Abstract
ATM is an apical kinase that governs cellular responses to DNA replication-associated double strand breaks (DSBs) accumulated by the topoisomerase inhibitor anti-cancer therapy. Here we identify that TIPIN, a major constituent of the fork protection complex in the replisome, plays a key role in coordinating ATM signaling tied to DNA replication stress. We demonstrate that TIPIN amplifies ATM signaling to promote DNA end resection and homology-directed repair. TIPIN itself is phosphorylated by ATM, which is required for the recruitment of MDC1 to stalled forks to promote ATM-dependent NF-κB activation. Inhibition of the NF-κB pathway by MDC1 depletion impairs upregulation of anti-apoptotic regulator c-FLIP, thus potentiating caspase-8 activation and cytotoxicity of topoisomerase inhibition. Together, our study defines TIPIN as a master regulator of ATM-dependent DSB repair and NF-κB signaling. We propose that targeting MDC1, a key effector of ATM-TIPIN signaling, acts as a chemosensitizer that suppresses therapy-induced senescence and augments the effectiveness of genotoxic therapy.
Similar content being viewed by others
Introduction
Cancer cells exhibit an increased burden of DNA replication stress and genome instability. Accordingly, exacerbating DNA breakage to obstruct DNA replication is an effective strategy for anti-cancer treatment1. Topoisomerase inhibitors and anthracycline derivatives are primary regimens to accumulate DNA replication-associated DSBs. Poly(ADP-ribose) polymerase (PARP) inhibitors generate a large amount of DSBs and compromise the integrity of DNA replication forks, which is exacerbated in the absence of BRCA1/2 function, conferring hypersensitivity in BRCA1/2-deficient tumors2. However, DNA damage signaling originating from DSBs often triggers a reversible form of growth arrest known as therapy-induced senescence (TIS), allowing cancer cells to evade the initial toxicity of drugs and repopulate in a more aggressive manner, leading to therapy resistance and disease recurrence3. Hence, modulating cellular responses to DNA damage in a way that potentiates the cytotoxicity of cancer cells while suppressing pro-survival/senescence signaling would be beneficial for increasing the effectiveness of DSB-inducing therapeutic regimens.
The Ataxia-telangiectasia mutated (ATM) serine/threonine protein kinase is a master regulator of DSB signaling4. ATM-dependent phosphorylation of key targets in the DNA damage response (DDR) coordinates diverse cellular stress responses including cell cycle checkpoint, DNA repair, and apoptosis, primarily through the regulation of CHK2 and p53 activities. Additionally, ATM engages in activation of the nuclear factor-kappa B (NF-κB) pathway, a transcriptional program that controls senescence-associated secretory phenotype (SASP) gene expression and establishment of senescence triggered by genotoxic stress5. This nuclear-initiated NF-κB signaling involves a series of ATM-dependent post-translational modifications on NF-κB essential modulator (NEMO)/IKKγ, which in turn facilitates the export of NEMO to the cytosol and subsequent degradation of IκB, an inhibitor of NF-κB, culminating in NF-κB-dependent transcriptional responses6. The genotoxic stress-induced NF-κB signal transduction is considered pro-survival as it establishes TIS and upregulates anti-apoptotic proteins7. Nevertheless, it remains largely elusive how the multifaceted ATM-dependent responses to DNA replication-associated DSBs are regulated to determine the fate of cells between survival and death under genotoxic therapy.
In this study, we identify a central role of TIPIN, a key component of the fork protection complex (FPC) within the DNA replication machinery8, in coordinating the ATM-dependent DNA damage checkpoint and NF-κB signaling. As a scaffold that tethers the CMG (CDC45, MCM2-7, GINS) helicase and polymerase activities, the FPC regulates both the progression and pausing of the replisome at active forks and its protection at stalled forks9,10. The interaction of the FPC with topoisomerase I (Top1) and PARP1 is known to help relieve torsional stress, resolve DNA replication-transcription conflicts, and suppress replication-associated DNA gaps11,12,13,14. Furthermore, TIPIN directly interacts with replication protein A (RPA), a single-stranded DNA (ssDNA)-binding protein, at stalled forks to promote Ataxia telangiectasia and Rad3-related (ATR)-dependent CHK1 phosphorylation to activate checkpoint under replication stress15,16,17. We herein provide evidence that TIPIN acts as both a regulator and substrate of ATM to integrate DNA replication-associated DSB signaling. Targeting TIPIN-mediated nuclear ATM-NEMO signaling dampens anti-apoptotic capacity by inhibiting NF-κB activation, thereby counteracting TIS and augmenting the cytotoxic effect of genotoxic therapy. Our study provides insight into the development of potential interventions to increase the sensitivity of DSB-inducing chemotherapy by targeting the DNA replication machinery and associated checkpoint responses that cancer cells rely on to alleviate their DNA replication vulnerability.
Results
Specific depletion of TIPIN in the FPC impairs cellular DSB responses to Top1 inhibition
TIMELESS (TIM) and TIPIN constitute an obligate heterodimer of the FPC, in which their stability is interdependent; siRNA-mediated knockdown of one leads to a depletion of the other (Fig. 1a)18,19. In line with previous studies, knockdown of either TIM or TIPIN reduced ATR-dependent CHK1 activation in response to hydroxyurea (HU), a chemotherapy that induces replication fork stalling and ssDNA accumulation (Fig. 1a)10,15. Unlike ionizing irradiation (IR) which generates DSBs in a cell cycle-independent manner, accumulation of the Top1 cleavage complex (Top1cc) stabilized by Top1 inhibitors (Top1i) results in the formation of one-ended DSB formation that are coupled with DNA replication20. Since the FPC is positioned at the front of the replisome and poised to encounter Top1cc lesions ahead of the fork19, we reasoned that the DSB signaling in response to Top1i may potentially be regulated by the FPC. Indeed, knockdown of TIPIN using two-independent siRNAs (siTIPIN#3 & #4) in U2OS cells led to a decrease in CHK2 phosphorylation (an ATM target) and RPA32 phosphorylation at S4/S8 (a marker for DNA end resection and ssDNA formation) in response to the Top1i camptothecin (CPT) (Fig. 1b and Supplementary Fig. S1a). In contrast, TIPIN depletion did not affect DSB responses against neocarzinostatin (NCS), an IR-mimetic compound (Supplementary Fig. S1b). TIM knockdown led to an effect opposite of TIPIN knockdown, exhibiting elevated pCHK2 and pRPA32 S4/S8, as loss of TIM is known to compromise the overall integrity of the replisome thus increasing fork breakage (Fig. 1b and Supplementary Fig. S1a). Notably, contrary to siRNA TIPIN#3, cells transfected with another siRNA, TIPIN#1, a more potent siRNA that destabilizes TIM to a greater extent, showed pKAP1 S824, pRPA32 S4/S8, and γH2AX levels comparable to cells depleted of TIM (Fig. 1c; a side-by-side comparison of two siRNAs is shown in Supplementary Fig. S1c). Release from CPT damage showed a similar result regarding the pRPA32 S4/S8 induction (Supplementary Fig. S1d). On the other hand, lowering the amount of transfecting siRNA TIPIN#1 revealed a defect in inducing pRPA32 S4/S8 as seen in siRNA TIPIN#3 or #4-transfected cells (Fig. 1d and Supplementary Figs. S1e and S1f). Depletion of TIPIN in HCT116 cells showed similar defects (Supplementary Fig. S1g). Ectopic expression of TIM did not rescue defects from siRNA TIPIN#3, confirming phenotypes specific to TIPIN knockdown (Supplementary Fig. S1h).
Western blotting (WB) of U2OS cells transfected with indicated siRNAs (vs. control) and treated with a 2 mM HU for 16 h or b 1 µM CPT for 1 h. c, d WB of U2OS cells transfected with indicated siRNAs and treated with 1 µM CPT for indicated times. TIPIN#1 (L: low) indicates 10 pmol siRNA transfection. e WB of U2OS cells transfected with indicated siRNA and treated with 1 µM CPT for 1 h. The amount of siRNA TIPIN#1 is indicated. Quantification of pRPA32 S4/S8: n = 3 from three biological replicates, mean ± s.d. **P < 0.01, *P < 0.05, ns: not significant, Student’s t test. f Schematic of TIPIN siRNA titration and its effect on DDR activation. g WB of U2OS Retro-X Flag-TIPIN N-terminus (amino acids 1-173) inducible cell line transfected with siRNA TIPIN#1 (H: high, 20 pmol transfection) for 64 h, Flag-TIPIN-N induced by 100 ng/ml doxycycline (Dox) for 48 h, and treated with 1 µM CPT for 1 h. h WB showing knockdown of TIM or TIPIN. Two different amounts of siRNA was transfected for TIPIN knockdown. i Representative images of DNA fiber tracks and dot plot of IdU track lengths from U2OS cells transfected with indicated siRNA. Red bars indicate median from at least 150 fiber tracks. ****P < 0.0001, ns: not significant, Mann-Whitney test.
Titrating the amount of siRNA TIPIN#1 transfection further showed that its ability to suppress the pRPA32 S4/S8 induction is best at the intermediate concentration (Fig. 1e); more specifically, too potent of a TIPIN knockdown depletes TIM, causing global fork instability and pRPA32 induction, whereas the phenotype associated with TIPIN deficiency is lost with its weaker efficiency. Hence, the phenotype specific to TIPIN knockdown is only revealed when the stability of TIM (i.e., the FPC integrity) is preserved (Fig. 1f). Indeed, by titrating TIM knockdown, we also identified an amount of siRNA that stabilizes TIM enough to reveal a defect in pRPA32 S4/S8 (Supplementary Fig. S1i). To substantiate this idea, we designed an inducible system to express only the N-terminal domain of TIPIN that can bind TIM and support its stability (Fig. 1g). This TIPIN fragment, when induced by doxycycline (dox) in siRNA TIPIN#1-transfected cells, was able to suppress the pRPA32 S4/S8 induction, indicating that the true TIPIN knockdown phenotype (i.e., a defect in pRPA32 S4/S8 upregulation) is masked when TIM is destabilized (Fig. 1g). Furthermore, while siRNA TIPIN#1 reduced DNA fiber track lengths comparably to TIM knockdown, using a lower dose of siRNA TIPIN#1 restored DNA replication fork progression similar to siRNA TIPIN#3, both of which did not hinder DNA replication (Fig. 1h, i). Indeed, S phase progression was largely unaffected when TIPIN is depleted by siRNA TIPIN#3 or a lower dose of TIPIN#1 (Supplementary Fig. S1j). All together, we conclude that TIPIN is necessary for DDR signaling against replication-associated DSBs, which has not been previously revealed due to the collateral degradation of its binding partner TIM that normally leads to global replication fork instability and heightened DDR.
TIPIN is required for amplifying ATM signaling to promote DSB repair
Top1cc trapped by Top1i leaves behind single-strand breaks (SSBs) that are converted into one-ended DSBs when they are encountered by ongoing DNA replication forks21. The bulky lesion formed by Top1 DNA-protein crosslinks (DPCs) needs to be processed by DPC proteases or the ubiquitin-dependent proteasome pathway, which allows for subsequent access and repair of broken DNA via homology-directed repair (Fig. 2a)22. We therefore asked what steps of the Top1-DPC repair pathway are affected by TIPIN depletion in the context of ATM-mediated DSB signaling that leads to a defect in pRPA32 S4/S8 induction. When cells were pulsed with CPT for 1 h and released, we observed immediate activation of ATM signaling, including pKAP1 and pCHK2, followed by a progressive increase of pRPA32 S4/S8 that represents DPC removal and DNA end resection necessary for DNA repair (Fig. 2b). MG132, a proteasome inhibitor, or KU-60019, an ATM kinase inhibitor, efficiently blocked DSB signaling and pRPA32 S4/S8 induction upon CPT treatment (Fig. 2c& Supplementary Fig. S2a). It also prevented NBS1 phosphorylation at S343, a marker for activation of the MRE11-RAD51-NBS1 (MRN) complex that initiates and amplifies ATM signaling. Similarly, knockdown of TIPIN impaired those DSB-associated signaling pathways, including ATM activation, indicating that TIPIN promotes ATM-dependent DSB signaling (Fig. 2d). On the other hand, RADAR (rapid approach to DNA adduct recovery) analysis showed that the removal of Top1cc following CPT treatment is not affected by TIPIN deficiency, suggesting that TIPIN functions downstream of DPC processing to promote ATM signaling (Fig. 2e). Additionally, ATM inhibition did not cause a defect in DPC processing (Supplementary Fig. S2b). While ATM inhibition led to a compensatory hyperactivation of pDNA-PKcs at S2056, pRPA32 S4/S8 levels were still downregulated, indicating that ATM plays a substantial role in inducing pRPA32 S4/S8 upon CPT damage (Supplementary Fig. S2c). In response to HU, however, TIPIN knockdown exacerbated pRPA32 S4/S8 levels especially at a later time point, presumably reflecting prolonged ssDNA exposure that leads to fork collapse, thus highlighting the differential responses against replication fork stalling versus breakage (Supplementary Fig. S2d).
a Schematic depicting the processing of Top1cc stabilized by a Top1 inhibitor (star). b WB of U2OS cells pulse-treated with 100 nM CPT for 1 h followed by release into fresh media for indicated times. c WB of U2OS cells pretreated with 10 µM MG132 for 1 h followed by co-treatment with 1 µM CPT for indicated times. d WB of U2OS cells transfected with siRNA TIPIN#3 and treated with 1 µM CPT. e Top: Slot-blot of the RADAR assay from U2OS cells transfected with siRNA TIPIN#3 and treated with 1 µM CPT for indicated times. dsDNA serves as a loading control. Bottom: RADAR assay samples were digested with micrococcal nuclease and analyzed by WB. f WB of U2OS cells transfected with siRNA TIPIN#1 or TIPIN#3 and treated with 1 µM CPT. g Representative images of pRPA32 S4/S8 immunofluorescence in U2OS cells transfected with indicated siRNA and treated with 1 µM CPT for 1 h. Scale bar: 10 µm. Quantification of pRPA32 S4/S8 positive cells defined as >50 foci is shown. h Representative images of RAD51 foci formation in siRNA-transfected cells pulse-treated with 1 µM CPT for 1 h and released to fresh media for indicated times. Scale bar: 10 µm. Quantification of Rad51 foci positive cells defined as >25 foci is shown. For (g, h), n = 3 from three biological replicates, mean ± s.d. **P < 0.01, *P < 0.05, ns: not significant, Student’s t test (g) and two-way ANOVA (h).
Decreased pRPA32 S4/S8 induction in the absence of TIPIN indicates impaired DNA end resection, which is initiated by the MRN complex and the CtIP endonuclease23. Depletion of CtIP or MRE11 to a lesser extent abrogated pRPA32 S4/S8 induction in response to CPT (Supplementary Fig. S2e). Notably, TIPIN knockdown dramatically reduced the CtIP phosphorylation known to be essential for the interaction with MRN and stimulation of the MRE11 activity (Fig. 2f)24. Accordingly, pRPA32 S4/S8 foci formation was significantly reduced in TIPIN-depleted cells (Fig. 2g). In contrast, siRNA TIPIN#1 or TIM resulted in increased ATM activation, CtIP phosphorylation, and induction of pRPA32 and its foci formation, representing exacerbated fork breakage and heightened DDR signaling (Fig. 2f, g and Supplementary Fig. S2f). Resected DNA at DSBs is subject to homology-directed repair to recover from stalled forks25. Indeed, we observed that TIPIN knockdown results in a significant decrease in RAD51 foci formation, a functional marker of homologous recombination (HR), indicating that DSB repair is compromised (Fig. 2h and Supplementary Fig. S2g). A neutral comet assay also revealed increased DSBs in TIPIN-depleted cells (Supplementary Fig. S2h). Together, these data indicate that TIPIN is required for the activation and amplification of ATM signaling, thereby promoting the downstream resection and repair processes of replication-associated DSBs.
TIPIN is a downstream target of the ATM kinase and is phosphorylated at serine 222
Since we have shown that TIPIN is tied to ATM signaling, we further sought to identify the interplay between ATM and TIPIN in response to replication-associated DSBs. We identified one S/TQ consensus motif within the C-terminal end of TIPIN that serves as a target of ATM/ATR-dependent phosphorylation and is conserved in higher vertebrates (Fig. 3a). This serine 222 phosphorylation site was previously identified in a phospho-proteome analysis under IR damage26. We raised an antibody to specifically recognize TIPIN S222 phosphorylation and were able to detect the phosphorylation of TIPIN at S222 after CPT treatment, which coincides with ATM-CHK2 activation but prior to pRPA32 S4/S8 induction (Fig. 3b). Antibody specificity was further confirmed with a TIPIN S222A mutant (Supplementary Fig. S3a). This phosphorylation was specifically dependent on ATM but not on ATR or DNA-PKcs (Fig. 3c and Supplementary Fig. S3b for quantification). To identify the role of ATM-dependent TIPIN phosphorylation, we implemented a Retro-X Tet-On system whereby cells were reconstituted with siRNA-resistant Flag-TIPIN WT or S222A in a doxycycline-dependent manner following siRNA-mediated depletion of endogenous TIPIN (Fig. 3d and Supplementary Fig. S3c)27. While TIM was destabilized by TIPIN knockdown, its levels were restored when Flag-TIPIN WT or S222A was expressed back to TIPIN-depleted cells (Fig. 3d). However, neither TIPIN WT nor S222A was able to rescue the defect in pRPA32 S4/S8 induction, indicating that introduction of TIPIN cDNA was not sufficient to complement a TIPIN-deficient background (Supplementary Fig. S3d). Since TIPIN is the obligate binding partner of TIM and does not have any obvious nuclear localization signals, we reasoned that additional TIM expression may be needed to restore the functionality of TIPIN in cells. Notably, a previous study described requirement of exogenous TIM in supporting TIPIN to complement a defect in CHK1 phosphorylation under HU damage15. Similarly, ectopic expression of GFP-TIM resulted in an increased stability of Flag-TIPIN, leading to its robust phosphorylation specific to S222 and restoration of pRPA32 S4/S8 induction as well as pATM and pKAP1 activation (Fig. 3e and Supplementary Fig. S3e, S3f). Moreover, increased TIPIN localization at the chromatin-enriched fraction was observed (Supplementary Fig. S3g). In this condition, both TIPIN WT and S222A were able to restore pKAP1 and pRPA32 S4/S8 levels, indicating that TIPIN S222 phosphorylation is not necessary for promoting ATM checkpoint signaling (Fig. 3f).
a Schematic of human TIPIN showing its phosphorylation site at S222 and sequence alignment among vertebrates. b Anti-Flag immunoprecipitation (IP) and anti-pTIPIN S222 WB from U2OS cells expressing Flag-TIPIN and treated with 1 µM CPT for indicated times. c Same as (B), except pretreated with indicated inhibitors; 10 µM KU-60019 (ATMi), 10 µM VE-821 (ATRi), or 5 µM KU-57788 (DNA-PKi), and co-treated with 1 µM CPT for 1 h. d Top: Schematic showing the Retro-X TetOn inducible system of Flag-TIPIN. Bottom: WB of Retro-X Flag-TIPIN WT or S222A cells transfected with TIPIN siRNA and treated with 100 ng/mL doxycycline (Dox) for 48 h. e Anti-Flag IP and WB of Flag-TIPIN WT or S222A induced from U2OS cells expressing GFP-TIM and treated with 1 µM CPT for 1 h. f WB of Retro-X cells reconstituted with Flag-TIPIN WT or S222A by siRNA transfection and Dox-induced expression of cDNA in the presence of GFP-TIM, followed by 1 µM CPT treatment for 1 h. g Schematic showing CRISPR/Cas9-mediated knock-in (KI) to introduce an S222A mutation at Exon 6. h WB of U2OS WT and S222A KI cells treated with 1 µM CPT for 1 h. i Slot-blot of the RADAR assay from U2OS WT and S222A KI cells treated with 1 µM CPT for indicated times. j Representative images of RAD51 foci immunofluorescence in U2OS WT and S222A KI cells pulse-treated with 1 µM CPT for 1 h and released to fresh media for indicated times. Scale bar: 10 µm. Quantification of RAD51 foci-positive cells: red bars indicate median. ****P < 0.0001, ns: not significant, two-way ANOVA.
To further substantiate our result, we employed a CRISPR/Cas9 knock-in approach to replace serine 222 of endogenous TIPIN with alanine in U2OS cells (Fig. 3g and Supplementary Fig. S3h). The obtained homozygous knock-in clone exhibited a similar cell cycle profile with a parent cell line (Supplementary Fig. S3i). Similar to the Retro-X system, ATM signaling was not compromised in the TIPIN S222A cells in response to CPT, further supporting that TIPIN phosphorylation does not directly affect upstream DSB signaling (Fig. 3h). Additionally, the S222A cells showed no defect in Top1cc DPC removal, and RAD51 foci formation was comparable between WT and S222A cells treated with CPT (Fig. 3i, j). Together, we conclude that the ATM-dependent phosphorylation of TIPIN at S222 does not contribute to the upstream DSB signaling for DNA repair driven by ATM.
TIPIN phosphorylation promotes MDC1-dependent activation of the NF-κB pathway
Our results thus far indicate that phosphorylated TIPIN may work as a downstream effector of ATM signaling that is independent of fork processing and repair. CPT is known to trigger ATM-dependent NF-κB activation to attenuate apoptotic responses28. We therefore determined whether TIPIN modification is connected to genotoxic stress-dependent NF-κB signaling activation. We first confirmed that ATM inhibition (ATMi) prevents the activation of NF-κB signaling in U2OS cells as assessed by phosphorylation of NF-κB and its upstream regulator IKKα/β (Fig. 4a). Pretreatment of ATMi somewhat elevated basal levels of pNF-κB in unchallenged conditions, indicating that there may be an ATM-independent sensing mechanism of DNA damage to active NF-κB. In line with its role in ATM signal transduction, TIPIN knockdown impaired the activation of NF-κB against CPT in both time- and dose-dependent manners (Fig. 4b and Supplementary Fig. S4a, S4b). TIPIN knockdown specifically disrupted NF-κB signaling but not ATM checkpoint signaling in response to doxorubicin (Doxo), which intercalates with DNA and inhibits topoisomerase II-mediated DNA repair, thus not considerably inducing pRPA32 S4/S8 via DNA resection (Fig. 4c). This suggests that TIPIN controls a separate arm of the NF-κB pathway independently of ATM checkpoint signaling. The TIPIN S222A knock-in cells exhibited a defect in induction of pNF-κB and pIKKα/β as well as NEMO phosphorylation at S85, a modification known to transduce nuclear ATM-NEMO signaling to the cytosol29, indicating that TIPIN phosphorylation is required to promote NF-κB signaling (Fig. 4d and Supplementary Fig. S4c). pKAP1 and pRPA32 S4/S8 levels were somewhat elevated in this cell line, highlighting that the signaling of DSB processing and NF-κB activation is independent.
a WB of U2OS cells pretreated with 5 µM ATMi for 1 h followed by 1 µM CPT for indicated times. b WB of siRNA-transfected U2OS cells treated with 10 µM CPT for indicated times. c Same as (B), but treated with 1 µM doxorubicin (Doxo) for 1 h. d WB of U2OS WT and S222A KI cells treated with 10 µM CPT for 1 h. e Top: Schematic showing the Flp-In T-REx HA-TIPIN-Flag-TIM inducible system. Expression of the TIPIN WT and S222A fusion proteins was induced by 200 ng/mL doxycycline (Dox). Bottom: Table listing top 5 hits from IP-MS. f WB of U2OS cells transfected with siRNA MDC1 and treated with 10 µM CPT for 1 h. g WB of U2OS cells transfected with siRNA NEMO and treated with 10 µM CPT, 2 µM Doxo, or 200 ng/ml neocarzinostatin (NCS) for 2 h. h NF-κB luciferase reporter activity in U2OS cells transfected with indicated siRNAs followed by treatment with 1 µM CPT for 24 h. Relative luciferase activity was calculated as fold change by normalizing against siRNA Ctrl-transfected DMSO control. n = 3 from three biological replicates. ****P < 0.0001, two-way ANOVA. i Quantification of EMSA analysis in U2OS cells transfected with indicated siRNAs followed by treatment with 1 µM CPT for 1 h. Relative NF-κB activity was calculated as fold change by normalizing against siRNA Ctrl-transfected cells treated with CPT. n = 3 from three biological replicates. ****P < 0.0001, ***P < 0.001, two-way ANOVA. j Representative images of proximity ligation assay between MDC1 and EdU-labeled replication forks following treatment with 1 µM CPT for 30 min in U2OS WT or S222A KI cells. Scale bar: 10 µm. Dot plot of MDC1-EdU PLA mean intensity (arbitrary unit, A.U.): red bars indicate median from >350 cells. A representative plot is shown from n = 3, ****P < 0.0001, two-way ANOVA.
DNA damage-dependent target phosphorylation is recognized by downstream effectors with specific binding activity toward phospho-epitopes to transduce signaling of the DDR30. We therefore performed immunoprecipitation-mass spectrometry (IP-MS) to identify proteins that specifically interact with phosphorylated TIPIN. To this end, we stably expressed HA-tagged TIPIN WT or S222A fused to TIM-Flag as a bait to achieve robust ATM-dependent TIPIN phosphorylation necessary for IP (Fig. 4e). We confirmed specific phosphorylation of engineered TIPIN at S222 following CPT (Supplementary Fig. S4d). One notable target from our proteomics analysis was MDC1 (mediator of DNA damage checkpoint protein 1), which is known for transmitting DSB signaling by forming a complex with γH2AX to recruit repair proteins to the sites of DNA lesions31,32,33. While the role of MDC1 in the replication stress response begins to be appreciated, its connection to the NF-κB pathway is unknown34. Intriguingly, depletion of MDC1 resulted in a decrease in pNF-κB and pIKKα/β in response to CPT and NCS, indicating that MDC1 is required for promoting DNA damage-induced NF-κB signaling (Fig. 4f and Supplementary Fig. S4e). Knockdown of NEMO, an upstream regulator NF-κB activation, also disrupted genotoxic stress-induced NF-κB signaling upon treatment of CPT, Doxo, or NCS (Fig. 4g). Furthermore, knockdown of TIPIN or MDC1 suppressed transcriptional activation of NF-κB as revealed by both luciferase reporter and electrophoretic mobility shift assays (Fig. 4h, i and Supplementary Fig. S4f). Notably, localization of MDC1 at stalled forks, as assessed by a proximity ligation assay (PLA) between MDC1 and EdU-labeled DNA replication forks, was significantly reduced in cells expressing TIPIN S222A, supporting the idea that TIPIN phosphorylation is necessary for enriching MDC1 at stalled forks (Fig. 4j and Supplementary Fig. S4g). Together, these data suggest that phosphorylated TIPIN recruits MDC1 to stalled forks to promote ATM-dependent NF-κB signaling.
Disruption of the ATM-TIPIN-MDC1 signaling axis sensitizes cells to apoptosis against Top1 inhibition
Given the pro-survival role of the NF-κB pathway in establishing TIS and preventing cell death, we tested whether disrupting ATM-TIPIN-MDC1 signaling counteracts NF-κB signaling, leading to chemosensitization of cells in response to Top1 inhibition. Treating cells with BAY 11-7082, an NF-κB inhibitor, was sufficient to suppress NF-κB activation by Top1 inhibition (Fig. 5a). This resulted in a decrease in the induction of senescence compared to control, as measured by senescence-associated β-galactosidase (SAβG) activity, and an increase in apoptosis, as quantified by Annexin V positivity (Fig. 5b, c). In both control and BAY 11-7082 conditions, apoptosis was substantially increased when CPT-challenged cells were subsequently treated with ABT-263, a BH3 mimetic that acts as a senolytic drug to trigger specific killing of senescent cells, further confirming that cells undergo TIS in response to CPT (Fig. 5c)35. This change in cell fate was recapitulated by MDC1 knockdown as well as TIPIN knockdown; MDC1-depleted cells exhibited less SAβG positivity and impaired transcriptional induction of p21/CDKN1A, a key inducer of TIS (Fig. 5d and Supplementary Fig. S5a). Conversely, knockdown of MDC1 or TIPIN led to an increase in apoptosis, leading to hypersensitization under CPT treatment (Fig. 5e, f). All in all, these data suggest that targeting TIPIN-MDC1 contributes to the change in cell fate toward death by inhibiting NF-κB signaling.
a WB of U2OS cells pretreated with 10 µM BAY 11-7082 (a NF-κB inhibitor) for 1 h followed by cotreatment with 10 µM CPT. b Flow cytometry analysis of senescence-associated β-galactosidase (SAβG)-positive U2OS cells pretreated with 10 µM BAY 11-7082 for 1 h followed by 1 µM CPT treatment for indicated times. c Flow cytometry analysis of Annexin V staining in U2OS cells treated with 1 µM CPT for 48 h followed by 1 µM ABT-263 for an additional 24 h. d Flow cytometry analysis of SAβG+ in U2OS cells transfected with siRNA TIPIN#3 or MDC1 and treated with 1 µM CPT for 48 h. n = 3, mean ± s.d. ***P < 0.001, two-way ANOVA. e Flow cytometry analysis of Annexin V staining in U2OS cells transfected with indicated siRNA and treated with 1 µM CPT for 48 h. n = 3, mean ± s.d. ***P < 0.001, *P < 0.05, two-way ANOVA. f Cytotoxicity of U2OS cells transfected with indicated siRNA and treated with CPT for 56 h as measured by luminescence-based ATP viability assay. n = 3, mean ± s.d. **P < 0.01, *P < 0.05, two-way ANOVA. g WB of U2OS cells treated with 100 nM CPT for indicated times. h RT-qPCR of c-FLIP mRNA in siRNA-transfected U2OS cells treated with 1 µM CPT for 24 h. i, j WB of U2OS cells transfected with indicated siRNA and treated with 1 µM CPT up to 48 h. Procaspase-8 is sequentially cleaved to generate active forms of p43/p41 and p18. For (b–e, h), n = 3 from three biological replicates, mean ± s.d. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05, ns: not significant, two-way ANOVA.
To further identify the mechanism through which modulation of NF-κB signaling dictates the chemosensitivity of Top1 inhibition, we investigated how cell death is prevented by NF-κB. We observed an upregulation of c-FLIP (cellular FLICE-like inhibitory protein) following CPT treatment, which is known for antagonizing death receptor-mediated extrinsic apoptosis by preventing activation of caspase-8 (Fig. 5g)36. In contrast, expression of antiapoptotic BCL-2 family proteins BCL-XL and BCL-2, which prevent mitochondrion-dependent intrinsic apoptosis, was largely unaffected. Transcriptional upregulation of c-FLIP was dependent on NF-κB signaling, namely MDC1 and NEMO (Fig. 5h). Prolonged treatment of CPT resulted in cleavage of procaspase-8 (i.e., caspase-8 activation), which was exacerbated by TIPIN or MDC1 depletion, leading to an increase in downstream effector caspase-3 activation (Fig. 5i). This indicates that suppression of NF-κB-dependent c-FLIP upregulation causes caspase-8 hyperactivation and sensitizes cells to apoptosis. We also observed increased activation of caspase-8 and caspase-3 in MDC1-depleted cells in both time- and dose-dependent manners (Fig. 5j and Supplementary Fig. S5b). Knockdown of both isoforms of c-FLIP or the long isoform alone was sufficient to produce cleaved caspase-8 p18 even in the absence of DNA damage, indicating that c-FLIP is crucial for supporting the survival of these cancer cells (Supplementary Fig. S5c). As such, changing the levels of c-FLIP by modulating the NF-κB pathway tips the balance of cell fate toward death. Together, we identify caspase-8-dependent apoptosis as a key determinant of chemosensitivity in response to Top1 inhibition. This process is counteracted by NF-κB-dependent c-FLIP expression, which is supported by the ATM-dependent TIPIN-MDC1 signaling cascade under Top1i therapy.
MDC1 is a target of chemosensitization for irinotecan therapy in colorectal cancer
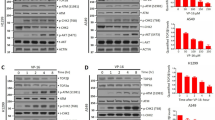
Finally, we determined whether TIPIN-MDC1 signaling that we identified in this study is relevant to chemosensitization in cancer therapy. Irinotecan (CPT-11) is a derivative of CPT used for the treatment of metastatic colorectal cancer (CRC), often combined with additional chemotherapy such as 5-fluorouracil or oxaliplatin37. We tested the effect of TIPIN-MDC1 deficiency, therefore compromised NF-κB signaling, on the chemotherapeutic response of CRC cells to SN-38, an active metabolite of the prodrug irinotecan. Dose-dependent activation of the NF-κB pathway was observed in HCT116 CRC cells treated with SN-38 (Supplementary Fig. S6a). Similar to U2OS, depletion of c-FLIP in HCT116 cells magnified caspase-8 activation and PARP1 cleavage, underscoring operation of the caspase-8 pathway for genotoxicity in this model (Supplementary Fig. S6b). Subsequently, knockdown of TIPIN or MDC1 was sufficient to inhibit SN-38-induced pNF-κB and pNEMO activation (Fig. 6a). This led to suppression of c-FLIP expression upregulated by SN-38 (Fig. 6b). MDC1-deficient HCT116 cells exhibited faster and higher activation of caspase-8 and caspase-3 (Fig. 6c and Supplementary Fig. S6c). Accordingly, cell death is enhanced in HCT116 cells depleted of MDC1 in response to SN-38 (Fig. 6d). We did not observe an increased cytotoxicity in TIPIN-depleted cells, which may be due to a cytostatic effect of replication fork instability caused by prolonged siRNA treatment.
a WB of HCT116 cells transfected with indicated siRNA and treated with 100 nM SN-38 for 1 h. b Same as (A), except SN-38 was treated for 24 h to assess c-FLIP expression. Quantification of c-FLIPL levels: n = 3 from three biological replicates, mean ± s.d. ****P < 0.0001, ***P < 0.001, two-way ANOVA. c WB of HCT116 cells transfected with siRNA MDC1#3 and treated with 100 nM SN-38 for indicated times. d Flow cytometry analysis of Annexin V staining in HCT116 cells transfected with indicated siRNA and treated with 100 nM SN-38 for 48 h. n = 3 from three biological replicates, mean ± s.d. **P < 0.01, ns: not significant, two-way ANOVA. e Working model depicting TIPIN’s roles in regulating replication fork dynamics and the DNA damage response. a The TIM-TIPIN complex is required for efficient replication fork progression. TIM acts as a scaffold of the replisome to interact with the CMG helicase while TIPIN stabilizes TIM. b At stalled forks caused by replisome uncoupling (e.g., HU), TIPIN interacts with RPA to recruit CLASPIN and promote ATR-dependent CHK1 phosphorylation. c In response to replication-associated DSBs (e.g., CPT, SN-38), TIPIN amplifies ATM-CHK2 checkpoint signaling, while TIPIN phosphorylation recruits MDC1 to promote ATM-dependent NF-κB signaling that establishes TIS and upregulates c-FLIP, which sustains cell survival by inhibiting caspase-8 activation. Accordingly, targeting MDC1 or potentially its interaction with TIPIN would act as a way to chemosensitize cells in cytotoxic anticancer therapy. Image created with Biorender.com.
TP53 mutations are a common occurrence in CRC38. While MDC1 knockdown led to increased levels of cleaved caspase-8 p18 when compared to control in both p53-proficient and -deficient HCT116 cells, subsequent caspase-3 activation was much more pronounced upon MDC1 knockdown in a p53-deficient background (Supplementary Fig. S6d; lane 2 and 4 vs. 6 and 8). This is largely due to a lack of p53 available to trigger caspase-3 activation through the intrinsic apoptotic pathway under genotoxic stress. This result suggests that a majority of CRC deficient in p53 may respond better to therapies when the extrinsic c-FLIP/caspase-8 axis is perturbed. Therefore, targeting the TIPIN-MDC1 pathway in NF-κB signaling could provide a means to sensitize CRC cells to irinotecan therapy regardless of the p53 status. Together, our data suggest that TIPIN, and its downstream effector MDC1, is a key regulator of ATM-dependent NF-κB signaling, which dictates the effectiveness of topoisomerase inhibition.
Discussion
Top1 inhibitors such as CPT and its derivatives irinotecan and topotecan, commonly used in the clinic, are known to cause an accumulation of one-ended DSBs at DNA replication forks, forming a unique lesion that originates from SSBs and replication fork stalling. The response to DSBs coupled to DNA replication is expected to be distinct from genome-wide DSB signaling; we herein identify TIPIN, within the replisome, as a new regulator of the ATM pathway in response to DNA replication-associated breaks. TIPIN coordinates ATM-dependent checkpoint and NF-κB signaling, thereby dictating the decision process between cancer cell survival versus death. The ATM-CHK2 checkpoint controls cell cycle and HR-mediated repair to facilitate recovery from fork damage, while the NF-κB axis engages pro-senescence and pro-survival pathways, both of which contribute to the evasion of cancer cells from cytotoxicity caused by clastogens. In addition to TIPIN’s known role, as part of the FPC, in ensuring efficient replisome progression at active forks and promoting ATR-dependent CHK1 phosphorylation under replication stress39, we now propose that TIPIN serves as a regulatory component of the ATM pathway in response to replication-associated DSBs (Fig. 6e). TIPIN amplifies MRN-dependent ATM activation, thereby promoting CHK2 phosphorylation and DNA end resection necessary for the repair of DSBs at stalled forks. Upon ATM activation, TIPIN itself becomes a target of phosphorylation, which helps engage ATM-dependent NF-κB activation. Given that components of the FPC are found to be upregulated in multiple cancers40,41,42, the roles of TIPIN in supporting ATM signaling may contribute to the survival of cancer cells under genotoxic chemotherapy.
Our study identifies several new regulators that determine the chemotherapeutic response to topoisomerase inhibitors, thus dictating fate decision of cancer cells. The first one is MDC1, which directly recognizes γH2AX and ATM through its BRCT and FHA phospho-binding domains and functions as an adaptor that regulates foci formation of DDR factors such as MRN, 53BP1, and BRCA1 at sites of DSBs to transduce DNA damage signals43. In this study, we reveal the interaction between TIPIN and MDC1 at stalled forks, which promotes ATM-dependent NF-κB activation. While a cryo-EM structure of the human replisome does not include the C-terminal region of TIPIN where S222 phosphorylation occurs, the position of the TIM-TIPIN complex at the leading edge of the replisome implies that the C-terminus of TIPIN should protrude from the replisome19. This configuration highlights TIPIN as a first responder that encounters replication barriers ahead of the fork (e.g., Top1cc) to propagate ATM signaling, while its modification may bring MDC1 to one-ended DSBs to engage NF-κB signaling.
Although exact mechanisms through which MDC1 supports nuclear-initiated NF-κB signaling are currently unclear, we envision that MDC1 enriched near DSB sites may help form a nuclear ATM-NEMO complex. Additionally, by recruiting downstream mediators, MDC1 may facilitate a series of enzymatic processes including poly(ADP-ribosyl)ation, ubiquitination, and SUMOylation of NEMO that are necessary for the export of NEMO into the cytosol and subsequent IKK and NF-κB activation44. NF-κB signaling is considered pro-survival under genotoxic stress as it allows for temporal activation of genes necessary for cancer cell survival, providing an opportunity for DNA repair45. This transcriptional program is also involved in cytokine responses associated with TIS, which accounts for chemotherapy resistance and disease recurrence46. We show that MDC1 deficiency leads to a decrease in the induction of TIS, but an increase in apoptotic cell death, indicating that TIPIN-dependent MDC1 regulation plays a key role in swaying the balance of survival versus death through modulation of the NF-κB pathway. Therefore, targeting MDC1 or ideally disrupting the TIPIN-MDC1 interaction could be a way of chemosensitization to suppress TIS and lower the threshold of apoptosis in response to DSB-inducing cancer therapy.
Another layer of regulation we have identified that underlies the cytotoxicity of topoisomerase inhibition is the c-FLIP/caspase-8 axis that determines the onset of apoptosis. c-FLIP is known as a major suppressor of the death receptor-mediated extrinsic apoptotic pathway36. c-FLIP heterodimerizes with procaspase-8 in the Fas-associated via death domain (FADD)-containing death-inducing signaling complex (DISC) to block caspase-8 activation upon the engagement of a death ligand47. c-FLIP also associates with a cytosolic signaling platform named the ripoptosome or complex IIb, which contains procaspase-8, FADD, and receptor-interacting serine/threonine kinase 1 (RIPK1) to activate caspase-8-dependent apoptosis48. In this regard, c-FLIP is considered as a major factor of chemotherapy resistance and poor prognosis in multiple cancers49,50. We herein show that NF-κB-dependent c-FLIP upregulation antagonizes apoptotic signaling in response to topoisomerase inhibitors. Targeting MDC1 shifts the balance of c-FLIP and caspase-8 to sensitize cells to death, therefore defining c-FLIP as a targetable vulnerability to enhance the cytotoxicity of cancer therapy. How nuclear ATM signaling coordinates with the ripoptosome to activate caspase-8 remains unknown. Interestingly, studies have revealed localization of caspase-8 into nucleus under DNA damage and roles of nuclear RIPK1 in chromatin remodeling that is necessary for inflammatory responses51,52. How replication-associated DSB lesions are transduced to the signaling of RIPK1/caspase-8 activation and how the replication machinery plays a role in coordinating these processes with ATM will be important questions to pursue. The present study identifies the replisome itself as a key determinant of chemosensitivity against replication-blocking genotoxic regimens via regulation of ATM signaling. In this regard, TIPIN modification or MDC1 levels may present new biomarkers of drug sensitivity. Further characterization of the crosstalk between the genotoxic stress-induced NF-κB and caspase-8 pathways will help develop strategies to target MDC1 or c-FLIP, as well as new regulators connected to the replication machinery such as the TIPIN-MDC1 interaction, in conjunction with standard chemotherapies to augment the effectiveness of anti-cancer treatment.
Methods
Plasmid construction
pcDNA3-Flag-TIPIN was a gift from Dr. Aziz Sancar (Addgene #22889). The pRetro-X TetOne and pcDNA5/FRT/TO Flp-In T-REx backbones were previously described10,14. pRetro-X TetOne Flag-TIPIN WT was constructed by subcloning Flag-TIPIN cDNA to the pRetro-X TetOne-puro backbone using EcoRI and BamHI restriction sites. cDNA resistant to siRNA TIPIN#1 was generated by introducing two silent mutations within Flag-TIPIN by site-directed mutagenesis (SDM). The construct was subsequently used as a template to generate pRetro-X TetOne Flag-TIPIN S222A. pRetro-X TetOne Flag-TIPIN-N was generated by introducing a premature stop codon after amino acid 173 in TIPIN cDNA. pcDNA5/FRT/TO-HA-TIPIN-Flag-TIM was constructed by subcloning HA-TIPIN cDNA lacking a stop codon followed by a short linker sequence (custom synthesis from Gene Universal) into pcDNA5/FRT/TO-Flag-TIM using HindIII and KpnI restriction sites. SDM was performed to generate pcDNA5/FRT/TO-HA-TIPIN S222A-Flag-TIM. All sequences were confirmed by Sanger DNA sequencing (Stony Brook Genomics Core Facility). DNA primer sequences can be found in Supplementary Table S1.
Cell culture and cell lines
U2OS and HCT116 cell lines were acquired from the American Tissue Culture Collection (ATCC). The HCT116 TP53 wild-type and knockout pair was acquired from Horizon Discovery (HD 104-001). The U2OS TIPIN S222A CRISPR knock-in cell line was established from the Genome Engineering & Stem Cell Center (GESC) at Washington University in St. Louis. The U2OS Flp-In T-REx cell line was provided by Dr. Daniel Durocher (Lunenfeld-Tanenbaum Research Institute). The Flp-In T-REx TIM cell line was described previously14. Cells were cultured in Dulbecco’s Modified Eagle’s Medium (U2OS) or McCoy’s 5A medium (HCT116) supplemented with 10% fetal bovine serum, 1% glutamine, and 1% penicillin/streptomycin, following standard culture conditions and procedures. U2OS pRetro-X TetOne Flag-TIPIN WT and S222A cell lines were generated by retroviral transduction followed by puromycin selection. The Flp-In U2OS HA-TIPIN-Flag-TIM cell line was generated by co-transfecting pcDNA5/FRT/TO-HA-TIPIN-Flag-TIM (TIPIN WT or S222A) and the pOG44 plasmid encoding the Flp recombinase into the host Flp-In T-REx U2OS cell line. Cells were then subjected to hygromycin selection. Doxycycline was used at 100 ng/mL in pRetro-X Flag-TIPIN cells and at 200 ng/mL in pcDNA5/FRT/TO-HA-TIPIN-Flag-TIM cells for cDNA expression.
DNA and siRNA transfection
Plasmid transfection was performed using GeneJuice (Millipore Sigma) according to the manufacturer’s protocol. Retroviral production for pRetro-X TetOne Flag-TIPIN was performed using X-tremeGENETM HP DNA transfection reagent (Millipore Sigma). siRNA duplexes were transfected at 20 nM, unless otherwise indicated, using Lipofectamine RNAiMAX (Thermo Fisher). siRNA targeting sequences can be found in Supplementary Table S1.
Materials and reagents
TIPIN pS222 antibody was produced through GenScript by immunizing a rabbit with a phospho-peptide KLLSN(pS)QTLGND. Chemical information can be found in Supplementary Table S2.
Cell lysis and Western blotting
Cell lysis and Western blotting were performed as previously described53. Cells were trypsinized, washed in ice-cold phosphate-buffered saline (PBS) and lysed in NETN300 (50 mM Tris-HCl pH 7.5, 300 mM NaCl, 0.2 mM EDTA, 1% NP40) lysis buffer supplemented with cOmpleteTM, EDTA-free protease inhibitor cocktails (Roche) and HaltTM phosphatase inhibitor cocktails (Thermo Fisher) for 40 min on ice. Lysates were then cleared by centrifugation at 14,000 rpm at 4 °C for 10 min. For subcellular fractionation, cells were lysed using lysis (S) buffer (10 mM HEPES pH 7.4, 10 mM KCl, 0.05% NP-40) for 15 min on ice and cleared by centrifugation at 14,000 rpm for 10 min (supernatant containing cytosolic proteins was saved as the S fraction). The pellet (nuclei) was washed with lysis (S) buffer, resuspended in P1 low salt buffer (10 mM Tris-HCl pH 7.5, 0.2 mM MgCl2, 1% Triton X-100), incubated on ice for 15 min and centrifuged (supernatant containing nuclear and nonchromatin-associated proteins was saved as the P1 fraction). The remaining pellet (chromatin) was washed with P1 low salt buffer, resuspended in 0.2 N HCl and incubated on ice for 20 min. After centrifugation, supernatant was transferred into a tube containing an equal volume of Tris-HCl pH 8.0 to neutralize acid (these acid-soluble chromatin-associated proteins were saved as the P2 fraction). Protein concentration was determined by Bradford assay (Bio-Rad) and 25–40 µg proteins were analyzed using SDS-PAGE. Protein gels were transferred onto PVDF membranes (Millipore Sigma), blocked in 5% milk or 5% bovine serum albumin (BSA), incubated with primary antibodies at 4 °C overnight, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies. HRP signals were detected by enhanced chemiluminescence (ECL) Western blotting substrates (Thermo Fisher) using the iBright Imaging System (CL1000; Thermo Fisher). Antibody information can be found in Supplementary Table S3.
RADAR assay
RADAR assay was performed according to the protocol previously described54. Briefly, 1 × 106 cells were seeded into 60 mm plates and treated with CPT the next day. Cells were washed once with ice-cold PBS and lysed on the plate using 600 µL DNAzol reagent supplemented with cOmpleteTM, EDTA-free protease inhibitor cocktails (Roche) for 10 min at 4 °C. 300 µL of 100% cold ethanol was added directly to the plate and lysates were transferred to microcentrifuge tubes. Nucleic acid aggregates were disrupted by repeated pipetting and pelleted (20,000 g for 15 min at 4 °C). The pellet was washed twice in 75% ethanol, air dried and dissolved in 0.1 mL ddH2O. Samples were then sonicated (30% amplitude for 10 s) using a QSonica sonicator system with a microtip probe attachment. DNA concentration was quantified using NanoDrop 1000 spectrophotometer and was normalized to 10 ng/µL in ddH2O. For slot blots, 3 µg DNA was loaded in duplicates and probed for Top1 or dsDNA. For SDS-PAGE analysis, 8 µg DNA was digested with 2000 gel units of micrococcal nuclease (NEB) supplemented with 1x nuclease reaction buffer (NEB) by incubation at 37 °C for 30 min. Digested samples were then run on SDS-PAGE gels followed by transferring onto PVDF membranes and Top1 levels detected using Top1 antibody. 8 µg dsDNA was run separately in slot blots as a loading control.
Immunofluorescence
Immunofluorescence was performed as previously described53. Cells were reverse transfected with siRNA and seeded onto multi-well plates. The next day, cells were trypsinized and 15,000 cells were seeded onto coverslips. Following treatments, cells were washed once with cold PBS on ice and fixed with 4% paraformaldehyde for 10 min at room temperature (RT). After three washes with PBS, cells were permeabilized with 0.3% Triton X-100 in PBS for 5 min on ice. Following permeabilization, cells were washed three times with cold PBS and blocked for 30 min at RT using 5% BSA in PBS and incubated with primary antibodies diluted in 1% BSA-PBS for 1 h at 37 °C. Following three PBS washes, cells were incubated with secondary antibodies coupled to fluorophores diluted in 1% BSA in PBS at 1:1000 for 1 h at 37 °C. Following three additional washes with PBS, coverslips were mounted onto glass microscope slides using VectaShield Antifade mounting medium containing DAPI (Vector Lab). Images were captured using an Eclipse Ts2R-FL inverted fluorescence microscope (Nikon) equipped with a Nikon DS-Qi2 digital camera.
Proximity ligation assay
Cells seeded on coverslips were pulsed with 70 µM EdU for 8 min, washed 3 times with PBS and treated with 1 μM CPT for 30 min. After treatment, cells were fixed with 4% paraformaldehyde for 10 min at RT. Cells were then permeabilized with 0.5% Triton X-100 in PBS for 5 min on ice, washed three times with PBS and washed with 1% BSA in PBS for 10 mins. Biotin azide was then conjugated to EdU via click reaction using the Click-iT reaction cocktail according to the manufacturer’s protocol (Thermo Fisher #C10269). Coverslips were then blocked with blocking solution (Millipore Sigma Duolink #DUO92101) for 1 h, followed by incubation with anti-MDC1 (1:1000) and anti-biotin (1:3000) primary antibodies for 1 h at 37 °C. Coverslips were then washed three times with wash buffer A for 5 min each and then incubated with 1x PLA MINUS and PLUS probes diluted in antibody diluent for 1 h at 37 °C. Coverslips were washed again with wash buffer A for 5 min and incubated with ligase diluted 1:40 in 1x ligation buffer for 30 min at 37 °C. Coverslips were then washed and incubated with rolling polymerase diluted 1:80 in 1x amplification buffer for 1 h 40 min at 37 °C. Coverslips were then washed three times with wash buffer B for 10 min each and mounted in mounting media containing DAPI. Images were captured using an Eclipse Ts2R-FL inverted fluorescence microscope (Nikon) equipped with a Nikon DS-Qi2 digital camera and PLA intensity per cell was analyzed using NIS-Elements, Research BR software (Nikon).
Flow cytometry
Flow cytometry was performed as previously described53. For Annexin V assays, cells were processed according to the manufacturer’s protocol (BD Biosciences, 556547). For the flow cytometric β-galactosidase senescence assay, cells were processed according to the manufacturer’s protocol (Thermo Fisher, C10840). All samples were analyzed with the Attune NxT acoustic focusing cytometer (Thermo Fisher) and analyzed with the Attune NxT software v2.7 (ThermoFisher).
RT-qPCR
RT-qPCR was performed as previously described53. RNA isolation was done using TRIzol (Invitrogen) and chloroform (Millipore Sigma). cDNA synthesis was performed using a high-capacity cDNA reverse transcription kit (Thermo Fisher, 4374967) according to the manufacturer’s protocol. Quantitative PCR was performed using Fast SYBR Green Master Mix (Thermo Fisher) and a QuantStudio 3 Real Time PCR system (Thermo Fisher). GAPDH mRNA levels were used as a control for normalization. Primers for RT-qPCR can be found in the Table S1.
Mass spectrometry
Samples were adjusted to 5% SDS, 100 mM triethyl ammonium bicarbonate (TEAB), 10 mM dithiothreitol (DTT) in 100 µL and incubated at 55 °C for 30 min. Proteins were alkylated in 25 mM iodoacetamide (IAc) at RT, for 30 min in the dark. Proteins were precipitated by addition 10 µL of 12% phosphoric acid, followed by 700 µL of S-Trap bind/wash buffer (90% methanol/ 50 mM TEAB) to produce a micro precipitate. Samples were then loaded on an S-Trap mini cartridge (cat. # K02-mini-10 Protifi), washed four times with 90% methanol, 100 mM TEAB, each followed by centrifugation at 4000 x g for 1 minute. The samples were digested with trypsin (20 µg) in 50 mM TEAB in a humified incubator overnight at 37 °C. Peptides were eluted by sequential addition of 80 µL 50 mM TEAB, 0.2% formic acid (FA), and 50% acetonitrile, each followed by centrifugation at 4000 x g for 1 minute. The samples were then dried by SpeedVac and resuspended in 0.1% formic acid and peptides analyzed by C18 reverse phase LC-MS/MS. HPLC C18 columns were prepared using a P-2000 CO2 laser puller (Sutter Instruments) and silica tubing (100 µm ID x 20 cm) and were self-packed with 3 µm Reprosil resin. Peptides typically were separated using a flow rate of 300 nL/minute, and a gradient elution step changing from 0.1% formic acid to 40% acetonitrile (ACN) over 90 min, followed 90% ACN wash and re-equilibration steps. Peptide identification and quantitation was performed using an orbital trap (Q-Exactive HF; Thermo Fisher) instrument followed by protein database searching using Proteome Discoverer 2.4. Replicate samples were analyzed, using two different HPLC gradient profiles (0-30% ACN over 90´ and 0-40% ACN over 90´). Electrospray ionization is achieved using spray voltage of ~2.3 kV. Information-dependent MS acquisitions are made using a survey scan covering m/z 375 – 1400 at 60,000 resolutions, followed by ‘top 20’ consecutive second product ion scans at 15,000 resolutions. AGC targets for MS and MS/MS are 1e6 and 2e5, maximum IT of 100 ms and 50 ms, an MS/MS loop size of 20 and dynamic exclusion for 20 s. Mass resolution cutoffs for MS and MS/MS are 10 ppm and 0.1 Da respectively. Data files were acquired with Xcalibur. Peptide alignments and quantitation were performed using Proteome Discoverer v2.4 software (Thermo Fisher). Protein false discovery rates experiments are binned at 0.01 and 0.05 FDR. Peptide and PSM FDR cutoffs are typically set to 0.01. Two missed tryptic cleavages were allowed and modifications considered included static cysteine derivatization, and variable deamidation (NQ), water loss (ST), oxidation (M), acetylation (K) and phosphorylation (STY). Pairwise peptide log2 ratios between samples allow label free abundance calculations and t-test, based on the background population of peptides. The human UniProt dataset (73,101 entries) is used for data alignment. Matched peptide-based label free quantitation and p values were calculated by Benjamini-Hochberg correction for FDR.
Cell viability ATP assays
To measure cell viability, 2000-4000 cells per well were seeded in 96 well plates and allowed to attach for 24 h. Drugs were treated as specified, and cell viability was determined using the CellTiter-Glo luminescent cell viability assay (Promega). ATP-derived luminescence was measured by the GloMax Explorer microplate reader (Promega).
DNA fiber analysis
Exponentially growing cells were pulse labeled with 50 μM 5-chloro-2´-deoxyuridine (CldU) for 30 min, washed with PBS, and then labeled with 250 μM 5-iodo-2´-deoxyuridine (IdU) for 30 min before harvesting. The pellet was then resuspended in PBS (~3000 nuclei/sample), and 2 μL of the cell suspension was spotted onto a glass slide and lysed with 6 μL fiber lysis solution (200 mM Tris-HCl [pH7.5], 50 mM EDTA, 0.5% SDS) for 5 min followed by tilting the slide to allow DNA fibers to spread. Slides were then air dried for 10-15 min and DNA was fixed in methanol/acetic acid solution (3:1) for 5 min at RT. Slides were then washed in PBS for 5 min followed by DNA denaturation by incubating in a 2.5 M HCl solution for one hour. Slides were then washed in PBS, blocked in 5% BSA for 30 min, and incubated 2 hours with rat monoclonal anti-BrdU (1:200) for CldU and mouse monoclonal anti-BrdU (1:20) for IdU. Following washes in PBS containing 0.025% Tween 20 (PBS-T), slides were incubated with secondary antibodies (Alexa Fluor 594 goat anti-rat and Alexa Fluor 488 goat anti-mouse, 1:400 each) and incubated for 1 h. After PBS-T washes, slides were mounted with ProLongTM Gold Antifade Mounting media (Thermo Fisher) and imaged with an Eclipse Ts2R-FL inverted fluorescence microscope (Nikon) equipped with a Nikon DS-Qi2 digital camera and analyzed using NIS-Elements, Research BR software (Nikon).
Electromobility shift assay (EMSA) and luciferase assay
EMSA was performed as previously described using the Odyssey EMSA kit (LICORBio, 829-07910) in which nuclear extract is incubated with IRDye intrared dye end-labeled NF-κB consensus oligonucleotide (829-07924)55. The reaction mixture was subjected to 4% native gel electrophoresis at 200 V, and the gel was imaged using LICOR Odyssey F imager (LICORBio). A luciferase assay was performed using Nano-Glo Dual-Luciferase Reporter (DLR) assay system (Promega, N1610) in cells co-transfected with the NanoLuc reporter vector with NF-κB response element (pNL3.2 NF-κB-RE NlucP; Promega, N1111) and the pGL4.54 [luc2/TK] firefly luciferase vector (Promega, E5061) as a control reporter according to manufacturer’s instructions. Reporter-derived luminescence was measured by a GloMax Explorer microplate reader, and the NanoLuc signals were normalized by the luc2 firefly signals (Promega).
DNA comet assay
DNA comet assay was performed using the CometAssay kit (4250-050-K, R&D Systems) as previously described53. Comets were imaged at 10× magnification with an Eclipse Ts2R-FL inverted fluorescence microscope (Nikon) equipped with a Nikon DSQi2 digital camera and analyzed using OpenComet on Fiji and Prism (GraphPad). Up to at least 100 individual nuclei were evaluated per group.
Statistics and Reproducibility
All statistical analyses were performed using Prism (GraphPad) and in all cases, ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. The comparison of two groups of variables that follow a normal distribution was done with an unpaired Student’s t test with a 95% confidence interval, using two-tailed p values on a set of at least 3 biological replicates. Where indicated, Student’s t tests were used to compare means of replicates. In experiments that involve two factors, a two-way ANOVA was performed followed by correcting of multiple comparisons using the Dunnett test. For DNA fiber assay, distribution of data was tested using a two-side Mann-Whitney U-test, with a 95% confidence interval.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are available within the Article and Supplementary Information. The raw and processed data from Mass Spectrometry are available through the ProteomeXchange Consortium via the MassIVE partner repository (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) under the dataset Identifier MSV000097385. Unprocessed Western blot images can be found in Supplementary Fig. S7. Numerical source data for graphs are provided in a Supplementary Data file. Correspondence and request for materials should be addressed to Hyungjin Kim, PhD.
References
da Costa, A., Chowdhury, D., Shapiro, G. I., D’Andrea, A. D. & Konstantinopoulos, P. A. Targeting replication stress in cancer therapy. Nat. Rev. Drug Discov. 22, 38–58 (2023).
Groelly, F. J., Fawkes, M., Dagg, R. A., Blackford, A. N. & Tarsounas, M. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer 23, 78–94 (2023).
Wang, L., Lankhorst, L. & Bernards, R. Exploiting senescence for the treatment of cancer. Nat. Rev. Cancer 22, 340–355 (2022).
Blackford, A. N. & Jackson, S. P. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell 66, 801–817 (2017).
Miyamoto, S. Nuclear initiated NF-kappaB signaling: NEMO and ATM take center stage. Cell Res. 21, 116–130 (2011).
Huang, T. T., Wuerzberger-Davis, S. M., Wu, Z. H. & Miyamoto, S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell 115, 565–576 (2003).
Janssens, S. & Tschopp, J. Signals from within: the DNA-damage-induced NF-kappaB response. Cell Death Differ. 13, 773–784 (2006).
Patel, J. A. & Kim, H. The TIMELESS effort for timely DNA replication and protection. Cell Mol. Life Sci. 80, 84 (2023).
Patel J. A. et al. Replisome dysfunction upon inducible TIMELESS degradation synergizes with ATR inhibition to trigger replication catastrophe. Nucleic Acids Res. https://doi.org/10.1093/nar/gkad363 (2023).
Rageul, J. et al. SDE2 integrates into the TIMELESS-TIPIN complex to protect stalled replication forks. Nat. Commun. 11, 5495 (2020).
Petropoulos, M. et al. Transcription-replication conflicts underlie sensitivity to PARP inhibitors. Nature 628, 433–441 (2024).
Saldanha, J. et al. The TIMELESS and PARP1 interaction suppresses replication-associated DNA gap accumulation. Nucleic Acids Res. 52, 6424–6440 (2024).
Shyian, M. et al. Fork pausing complex engages topoisomerases at the replisome. Genes Dev. 34, 87–98 (2020).
Rageul, J. et al. Poly(ADP-ribosyl)ation of TIMELESS limits DNA replication stress and promotes stalled fork protection. Cell Rep. 43, 113845 (2024).
Kemp, M. G. et al. Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J. Biol. Chem. 285, 16562–16571 (2010).
Unsal-Kacmaz, K. et al. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol. Cell Biol. 27, 3131–3142 (2007).
Yoshizawa-Sugata, N. & Masai, H. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J. Biol. Chem. 282, 2729–2740 (2007).
Chou, D. M. & Elledge, S. J. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc. Natl. Acad. Sci. USA 103, 18143–18147 (2006).
Jones, M. L., Baris, Y., Taylor, M. R. G. & Yeeles, J. T. P. Structure of a human replisome shows the organisation and interactions of a DNA replication machine. EMBO J. 40, e108819 (2021).
Pommier, Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer 6, 789–802 (2006).
Pommier, Y., Leo, E., Zhang, H. & Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 17, 421–433 (2010).
Sun, Y., Saha, L. K., Saha, S., Jo, U. & Pommier, Y. Debulking of topoisomerase DNA-protein crosslinks (TOP-DPC) by the proteasome, non-proteasomal and non-proteolytic pathways. DNA Repair 94, 102926 (2020).
Makharashvili, N. & Paull, T. T. CtIP: A DNA damage response protein at the intersection of DNA metabolism. DNA Repair 32, 75–81 (2015).
Anand, R., Ranjha, L., Cannavo, E. & Cejka, P. Phosphorylated CtIP functions as a co-factor of the MRE11-RAD50-NBS1 endonuclease in DNA end resection. Mol. Cell 64, 940–950 (2016).
Arnaudeau, C., Lundin, C. & Helleday, T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 307, 1235–1245 (2001).
Matsuoka, S. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 (2007).
Heinz, N. et al. Retroviral and transposon-based tet-regulated all-in-one vectors with reduced background expression and improved dynamic range. Hum. Gene Ther. 22, 166–176 (2011).
Huang, T. T. et al. NF-kappaB activation by camptothecin. A linkage between nuclear DNA damage and cytoplasmic signaling events. J. Biol. Chem. 275, 9501–9509 (2000).
Wu, Z. H., Shi, Y., Tibbetts, R. S. & Miyamoto, S. Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science 311, 1141–1146 (2006).
Mohammad, D. H. & Yaffe, M. B. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair 8, 1009–1017 (2009).
Lou, Z., Minter-Dykhouse, K., Wu, X. & Chen, J. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature 421, 957–961 (2003).
Stewart, G. S., Wang, B., Bignell, C. R., Taylor, A. M. & Elledge, S. J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421, 961–966 (2003).
Goldberg, M. et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421, 952–956 (2003).
Liptay M. et al. MDC1 counteracts restrained replication fork restart and its loss causes chemoresistance in BRCA1/2-deficient mammary tumors. bioRxiv, 2022.2008.2018.504391 (2022).
Shahbandi, A. et al. BH3 mimetics selectively eliminate chemotherapy-induced senescent cells and improve response in TP53 wild-type breast cancer. Cell Death Differ. 27, 3097–3116 (2020).
Ivanisenko, N. V. et al. Regulation of extrinsic apoptotic signaling by c-FLIP: towards targeting cancer networks. Trends Cancer 8, 190–209 (2022).
Fuchs, C., Mitchell, E. P. & Hoff, P. M. Irinotecan in the treatment of colorectal cancer. Cancer Treat. Rev. 32, 491–503 (2006).
Iacopetta, B. TP53 mutation in colorectal cancer. Hum. Mutat. 21, 271–276 (2003).
Saldanha J., Rageul J., Patel J. A. & Kim H. The adaptive mechanisms and checkpoint responses to a stressed DNA replication fork. Int. J. Mol. Sci. 24, 10488 (2023).
Elgohary, N. et al. Protumorigenic role of timeless in hepatocellular carcinoma. Int. J. Oncol. 46, 597–606 (2015).
Yoshida, K. et al. TIMELESS is overexpressed in lung cancer and its expression correlates with poor patient survival. Cancer Sci. 104, 171–177 (2013).
Bianco, J. N. et al. Overexpression of claspin and timeless protects cancer cells from replication stress in a checkpoint-independent manner. Nat. Commun. 10, 910 (2019).
Lou, Z. et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 21, 187–200 (2006).
McCool, K. W. & Miyamoto, S. DNA damage-dependent NF-kappaB activation: NEMO turns nuclear signaling inside out. Immunol. Rev. 246, 311–326 (2012).
Guo, Q. et al. NF-kappaB in biology and targeted therapy: new insights and translational implications. Signal. Transduct. Target Ther. 9, 53 (2024).
Schmitt, C. A., Wang, B. & Demaria, M. Senescence and cancer—role and therapeutic opportunities. Nat. Rev. Clin. Oncol. 19, 619–636 (2022).
Schleich, K. et al. Molecular architecture of the DED chains at the DISC: regulation of procaspase-8 activation by short DED proteins c-FLIP and procaspase-8 prodomain. Cell Death Differ. 23, 681–694 (2016).
Tenev, T. et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol. Cell 43, 432–448 (2011).
McLaughlin, K. A. et al. FLIP: a targetable mediator of resistance to radiation in non-small cell lung cancer. Mol. Cancer Ther. 15, 2432–2441 (2016).
Longley, D. B. et al. c-FLIP inhibits chemotherapy-induced colorectal cancer cell death. Oncogene 25, 838–848 (2006).
Muller, I. et al. Cancer cells employ nuclear caspase-8 to overcome the p53-dependent G2/M checkpoint through cleavage of USP28. Mol. Cell 77, 970–984.e977 (2020).
Li, W. et al. Nuclear RIPK1 promotes chromatin remodeling to mediate inflammatory response. Cell Res. 32, 621–637 (2022).
Patel J. A. et al. The DNA-PKcs/JNK/p53 pathway underlies changes in cell fate decision toward death during DNA replication catastrophe. Nucleic Acids Res. 53, gkaf573 (2025).
Sun Y. Quantitative Detection of DNA-protein crosslinks and their post-translational modifications. J. Vis. Exp. 194, e65315 (2023).
Weinheimer, A. S. et al. Extended DNA-binding interfaces beyond the canonical SAP domain contribute to the function of replication stress regulator SDE2 at DNA replication forks. J. Biol. Chem. 298, 102268 (2022).
Acknowledgements
We would like to thank the Biological Mass Spectrometry Shared Resource at Stony Brook University for IP-MS analysis and the Genome Engineering & Stem Cell Center (GESC) at Washington University School of Medicine for CRISPR/Cas9 gene editing service. H.K. discloses support for the research of this work from National Institutes of Health R01GM144399 & R01CA285515, the Center for Healthy Aging comprehensive grant, and Carol Baldwin Breast Cancer Research Award.
Author information
Authors and Affiliations
Contributions
A.K. designed and performed all of the molecular and cellular experiments with the help of J.F.L. N.L., and J.A.P. provided technical assistance. J.H. performed LC-MS/MS and analyzed data. H.K. conceptualized and supervised the project. H.K. wrote the manuscript with the assistance of A.K.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Shilpee Dutt and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Valeria Naim and Mengtan Xing. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Khan, A., Lim, J.F., Lo, N. et al. TIPIN coordinates ATM-dependent checkpoint and NF-κB signaling to counteract DNA replication damage from topoisomerase inhibition. Commun Biol 8, 1683 (2025). https://doi.org/10.1038/s42003-025-09084-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09084-7