 expression, which activated the AMPK-mTOR signaling pathway and thereby upregulating IRG-47 expression. However, this effect was attenuated upon ATP5A1 knockdown using siATP5A1-1 (Fig. 8G), suggesting that Ben_0691 upregulated ATP5A1 to activate the AMPK-mTOR signaling pathway, which in turn promotes IRG-47 expression.
IRG-47 is an IFN-γ-inducible protein. To examine the relationship between the ATP5A1/AMPK/mTOR axis and IFN-γ induction, we measured IFN-γ mRNA levels in ATP5A1-overexpressing cells. IFN-γ mRNA levels were significantly elevated in these cells compared to control plasmid-transfected cells (Fig. 8H). However, this upregulation was abolished when the AMPK/mTOR pathway was inhibited with AICAR or RAPA (Fig. 8H), suggesting that the ATP5A1/AMPK/mTOR axis functions upstream to induce IFN-γ production, and thereby promoting IRG-47 expression.
Summarily, these results indicate that Ben_0691-induced IRG-47 expression relies on the ATP5A1-AMPK-mTOR-IFN-γ signaling pathway.
Discussion
Interferon-γ (IFN-γ) is a crucial cytokine for controlling infections and regulating the immune system40. Animals with disrupted IFN-γ or its receptor gene are highly susceptible to infectious diseases and die rapidly41. IFN-γ primarily functions by activating IFN-γ-responsive genes42. Among these, GTPase family proteins are notable for their anti-pathogen infection effects43. Current evidence supports that the 47 KDa immunity-related GTPase (IRG) family proteins are typically induced during infection and mediate resistance to pathogens infection, especially intracellular bacterial and protozoan infections44. As demonstrated in multiple systems: (a) IRGM1 (LRG-47) mediates macrophage resistance to Mycobacterium tuberculosis infection16; (b) IRGM3 (IGTP) provides resistance to early liver infection by Leishmania donovani45; and (c) IRGB10 or IRGM3 protein mediates embryonic fibroblasts resistance to Chlamydia trachomatis infection20. In most experimental systems, IRG proteins induction strongly correlates with host resistance to intracellular bacterial and protozoan infections41. However, the role of IRG proteins in extracellular or facultative intracellularly bacterial infections is currently unknown. In this study, we demonstrate for the first time that increased IRG-47 protein significantly reduces the infection of bovine lung epithelial cells by the predominantly extracellularly resident Mmm, thus mediating host cell resistance to Mmm infection. Notably, our findings suggest that Ben-181-vacc specifically upregulates host cell IRG-47, which may be closely related to its immunoprotection against Mmm infection.
GTPase family proteins exhibit pathogen-specific resistance to infection46,47,48,49. For instance, IRGM3 mediates a significant host resistance to Toxoplasma gondii but not Listeria monocytogenes infection50. While IRGM3- and LRG-47-deficient mice are killed rapidly during the acute phase of Salmonella typhimurium infection, IRG-47-deficient mice show only a partial decrease in their resistance51. Consistent with these findings, IRG-47 also demonstrates pathogen-specific resistance to infection. Just as IRG-47 protected the host cell against Mmm infection in our study, but a previous study indicated it only inhibited inflammation triggered by Schistosoma japonicum infection without conferring resistance to the infection52.
Ben-181-vacc specifically upregulated host cell IRG-47 expression, whereas Ben-468-lowprot did not. We reasoned that one or more of the 35 antigens lost in Ben-468-lowprot may specifically elevate IRG-47, which in turn mediates host cell resistance to Mmm infection. These antigens are therefore likely to be protective. Considering the limitations of currently available attenuated Mmm vaccines, a deeper understanding Mmm protective antigens and their immunoprotection mechanism is crucial for improving existing vaccines or developing novel subunit vaccines. In this study, we identified Ben_0691 as a protective antigen that specifically upregulates IRG-47 expression to reduce Mmm infection. Further analysis showed that increased IRG-47 protein levels diminished early cell-associated Mmm, likely by disturbing bacterial adherence, resulting in an anti-Mmm infection effect. Notably, our results support that IRG-47 protein level was associated with its resistance to Mmm infection, although the exact mechanism warrants further investigation.
The amino acid sequence of Ben_0691 was analyzed against the NCBI non-redundant (nr) database using BLASTp, it was annotated as an ABC three-component system protein. Across the full-length sequence, Ben_0691 displays high sequence identity (≥89%) with the ABC three-component system protein from other members of the Mycoplasma mycoides cluster, indicating that it is highly conserved within this cluster. Outside the cluster, Ben_0691 shares moderate identity with the corresponding proteins in M. feriruminatoris (~65% identity, 100% coverage), M. agalactiae (~53% identity, 95% coverage) and M. yeatsii (~39% identity, 94% coverage). InterPro analysis further revealed the presence of a conserved C-terminal domain 10 (CTD10) of ABC three-component systems53, supporting that Ben_0691 retains conserved structural features among these Mycoplasma species.
ATP5A1 is one of the ATP synthase F1 subunits54. ATP synthase is a ubiquitous multimeric protein complex crucial for cellular ATP production. While ATP synthase is generally embedded in the inner mitochondrial membrane, its various subunits have been shown to be localized to the plasma membrane of many cell lines, including endothelial cells, adipocytes, keratinocytes, hepatocytes, and certain cancer cells37. In our study, ATP5A1 was also found on the membrane surface of bovine lung epithelial cells. Ben_0691 interacted directly with the membrane localized ATP5A1, and this interaction was necessary for Ben_0691 to upregulate IRG-47, thereby resisting Mmm infection. Moreover, many studies have documented the critical role of ATP5A1 in mitochondrial health. For instance, Toxoplasma gondii GRA8 induces ATP5A1-SIRT3 to mediate host mitochondrial metabolic resuscitation38, and emodin ameliorates mitochondrial dysfunction by upregulating ATP5A155. We speculate that Ben_0691-enhanced ATP5A1 expression may also improve mitochondrial health and contribute to anti-Mmm infection response.
One limitation of our study is the inability to perform animal experiments to validate our findings. Specifically, research involving live Mmm cells in cattle require containment in a Biosafety Level 4 (P4) laboratory. Unfortunately, our application for approval from the Ministry of Agriculture and Rural Affairs was not granted, preventing us from conducting these crucial validations. We aim to conduct these in vivo experiments in future studies pending regulatory approval.
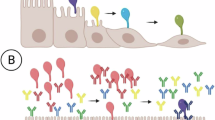
In conclusion, our study reveals the critical role of IRG family proteins in resisting Mmm infection, and further proposes a molecular mechanism model by which the Mmm potential immunoprotective protein Ben_0691 utilizes IRG family proteins to mediate this resistance (Fig. 9). We hope this study will provide a theoretical foundation for improving or developing vaccines against Mmm infections, and may lead to the development of new therapies for Mmm-related disorders.
Model depicting the inhibition of Mmm epithelial infection by Ben_0691 promoting IRG-47 expression (the figure was drawn by Figdraw, www.figdraw.com, ID:YYTYUdd2a2). The vaccine strain Ben-181-vacc attaches to host cells, and its membrane protein Ben_0691 binds to host ATP5A1, which triggers the AMPK-mTOR-IFN-γ signaling pathway cascade, promoting IRG-47 expression to inhibit Mmm infection.
Methods
Mycoplasma strains, cells, and culture conditions
Mmm strains Ben-1 and Mu-1 were isolated from the lung of a diseased cattle with pneumonia in Benxi and Inner Mongolia of China, respectively56, and maintained in our laboratory. The strains were grown in PPLO medium (Basal Media, Shanghai, China) and used at low passage number (<10). To estimate the number of colony-forming unit (CFU) in the cultures, serial dilutions were plated on a modified PPLO medium containing 1.5% agarose (V2111; Promega) and incubated at 37 °C. CFU was counted 7–10 days later using a microscope57. Embryonic bovine lung epithelial (EBL) cells were kindly provided by Prof. Fei Xue of State Key Laboratory for Animal Disease Control and Prevention, Harbin Veterinary Research Institute (HVRI), Chinese Academy of Agricultural Sciences (CAAS), Harbin, China. HeLa cells were purchased from the American Type Culture Collection (ATCC). These cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco), 100 μg/ml streptomycin (Gibco), 100 U/ml penicillin (Gibco), and 10 mM HEPES (Invitrogen). The cells were incubated at 37 °C in 5% CO2.
For infection experiments, EBL cells were maintained in DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin, inoculated with Mmm cells at a multiplicity of infection (MOI) of 50. All experiments with infectious Mmm were performed in the biosafety level 4 and animal biosafety level 4 facilities in the HVRI of CAAS.
Plasmids or siRNAs construction and transfection
To construct eukaryotic expression plasmids for Ben_0691 or ATP5A1, the nucleotide sequence encoding the Ben_0691 from Mmm Ben-1 strain (CP011260.1) was codon-optimized and synthesized by Beijing Genomics Institute (BGI, Beijing, China). The Ben_0691 nucleotide sequence was cloned into the pCAGGS-Flag or pEGFP-C1 vector with the restriction enzyme Sal I or EcoR I using a ClonExpress II One Step Cloning Kit (Vayzme, Nanjing, China). The bovine ATP5A1 gene (NM_174684.2) was codon-optimized and synthesized by the BGI and cloned into the pCAGGS-Myc vector with the restriction enzyme EcoR I and Kpn I using a ClonExpress II One Step Cloning Kit (Vayzme).
For the identification of the key amino acid sites in Ben_0691 that interact with the ATP5A1, three truncated Ben_0691 constructs based on the plasmid pEGFP-C1 (expressing the aa 1–52, 53–103 and 104–154 of Ben_0691, named Ben_0691-P1-GFP, Ben_0691-P2-GFP and Ben_0691-P3-GFP, respectively), and the mutated constructs based on the plasmid Ben_0691-P2-GFP (with the aa 53–62, 63–72, 73–82, 83–92, 93–103, 63–65, 66–68 or 69–72 mutated) were synthesized by Genesoul Technology (Harbin, China). To verify whether the interaction with ATP5A1 is necessary for Ben_0691 to upregulate IRG-47, the mutated construct based on Ben_0691-GFP/Flag with the aa 66–68 mutated was synthesized by Genesoul Technology. To determine the key domain in ATP5A1 that interacts with Ben_0691, the four truncated ATP5A1 constructs based on the plasmid pCAGGS-Myc (expressing the aa 1–194, 1–421, 195–553, and 422–553 of ATP5A1, named ATP5A1-D1-Myc, ATP5A1-D2-Myc, ATP5A1-D3-Myc and ATP5A1-D4-Myc, respectively) were synthesized by Genesoul Technology.
To construct the eukaryotic expression plasmid for IRG-47, the bovine IRG-47 gene (NM_001034545.2) was PCR amplified from EBL cell genome and cloned into the pCAGGS-Myc vector with the restriction enzyme EcoR I using a ClonExpress II One Step Cloning Kit (Vayzme). To knockdown IRG-47 or ATP5A1 in EBL cells, siIRG-47-1 (sense strand: 5’-CCAAAUUUGUCUGAAUCUUdTdT-3’ and antisense strand: 5’-AAGAUUCAGACAAAUUUGGdTdT-3’), siIRG-47-2 (sense strand: 5’-GAAACAGUAUCAGAGCCAUdTdT-3’ and antisense strand: 5’- AUGGCUCUGAUACUGUUUCdTdT-3’), siIRG-47-3 (sense strand: 5’-GUGGAUAGUGAUUUAUAUAdTdT-3’ and antisense strand: 5’-UAUAUAAAUCACUAUCCACdTdT-3’), siATP5A1-1 (sense strand: 5’-CAGUAUUCUCCCAUGGCUAdTdT-3’ and antisense strand: 5’-UAGCCAUGGGAGAAUACUGdTdT-3’), siATP5A1-2 (sense strand: 5’-GUUUAAAGGGUAUGUCUUUdTdT-3’ and antisense strand: 5’-AAAGACAUACCCUUUAAACdTdT-3’) and siATP5A1-3 (sense strand: 5’-GCAAUUGACACAAUCAUUAdTdT-3’, and antisense strand: 5’-UAAUGAUUGUGUCAAUUGCdT dT-3’) were synthesized by Sigma-Aldrich.
PolyJet DNA in vitro transfection reagent (SignaGen, USA) was used to transfect recombinant plasmids into EBL or Hela cells, and Lipofectamine 3000 Transfection Reagent (lipo3000, Invitrogen, USA) was used to transfect siRNAs into EBL cells.
Western blotting
To detect the expression level of the target protein, cells were harvested at the indicated time points. An equal number of cells were lysed in RIPA buffer with protease inhibitors (Roche, 4693159001) for 30 min at 4 °C, and the protein concentration was determined using a BCA Protein Assay Kit (Beyotime, Shanghai, China). The protein lysates were mixed with 1 × SDS loading buffer (Biosharp, Wuhan, China) and denatured at 100 °C for 10 min. Equal amounts of total cell lysates were separated by SDS-PAGE. The proteins in the gel were transferred onto nitrocellulose membranes (Pall Corporation, USA), which were then blocked with 5% cold-water fish skin gelatin in TBST (Solarbio, Beijing, China) at 4 °C overnight and then incubated for 2 h with different primary antibodies at room temperature (RT). A summary of antibodies used in this study is listed in Supplementary Table 2. After washing with TBST three times (10 min each), DyLight 800-labeled goat anti-mouse or -rabbit IgG (H + L) (1:10,000, Kirkegaard & Perry Laboratories, Gaithersburg, USA) was used for detection. The membrane was scanned using an Odyssey infrared imaging system, and the fluorescence intensity of each band was measured using Odyssey 2.1 software (LI-COR Biosciences).
Cell viability measurements
Cell viability was detected using a Cell Counting Kit-8 (CCK-8) according to the manufacturer’s protocol (Vazyme, Nanjing, China). EBL cells were seeded at a density of 2 × 103 cells per well in 96-well plates and subjected to different treatments. After incubation at 37 °C for the indicated time, 10 μl of CCK-8 solution was added to each well and incubated for another 2 h. Cell viability was determined by measuring the absorbance at 450 nm via a microplate reader (BioTek).
Real-time quantitative RT-PCR and TaqMan probe assay
To measure ATP5A1 or IFN-γ mRNA levels in different treatment EBL cells, total RNA was extracted from the cells using a RNeasy Mini kit (Qiagen Sciences, Hilden, Germany) according to the manufacturer’s instructions. RNA was reverse transcribed using a Transcriptor First-Strand cDNA Synthesis Kit (Roche Diagnostics, Indianapolis, USA). qPCR was performed in triplicate using FastStart Universal SYBR Green Master Mix (Rox) (Roche Diagnostics, Indianapolis, USA). All data were acquired using a QuantStudio 5 real-time PCR system (Applied Biosystems, Carlsbad, USA). The expression value of each gene was normalized to that of GAPDH, and final values were calculated using the ΔΔCt method. The results were analyzed using QuantStudio Design & Analysis software v1.4 (Applied Biosystems). The ATP5A1, IFN-γ or GAPDH primer sequences used in this study are provided in Supplementary Table 3.
The level of cell-associated mycoplasmas in Mmm-infected EBL cells were determined by using a specific TaqMan probe assay: the forward primer (F: 5’-AACCAGATGAATCAAAACAACCA-3’), reverse primer (R: 5’-CATTGGTTGGTCTGATTGTGGA-3’), and TaqMan probe (5’-VIC-ACCCAGAAATTAAGCCTGATCC-BHQ1-3’). This method was developed and validated in our laboratory. Briefly, EBL cells were transfected with pCAGGS-Myc, pEGFP-C1, IRG-47-Myc, Ben_0691-GFP, Ben_0691-M(66–68)-GFP or siRNAs (siNC, siIRG-47-1) and then infected with Mmm at an MOI of 50. EBL cells were washed twice with PBS to remove unadhered Mmm following exposure to Mmm for 12 h, and the cells along with Mmm were harvested using a cell scraper. The genomic DNA of cell-associated Mmm was extracted and its copy number was quantitated by TaqMan qPCR. The composition of the TaqMan qPCR assay mixture was according to the manufacturer’s protocol (Premix Ex Taq (Probe qPCR, TaKaRa, China). Amplification was performed using a QuantStudio 5 real-time PCR system. Each sample was assayed three times.
Immunofluorescence
To assess Mmm infection levels in different treated EBL cells, EBL cells were transfected with vectors pCAGGS-Myc/Flag, IRG-47-Myc, Ben_0691-GFP/Flag, Ben_0691-M(66–68)-Flag or siRNAs (siNC, siIRG-47-1) and then infected with Mmm at an MOI of 50. These cells were rinsed thrice with PBS to remove unadhered Mmm and fixed in 4% paraformaldehyde (PFA) for 30 min. The fixed cells were permeabilized with 0.1% Triton X-100 for 15 min and blocked for 1 h in 5% cold-water fish skin gelatin. Cells were then incubated with mouse anti-Mmm monoclonal antibodies 1D9 (produced in our laboratory) for 1 h at RT, washed thrice with PBS and incubated with goat anti-mouse secondary antibodies coupled to Alexa Fluor 488 (Invitrogen) for 1 h. The cell nuclei were labeled with DAPI (Sigma-Aldrich). Fluorescence images were acquired using a fluorescence microscope (EVOS FL Auto, Invitrogen).
Protein expression and purification
To generate the fusion protein rBen_0691, the Ben_0691 nucleotide sequence was cloned into the pMAL-c5X (New England Biolabs) vector with the restriction enzyme Bam HI using a ClonExpress II One Step Cloning Kit (Vayzme). The recombinant plasmid was transformed into E. coli BL21(DE3) cells. The expression of the recombinant fusion proteins was induced in a logarithmic-phase culture of the transformants by the addition of isopropyl-β-D-1-thiogalactopyranoside (IPTG, Sigma) to a final concentration of 0.3 mM at 16 °C for 20 h. Harvest the cells by centrifugation at 4000 g for 20 min and discard the supernatant. Resuspend the cells in 25 ml Column Buffer (20 mM Tris-HCl, 200 mM NaCl and 1 mM EDTA). Place the sample in an ice-water bath and sonicate in short pulses of 15 s. Centrifuge at 8000 g for 20 min to obtain the supernatant (crude extract). Dilute the crude extract 1:6 with Column Buffer. The rBen_0691 proteins were purified by using amylose agarose resin (New England Biolabs) from the crude extract according to the manufacturer’s instructions. rBen_0691 protein was concentrated and resuspended in PBS using a 3 kDa ultrafiltration tube (Millipore). rBen_0691 protein purification was analyzed by SDS-PAGE, and the protein concentration was determined with a BCA Protein Quantitation Kit (Beyotime, Shanghai, China).
Animals experiments and ethics statement
To obtain anti-Ben_0691 rabbit serum, the 8-week-old female Japanese large-eared white rabbits used in the experiments were obtained from Liaoning Changsheng Biotechnology (Benxi, China). Each rabbit was housed individually in a standard animal room with unrestricted access to food and water. Rabbits (n = 3) were immunized subcutaneously three times, 2 weeks apart, with 200 µg/rabbit of Ben_0691 protein mixed 1:1 v/v with complete or incomplete Freund’s adjuvant (Sigma Adjuvant System, SAS), or SAS alone as a control. At the end of the experiment, the rabbits were euthanized by intravenous injection of sodium pentobarbital (30 mg/kg) followed by intravenous injection of air. Rabbit blood was collected and processed to separate serum. The animal experiment for preparing anti-Ben_0691 rabbit serum was conducted in compliance with the Animal Welfare Act and Guide for the Care and Use of Laboratory Animals, approved by the Laboratory Animal Welfare Committee of Harbin Veterinary Research Institute (HVRI) of the Chinese Academy of Agricultural Sciences (approval number 231128-01-GR).
Purifying antibodies against Ben_0691
Anti-Ben_0691 antibodies were purified from the rabbit serum anti-Ben_0691 as described previously58. The antiserum was mixed with sodium acetate and caprylic acid, then centrifuged at 5100 g for 20 min to collect the supernatant (total IgG). Amylose resin was added to a column and incubated with 5 mg rBen_0691 protein. After washing the resin, the total IgG was then passed through the column three times and then eluted with potassium thiocyanate. Finally, the eluted antibody was loaded onto a desalting column to remove buffer salts and small molecules. The IgG concentration was determined by UV spectroscopy (275 nm).
Blocking assay
To confirm the role of Mmm cell surface Ben_0691 in promoting IRG-47 expression in EBL cells, Ben-181-vacc were washed thrice with PBS and preincubated with the purified antibodies against Ben_0691 (10, 20, and 50 μg/ml) or isotype control IgG (Abcam, 50 μg/ml) at 37 °C for 30 min. These bacteria were washed and then added to EBL cells (MOI = 50). Following incubation for 12 h, the cells were washed three times with PBS and subjected to Western blotting analysis.
Confocal microscopy
To investigate whether Ben_0691 altered the intracellular location of IRG-47, EBL cells were transfected with IRG-47-Myc and Ben_0691-GFP or pEGFP-C1 for 48 h; To observe the co-localization of Ben_0691 and ATP5A1, Hela cells were transfected with Ben_0691-Flag and/or ATP5A1-Myc for 48 h. These cells were fixed with 4% PFA in PBS for 30 min at RT. After permeabilizing with 0.1% Triton X-100 for 15 min, the cells were blocked for 1 h in 5% cold-water fish skin gelatin. The cells were then incubated with rabbit anti-Flag or mouse anti-Myc antibodies for 1 h at RT, washed thrice with PBS, and incubated with secondary antibodies coupled to Alexa Fluor 488 or 633 (Invitrogen) for 1 h. The cell nuclei were labeled with DAPI. Fluorescence was observed by confocal microscopy (LSM980, Zeiss).
MBP pull-down assay
To identify the key host molecules involved in Ben_0691 promoting IRG-47 expression, EBL cells were harvested and resuspended in 1 ml of RIPA lysate containing protease inhibitor. The cells were lysed on ice for 1 h and centrifuged at 12,000 r/min at 4 °C for 15 min to collect the supernatant. 50 μl anti-MBP magnetic beads (New England Biolabs) were washed with PBS twice. The 100 μg MBP or rBen_0691 protein produced in E. coli BL21(DE3) cells was conjugated to anti-MBP magnetic beads for 2 h at 4 °C. The beads were then washed with PBS thrice and incubated with the supernatant from the lysed EBL cells on an MACSmix Tube rotator for 4 h at 4 °C. The beads were collected with a magnetic separator (Bimake) and then washed five times with PBS to remove the unbound proteins, the protein complexes binding to the beads were eluted with 75 μl of 1 × SDS elution buffer. Eluted proteins were separated by SDS-PAGE and silver-stained (Beyotime, Shanghai, China). The specific protein bands pulled down by rBen_0691 rather than MBP protein were excised and sent to BGI (Beijing, China) for mass spectrometry analysis.
Co-IP assays
To verify the interaction between two proteins, the plasmid expressing ATP5A1-Myc protein was transfected with the plasmids expressing Mmm proteins (Ben_0691, Ben_0691 truncates, or Ben_0691 mutations) into Hela cells, respectively. Hela cells are frequently utilized for protein expression and the examination of protein-protein interactions in vitro59. At 48 h posttransfection, the cells were lysed with ice-cold lysis buffer for 30 min at 4 °C. After centrifugation (8000 g, 10 min) at 4 °C, the clarified extracts were incubated with anti-GFP magnetic beads (Beyotime, Shanghai, China) or anti-c-Myc magnetic beads (MedChemExpress, USA) on a MACSmix Tube rotator at 4 °C overnight. The incubated beads were collected with a magnetic separator (Bimake) and then washed five times with lysis buffer to remove the unbound proteins. The protein complexes binding to the beads were eluted with 75 μl of 1 × SDS elution buffer, followed by SDS-PAGE and Western blotting.
To validate the interaction between endogenous ATP5A1 and Ben_0691, Mmm Ben-181-vacc (MOI = 50) were washed thrice with PBS and then added to EBL cells. Following incubation for 12 h, the cells were washed three times with PBS and lysed with ice-cold lysis buffer for 30 min at 4 °C. After centrifugation (8000 g, 10 min) at 4 °C, the clarified extracts were incubated with magnetic beads conjugated with anti-ATP5A1 or -IgG antibody on a MACSmix Tube rotator at 4 °C overnight. The incubated beads were washed five times with lysis buffer to remove the unbound proteins. The protein complexes binding to the beads were eluted with 75 μl of 1 × SDS elution buffer, followed by Western blotting.
Flow cytometric analysis
EBL cells were harvested to detect the cell surface ATP5A1 expression. Cells were collected by centrifugation at 1000 rpm for 5 min, rinsed thrice with PBS, and then incubated with the rabbit anti-ATP5A1 polyclonal antibody or rabbit IgG isotype control polyclonal antibody (ab37415; Abcam) for 1 h at 37 °C. Subsequently, the cells were incubated with goat anti-rabbit IgG-Alexa Fluor 488 (Invitrogen) for 1 h at 37 °C in the dark. Finally, the cell samples were fixed with 4% PFA for 30 min. To determine the percentage of infected cells per condition, 10,000 cells were scored and analyzed using the flow cytometry (A60-Universal, Apogee, Britain). The isotype control (isotype antibody + secondary antibody) or secondary antibody control (secondary antibody only) was used to determine background staining. All flow cytometry data were analyzed using the FlowJo software (version X10.0; FlowJo, LLC, Ashland, OR). The gating strategy is shown in Supplementary Fig. 3.
Far-Western blotting
To explore whether Ben_0691 could directly bind to ATP5A1, a Far-Western blotting was performed. 1 μg of recombinant protein rBen_0691 or negative control protein MBP was separated by SDS-PAGE and transferred onto nitrocellulose membranes. After blocking with 5% (w/v) cold-water fish skin gelatin, the membrane was incubated with PBS, 1 μg/ml ATP5A1-His or His-tag protein. This followed by incubation with the primary antibody including mouse anti-MBP (NEB), rabbit anti-ATP5A1 (Proteintech) or mouse anti-His antibody (Zhongshan Goldenbridge-Bio), and then with DyLight 800-labeled goat anti-mouse or -rabbit IgG (H + L) (1:10,000 dilution, Kirkegaard & Perry Laboratories). Finally, the membranes were scanned using an Odyssey infrared imaging system.
Statistical and reproducibility
GraphPad Prism software (version 9.0; GraphPad Software Inc.) was used for all statistical analyses. Data obtained from a minimum of three independent experiments are reported as the mean ± standard deviation (SD). The significance of differences between the two groups was determined with a two-tailed Student’s t test (Figs. 2G, H, 3A, D, 8C, D). For multigroup comparisons, one-way analysis of variances (ANOVA) with Dunnett’s test (Figs. 1C, 4F and S2B), or with Tukey’s test (Figs. 1D, E, 7B, C and 8H) was employed. For all analyses, a probability (p) value of < 0.05 was considered statistically significant. In some cases, the relative band intensity of the Western blotting was quantitated by the ImageJ software in comparison with the β-actin control and normalized with the control.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Data files. The mass spectrometry proteomics data have been provided in Supplementary Data 1. The source data behind the graphs are provided in the main manuscript can be found in Supplementary Data 2. All uncropped blot images are included in Supplementary information as Supplementary Fig. 4.
References
Nielsen, S. S. et al. Assessment of the control measures for category A diseases of Animal Health Law: contagious bovine pleuropneumonia. EFSA J. 20, https://doi.org/10.2903/j.efsa.2022.7067 (2022).
Di Teodoro, G. et al. Contagious bovine pleuropneumonia: a comprehensive overview. Vet. Pathol. 57, 476–489 (2020).
Pan, Q. et al. Mycoplasma glycine cleavage system key subunit GcvH is an apoptosis inhibitor targeting host endoplasmic reticulum. PLOS Pathogens 20, https://doi.org/10.1371/journal.ppat.1012266 (2024).
Xin, J. et al. A history of the prevalence and control of contagious bovine pleuropneumonia in China. Vet. J. 191, 166–170 (2012).
Wu, Q. et al. A candidate competitive ELISA based on monoclonal antibody 3A8 for diagnosis of contagious bovine pleuropneumonia. Appl. Microbiol. Biotechnol. 108, https://doi.org/10.1007/s00253-024-13127-0 (2024).
Provost, A. et al. Contagious bovine pluropneumonia. Rev. Sci. Tech. 6, 565–679 (1987).
Onono, J. O., Wieland, B. & Rushton, J. Estimation of impact of contagious bovine pleuropneumonia on pastoralists in Kenya. Prevent. Vet. Med. 115, 122–129 (2014).
Tambi, N. E., Maina, W. O. & Ndi, C. An estimation of the economic impact of contagious bovine pleuropneumonia in Africa. Rev. Sci. Tech. 25, 999–1011 (2006).
Jores, J. et al. Contagious Bovine and Caprine Pleuropneumonia: a research community’s recommendations for the development of better vaccines. npj Vaccines 5, https://doi.org/10.1038/s41541-020-00214-2 (2020).
Dedieu, L. et al. Gamma interferon-producing CD4 T-cells correlate with resistance to Mycoplasma mycoides subsp. mycoides SC infection in cattle. Vet. Immunol. Immunopathol. 107, 217–233 (2005).
Taylor, G. A., Feng, C. G. & Sher, A. Control of IFN-γ-mediated host resistance to intracellular pathogens by immunity-related GTPases (p47 GTPases). Microbes Infect. 9, 1644–1651 (2007).
MacMicking, J. D. Immune control of phagosomal bacteria by p47 GTPases. Curr. Opin. Microbiol. 8, 74–82 (2005).
Zhao, Y. O., Khaminets, A., Hunn, J. P. & Howard, J. C. Disruption of the Toxoplasma gondii parasitophorous vacuole by IFNγ-inducible immunity-related GTPases (IRG proteins) triggers necrotic cell death. PLoS Pathog. 5, e1000288 (2009).
Martens, S. et al. Mechanisms regulating the positioning of mouse p47 resistance GTPases LRG-47 and IIGP1 on cellular membranes: retargeting to plasma membrane induced by phagocytosis. J. Immunol. 173, 2594–2606 (2004).
Collazo, C. M. et al. Inactivation of LRG-47 and IRG-47 reveals a family of interferon γ–inducible genes with essential, pathogen-specific roles in resistance to infection. J. Exp. Med. 194, 181–188 (2001).
MacMicking, J. D., Taylor, G. A. & McKinney, J. D. Immune control of tuberculosis by IFN-γ-inducible LRG-47. Science 302, 654–659 (2003).
Boehm, U. et al. Two families of GTPases dominate the complex cellular response to IFN-γ. J. Immunol. 161, 6715–6723 (1998).
Dockterman, J., Fee, B. E., Taylor, G. A. & Coers, J. Murine Irgm paralogs regulate nonredundant functions to execute host defense to Toxoplasma gondii. Infect. Immun. 89, https://doi.org/10.1128/iai.00202-21(2021).
Pradipta, A. et al. Cell-autonomous Toxoplasma killing program requires Irgm2 but not its microbe vacuolar localization. Life Sci. Alliance 4, https://doi.org/10.26508/lsa.202000960(2021).
Bernstein-Hanley, I. et al. The p47 GTPases Igtp and Irgb10 map to the Chlamydia trachomatis susceptibility locus Ctrq-3 and mediate cellular resistance in mice. Proc. Natl. Acad. Sci. USA 103, 14092–14097 (2006).
Taylor, G. A., Feng, C. G. & Sher, A. p47 GTPases: regulators of immunity to intracellular pathogens. Nat. Rev. Immunol. 4, 100–109 (2004).
Dockterman, J. & Coers, J. How did we get here? Insights into mechanisms of immunity-related GTPase targeting to intracellular pathogens. Curr. Opin. Microbiol. 69, https://doi.org/10.1016/j.mib.2022.102189 (2022).
Al-Zeer, M. A., Al-Younes, H. M., Braun, P. R., Zerrahn, J. & Meyer, T. F. IFN-γ-inducible Irga6 mediates host resistance against Chlamydia trachomatis via autophagy. PLoS ONE 4, e4588 (2009).
Tian, B. et al. Interferon-inducible GTPase 1 impedes the dimerization of rabies virus phosphoprotein and restricts viral replication. J. Virol. 94, https://doi.org/10.1128/jvi.01203-01220 (2020).
Man, S. M. et al. IRGB10 liberates bacterial ligands for sensing by the AIM2 and caspase-11-NLRP3 inflammasomes. Cell 167, 382–396.e317 (2016).
Coers, J. et al. Chlamydia muridarum evades growth restriction by the IFN-γ-inducible host resistance factor Irgb10. J. Immunol. 180, 6237–6245 (2008).
Zhou, Y. et al. P19 contributes to Mycoplasma mycoides subsp. mycoides adhesion to EBL cells. Micro. Pathog. 93, 13–21 (2016).
Li, Y. et al. Changes in pathogenicity and immunogenicity of Mycoplasma mycoides subsp. mycoides strains revealed by comparative genomics analysis. Sci. Rep. 6, https://doi.org/10.1038/srep19081 (2016).
Zhao, G., Lu, D., Li, M. & Wang, Y. Gene editing tools for mycoplasmas: references and future directions for efficient genome manipulation. Front. Microbiol. 14, 1191812 (2023).
March, J. B., Clark, J. & Brodlie, M. Characterization of strains of Mycoplasma mycoides subsp. mycoides small colony type isolated from recent outbreaks of contagious bovine pleuropneumonia in Botswana and Tanzania: evidence for a new biotype. J. Clin. Microbiol. 38, 1419–1425 (2000).
Hegde, S. et al. In vitro and in vivo cell invasion and systemic spreading of Mycoplasma agalactiae in the sheep infection model. Int. J. Med. Microbiol. 304, 1024–1031 (2014).
Dowling, A. J., Hill, G. E. & Bonneaud, C. Multiple differences in pathogen-host cell interactions following a bacterial host shift. Sci. Rep. 10, 6779 (2020).
Totté, P., Bonnefois, T. & Manso-Silván, L. Interactions between Mycoplasma mycoides subsp. mycoides and bovine macrophages under physiological conditions. Plos ONE 19, e0305851 (2024).
Consortium, U. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019).
Xing, M. et al. Fucoidan from Fucus vesiculosus prevents the loss of dopaminergic neurons by alleviating mitochondrial dysfunction through targeting ATP5F1a. Carbohydr. Polym. 303, https://doi.org/10.1016/j.carbpol.2022.120470 (2023).
Wang, X. J. et al. A novel crosstalk between two major protein degradation systems: regulation of proteasomal activity by autophagy. Autophagy 9, 1500–1508 (2013).
Chang, Y.-W. et al. Spatial and temporal dynamics of ATP synthase from mitochondria toward the cell surface. Commun. Biol. 6, https://doi.org/10.1038/s42003-023-04785-3 (2023).
Kim, Y.-R. et al. Toxoplasma gondii GRA8 induces ATP5A1–SIRT3-mediated mitochondrial metabolic resuscitation: a potential therapy for sepsis. Exp. Mol. Med. 50, e464 (2018).
Song, Y., Wang, F., Wei, Y., Chen, D. & Deng, G. ATP5A1 participates in transcriptional and posttranscriptional regulation of cancer-associated genes by modulating their expression and alternative splicing profiles in HeLa cells. Technol. Cancer Res. Treat. 20, 15330338211039126 (2021).
Schoenborn, J. R. & Wilson, C. B. Regulation of interferon-γ during innate and adaptive immune responses. Adv. Immunol. 96, 41–101 (2007).
Kak, G., Raza, M. & Tiwari, B. K. Interferon-gamma (IFN-γ): exploring its implications in infectious diseases. Biomol. Concepts 9, 64–79 (2018).
Bach, E. A., Aguet, M. & Schreiber, R. D. The IFNγ receptor: a paradigm for cytokine receptor signaling. Annu. Rev. Immunol. 15, 563–591 (1997).
Rafeld, H. L., Kolanus, W., Van Driel, I. R. & Hartland, E. L. Interferon-induced GTPases orchestrate host cell-autonomous defence against bacterial pathogens. Biochem. Soc. Trans. 49, 1287–1297 (2021).
Pilla-Moffett, D., Barber, M. F., Taylor, G. A. & Coers, J. Interferon-inducible GTPases in host resistance, inflammation and disease. J. Mol. Biol. 428, 3495–3513 (2016).
Murray, H. W., Mitchell-Flack, M., Taylor, G. A. & Ma, X. IFN-γ-induced macrophage antileishmanial mechanisms in mice: a role for immunity-related GTPases, Irgm1 and Irgm3, in Leishmania donovani infection in the liver. Exp. Parasitol. 157, 103–109 (2015).
Wandel, M. P. et al. GBPs inhibit motility of Shigella flexneri but are targeted for degradation by the bacterial ubiquitin ligase IpaH9. 8. Cell Host Microbe 22, 507–518.e505 (2017).
Haldar, A. K., Nigam, U., Yamamoto, M., Coers, J. & Goyal, N. Guanylate binding proteins restrict Leishmania donovani growth in nonphagocytic cells independent of parasitophorous vacuolar targeting. MBio 11, https://doi.org/10.1128/mbio.01464-01420 (2020).
Kim, B.-H. et al. A family of IFN-γ–inducible 65-kD GTPases protects against bacterial infection. Science 332, 717–721 (2011).
Santos, J. C. et al. Human GBP1 binds LPS to initiate assembly of a caspase-4 activating platform on cytosolic bacteria. Nat. Commun. 11, 3276 (2020).
Taylor, G. A. et al. Pathogen-specific loss of host resistance in mice lacking the IFN-gamma-inducible gene IGTP. Proc. Natl. Acad. Sci. USA 97, 751–755 (2000).
Henry, S. C. et al. Impaired macrophage function underscores susceptibility to Salmonella in mice lacking Irgm1 (LRG-47). J. Immunol. 179, 6963–6972 (2007).
Chen, X. et al. IFN-inducible p47 GTPases display differential responses to Schistosoma japonicum acute infection. Cell. Mol. Immunol. 7, 69–76 (2009).
Hunter, S. et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 37, D211–D215 (2009).
Nůsková, H. et al. Mitochondrial ATP synthasome: expression and structural interaction of its components. Biochem. Biophys. Res. Commun. 464, 787–793 (2015).
Di, T. et al. Emodin blocks mPTP opening and improves LPS-Induced HMEC-1 cell injury by upregulation of ATP5A1. Chem. Biodivers. 21, https://doi.org/10.1002/cbdv.202301916 (2024).
Li, Y. et al. Strains of Mycoplasma mycoides subsp. mycoides small colony type isolated in China between 1953 and 1960 show close similarity to strains of the Africa/Australia cluster. Vet. J. 179, 137–141 (2009).
Vogl, G. et al. Mycoplasma gallisepticum invades chicken erythrocytes during infection. Infect. Immun. 76, 71–77 (2008).
Piccinni, M. Z. & Guille, M. J. Purifying antibodies raised against Xenopus peptides. Cold Spring Harb. Protoc. 2020, pdb. prot105619 (2020).
Federico, A. & Monti, S. Contextualized protein-protein interactions. Patterns 2, 100153 (2021).
Acknowledgements
This study was supported by the National Key Research and Development Program of China: Research on New Quarantine and Detection Technologies for Important Emerging and Exotic Animal Diseases at Ports (2021YFD1800503 to J.X.). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization, J.X. and Q.P.; methodology, T.L. and Q.P.; investigation, T.L., H.Z., X.W., Q.X., Y.L., Q.W., and Y.W.; formal analysis, T.L. and Q.P.; visualization, T.L. and Q.P.; writing-original draft, T.L. and Q.P.; writing-review & editing, T.L. and Q.P.; project administration, J.X. and Q.P.; funding acquisition, J.X.; supervision, J.X. and Q.P.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Takuya Kanda and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editor: Tobias Goris. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, T., Zhao, H., Wang, X. et al. Ben_0691 functions as an IRG-47 mycoplasma stimulator that mediates host cell resistance to Mycoplasma mycoides subsp. mycoides infection. Commun Biol 8, 1571 (2025). https://doi.org/10.1038/s42003-025-08950-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08950-8