Abstract
Obesity is associated with impaired spermatogenesis and decreased sperm quality, in part through reducing Sertoli cells (SCs) lactate and nicotinamide adenine dinucleotide (NAD+) production. It is not known whether nicotinamide mononucleotide (NMN) treatment improves spermatogenesis. In the present study, NMN improved lipid metabolism and enhanced spermatogenesis in obese mice, alleviated SCs dysfunction in vivo and in vitro. Label-free quantitative acetylomics analysis of mouse testes suggested that protein acetylation influenced both the structural and functional properties of metabolic proteins. The beneficial effects of NMN were due in part to changes in the acetylation of glycolysis-related proteins. Furthermore, multi-omics and correlation analyses demonstrated that interactions among the gut microbiota, metabolites, spermatogenesis, and LDHC acetylation mediated the beneficial effects of NMN. Importantly, we found that NMN treatment reduced acetylation of the lysine residues 5, 17, and 90 of LDHC, which plays a critical role in SC lactate production in obesity. Collectively, our findings show that NMN supplementation improves sperm quality in obese mice by decreasing LDHC acetylation, thereby increasing Sertoli cell lactate and NAD+ production.

Similar content being viewed by others
Introduction
Obesity is a chronic metabolic disease caused by multiple factors that has been defined as a 21st century epidemic by the World Health Organization (WHO)1. Obesity is associated with a variety of chronic conditions such as hyperlipidemia, hyperglycemia, diabetes, and low-grade inflammation2,3. In addition, obesity has been shown to reduce fertility in men, as well as in animal models. A large Danish study involving around 48,000 male participants reported a dose-response relationship between body mass index (BMI) and infertility, and a more recent clinical study showed that overweight or obese men have decreased sperm quality, including lower sperm concentrations and reduced sperm motility4. In addition, high fat diet (HFD)-induced animal models of obesity exhibit impaired spermatogenesis, such as a disrupted testicular structure, decreased sperm concentration, and reduced sperm motility5.
As well as affecting sperm parameters, obesity has also been shown to influence the structure and function of Sertoli cells (SCs). SCs are somatic cells in the testes that provide the energy needed for germ cells to develop into spermatozoa and form a blood-testis barrier (BTB) to protect the spermatogenic milieu6. Studies have shown that an HFD disrupts the glucose-lactate metabolism cycle by inhibiting the expression of GLUT-1/3 and MCT-4 in the SCs of the testes, thereby affecting sperm energy supply found that the number of SCs decreased, and the remaining SCs were atrophied, in an HFD-induced rat model of obesity7. In addition, the integrity of the BTB was disrupted in mice fed an HFD8. A 2024 Nature study showed that a short-term (2-week) HFD exposure in paternal rats can induce mitochondrial tRNA (mt-tRNA) fragmentation in sperm, leading to glucose intolerance in offspring. These mt-tRNA fragments are integrated into the embryonic genome through epigenetic mechanisms, disrupting early embryonic development and metabolic regulation in adult offspring. Similarly, human cohort studies have demonstrated that paternal BMI is significantly associated with the risk of insulin resistance in the offspring, independent of the metabolic status of the mother9.
Obesity not only affects the ability of SCs to maintain the integrity of the BTB, but also impairs SCs-mediated energy production. The testis is a naturally oxygen-deprived organ, and SCs provide the energy needed for male germ cell differentiation by generating lactate and pyruvate via glycolysis10,11. Obesity causes abnormal glucose metabolism, which manifests as decreased lactate levels and lactate dehydrogenase (LDH) activity in SCs12,13 and results in impaired spermatogenesis14. LDH catalyzes the reduction of pyruvate to lactate with the concomitant oxidation of NADH to NAD+, which is positively correlated with lactate levels in both tissues and cells15.
NAD+ is a central metabolite involved in protein acetylation and regulation of energy homeostasis16. NAD+ biosynthesis in the metabolic organs is severely compromised in HFD-induced animal models of obesity17, and decreased NAD+ levels and increased protein acetylation have been observed in multiple tissues from obesity models18. Furthermore, low NAD+ levels have been shown to impair glycolysis19, thereby preventing cells from rapidly generating ATP. Administration of NAD+ precursors to obese mice effectively increases NAD+ levels and sirtuin activity in multiple tissues20,21, which in turn decreases global protein acetylation and improves metabolic homeostasis22. In addition, increasing NAD+ levels has been shown to protect animals from HFD-induced liver disease and insulin resistance20. Nicotinamide mononucleotide (NMN) is an essential precursor molecule of NAD+ and plays an important role in the prevention of obesity-related complications22. Supplementation with NMN increases hepatic NAD+ levels and prevents hepatic lipid accumulation in pre-clinical models of NAFLD23, as well as improving HFD-induced glucose intolerance and dyslipidemia24. Thus, increasing NAD+ levels via NMN administration can improve several symptoms of obesity; however, it remains unclear whether this approach could help reverse impaired spermatogenesis.
The aim of this study was to determine whether increasing NAD+ levels in an HFD-induced mouse model of obesity would improve spermatogenesis and, if so, to explore the underlying mechanisms. We found that supplementation with NMN, an NAD+ precursor, decreased body weight, improved lipid metabolism, and ameliorated sperm quality in obese mice. Importantly, we found that NMN treatment decreased acetylation of LDHC, a testis-specific LDH, thereby increasing SCs lactate and NAD+ production, enhancing the expression of tight junction proteins by SCs, and improving spermatogenesis in these obese mice. Our findings provide new insight into the mechanisms responsible for reduced fertility in men and suggest potential treatment targets.
Results
NMN treatment decreases body weight and enhances spermatogenesis in obese mice
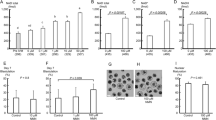
To examine the effects of NMN on obesity and spermatogenesis, we first established a mouse model of obesity by feeding wild-type mice normal chow or an HFD. After 12 weeks, the mice fed an HFD (the Obe group) exhibited significantly higher body weights than the Ctrl mice (Fig. 1A, B). Starting on week 13, half of the mice fed an HFD also received NMN daily by oral gavage (the Obe+NMN group). Eight weeks of treatment with NMN significantly reduced the body weight of the mice in the Obe+NMN group (Fig. 1C).
A The general appearance of the mice at end of the study. B Mice body weight in the HFD and CD groups (n = 10). C Mice body weight of the Ctrl, Obe, and Obe+NMN groups in response to each diet and treatment (n = 12). D Testicular index of mice in the Ctrl, Obe, and Obe+NMN groups (n = 12). E Epididymal index (1); sperm count (2); abnormal sperm rate (3); and sperm motility (4) of mice in the Ctrl, Obe, and Obe+NMN groups (n = 12). F HE staining of the epididymis in the Ctrl, Obe, and Obe+NMN groups; Scale bars 100 μm (upper) and 50 μm (lower). G HE staining of the testis in the Ctrl, Obe, and Obe+NMN groups; Scale bars 100 μm (upper) and 50 μm (lower). H Seminiferous tube area (1); and seminiferous tube diameter (2) in the Ctrl, Obe, and Obe+NMN groups. All data are presented as median with interquartile range or mean ± SEM. *p < 0.05; **p < 0.01.
Next, we performed HE staining of adipose tissue to detect differences in adipocyte structure and size among the three groups. Compared with the Ctrl group, adipocytes in adipose tissue of the obese mice were larger, but their size decreased dramatically after NMN treatment (Supplementary Fig. 1A). Consistent with this, NMN treatment also markedly reduced fat mass, Lee’s obesity index, fat diameter, and fat area in obese mice (Supplementary Fig. 1B).
To assess the effects of the HFD on mouse spermatogenesis, we analyzed the epididymal sperm count, sperm motility, morphologically abnormal sperm count, and cellular structure of the testes in each group of mice. The sperm count and sperm motility were lower, and the abnormal sperm count was higher, in the obese mice compared with the Ctrl mice, and these changes were reversed by treatment with NMN (Fig. 1D, E). Furthermore, morphological analysis showed that the cells in the epididymal duct wall of the obese mice were loosely and irregularly arranged, with a small number of vacuoles in the cytoplasm (Fig. 1F), while the cellular organization and vacuoles in Obe+NMN mice were similar to those seen in the Ctrl group (Fig. 1G). Importantly, NMN treatment also increased the mean area and diameter of the seminiferous tubules compared with the Obe group (Fig. 1H).
Taken together, these results suggest that NMN treatment decreases body weight, adipocyte size, and obesity indices, as well as improving sperm quality and testicular structure, in obese mice.
Treatment with NMN ameliorates disrupted lipid metabolism in obese mice
Given that obesity increases lipid accumulation in the body25, and NMN decreased obesity in our mouse model, we next asked whether NMN would affect lipid deposition (also known as steatosis), serum lipid levels, and serum lipid metabolite levels in obese mice.
To assess steatosis, mice from all three groups were sacrificed after NMN intervention, and their livers were removed and stained with HE and oil red O to detect lipid droplets. Compared with the Ctrl group, there were significantly more lipid droplets in the obese group, indicating that obesity causes liver steatosis in mice (Fig. 2A). In contrast, fewer intracellular lipid droplets were observed in the Obe+NMN group than in the Obe group (Fig. 2B). These results suggest that NMN treatment reduced hepatic steatosis in obese mice.
A Levels of the lipid parameters TC (1), TG (2), LDL (3), and HDL (4) in the Ctrl, Obe, and Obe+NMN groups (n = 5). B HE staining of the liver. The black arrows indicate fat vacuoles. C Oil red O staining of liver. The green arrows indicate fat droplets. D Partial least squares discrimination analysis (PLS-DA) score plots for the Ctrl and Obe groups in positive ion mode (1) and negative ion mode (2) and the Obe and Obe+NMN groups in positive ion mode (3) and negative ion mode (4) (Ctrl, n = 10; Obe, n = 10; Obe+NMN, n = 10). E Volcano plot of differentially expressed metabolites between the Obe and Ctrl groups (1) and the Obe and Obe+NMN groups (2). F Heatmap of metabolites associated with lipid metabolism whose expression levels differed in serum samples from the different groups. G KEGG classification map and KEGG enrichment bubble map of differentially expressed metabolites in the Ctrl and Obe groups (1,2) and in the Obe and Obe+NMN groups (3,4). H Correlations among differentially expressed metabolites and fat mass, lipid metabolism indicators, and male reproductive function indicators (1); correlation network diagram of lipid metabolism indicators and differentially expressed metabolites (2). All data are presented as median with interquartile range or mean ± SEM. *p < 0.05; **p < 0.01.
Next, we analyzed lipid levels in serum samples taken from all three groups after 8 weeks of treatment. As shown in Fig. 2C, triglyceride (TG), total cholesterol (TC), and low-density lipoprotein (LDL) levels were significantly decreased, and the high-density lipoprotein (HDL) level was significantly increased, in obese mice compared with Ctrl mice. Furthermore, treatment with NMN restored the levels of all four lipids to levels comparable to those seen in NC mice.
To analyze the serum levels of metabolites associated with lipid metabolism, we performed serum metabolomics analysis of blood samples taken from mice on week 18 of the experiment. PLS-DA confirmed the reliability of the results (Fig. 2D, Supplementary Fig. 2A), which were then used to construct volcano plots of the differentially expressed metabolites. In total, 72 metabolites were upregulated and 143 metabolites were downregulated in the serum of Obe mice compared to control mice, and NMN treatment partially reversed these changes (Fig. 2E). Next, differentially expressed metabolites identified by carrying out pairwise comparisons between different groups were subjected to hierarchical clustering analysis to determine differences in metabolic expression patterns between and within each pair of groups. The results showed that the expression levels of the differentially expressed metabolites in the Obe group were significantly different from those in the Ctrl and Obe+NMN groups, and were similar between the Obe+NMN group and the Ctrl group (Fig. 2F, Supplementary Fig. 2B). Interestingly, we found that the expression levels of CAR 16:1, CAR 18:2, CAR 18:0, CAR 17:0, and CAR 20:2, metabolites that affect reproductive function, were elevated in the Obe group and decreased after NMN treatment (Fig. 2F). Pathway analysis of co-expressed differentially expressed metabolites showed that they were mainly enriched in the lipid metabolism and energy metabolism pathways (Fig. 2G (1), (2)), and that HFD may affect mouse metabolism through the metabolism, amino acid biosynthesis, tyrosine metabolism, phenylalanine metabolism, tryptophan metabolism, and TCA cycle pathways (Fig. 2G (3)).
Subsequently, we assessed the interactions among the differentially expressed metabolites and various indicators of spermatogenesis and lipid metabolism. Spearman correlation analysis showed that PC 38:5, which promotes fat metabolism26, prevents fatty liver, and lowers serum cholesterol levels, was positively correlated with sperm motility, sperm count, epididymal and testicular indices, and HDL levels (Fig. 2H (1)). In addition, network correlation analysis showed that HDL was negatively correlated with 11-deoxy prostaglandin F1a, PC 38:5, 3-methylindole, and methyl isoquinoline-3-carboxylate that, in turn, were positively correlated with LDL, TG, and TC (Fig. 2H (2)).
Taken together, these results show that impaired spermatogenesis in obese mice is associated with changes in lipid metabolism and that these changes can be reversed by treatment with NMN.
NMN treatment partially reverses the negative effects of obesity on the gut microbiota
Clinical studies have shown that the gut microbiota is structurally dysregulated in obese patients and that disruption of the gut microbiota accelerates abnormal lipid metabolism and impairs spermatogenesis. Therefore, we next asked whether NMN improves spermatogenesis in obese mice by altering the gut microbiota. UniFrac-based principal coordinate analysis (PCoA) (Supplementary Fig. 3A) and non-metric multidimensional scaling (NMDS) (Supplementary Fig. 3B) analyses showed significant differences in the composition of the gut microbiota between the Obe group and the control group and indicated that NMN treatment altered the gut microbiota in the Obe group, but not significantly. Venn diagrams were constructed to visualize the abundance of common and unique amplicon sequence variants across the groups (Supplementary Fig. 3C).
To further evaluate the effects of NMN treatment on the composition of the mouse gut microbiota, we compared the composition of the gut microbiota from each group at the phylum, genus, and family level. As shown in Supplementary Fig. 3D, the gut microbiota in all three groups was mainly composed of the phyla Firmicutes, Deferribacterota, Bacteroidetes, Actinobacteria, and Verrucomicrobiota. The Firmicutes/Bacteroidota ratio, which is increase in obesity27, was clearly higher in the Obe group than in the Ctrl group, and decreased after NMN treatment, although this difference was not significant (Supplementary Fig. 3E). At the phyla level, the relative abundance of Bacteroidota was reduced, and that of Proteobacteria was increased, in the Obe group compared with the control group, while NMN treatment further reduced the levels of Bacteroidota and Proteobacteria (Supplementary Fig. 3D (1), H). At the genus level, the relative abundance of Ligilactobacillus and Escherichia-Shigella was increased in the Obe group compared with the Ctrl group and improved after NMN treatment, and the relative abundance of Colidextribacter and Lachnospiraceae NK4A136 group was decreased in the Obe group compared with the NC group and improved after NMN treatment (Supplementary Fig. 3D (2)). We also found that Bacteroides caecimuris, which has hepatoprotective effects28, and Lachnospiraceae bacterium 28-4, which is associated with body weight reduction29, were the dominant species in Obe+NMN mice, while Lachnospiraceae bacterium 10-1, which is associated with obesity, and the pathogenic bacterium Mucispirillum schaedleri30 were the dominant species in Obe mice (Supplementary Fig. 3I).
Given that the diversity of the gut microbiota is also an important determinant of obesity, next we assessed the alpha and beta diversity of the intestinal flora in the three groups of mice. The Simpson index, a measure of alpha diversity, was significantly higher in the Obe group than in the Ctrl group, but the Shannon index (another alpha diversity metric) was not; however, both the Simpson and Shannon indices were significantly higher in the NMN group than in the Ctrl group (Supplementary Fig. 3F), indicating that obesity markedly alters gut microbiota diversity in correlation with alterations in indicators of lipid metabolism and spermatogenesis, and that treatment with NMN partly reversed these effects. Furthermore, analyzing beta diversity by measuring the Bray–Curtis distance showed that obesity increased species abundance, and treatment with NMN further increased the abundance of species within the gut microbiota (Supplementary Fig. 3G, Supplementary Table 3).
In order to identify specific groups of bacteria that differed between groups, linear discriminant analysis effect size (Lefse) analysis was performed, using an LDA score of 4.0. Lefse to detect bacterial branching from phylum to genus, and statistical and correlation analyses of the microbiota were carried out. In total, the abundance of 12, 8 and 9 species differed significantly between the Ctrl, Obe, and Obe+NMN groups, respectively (Supplementary Fig. 3J). The Obe group was mainly enriched to Campylobacteria and Helicobacter, and the abundance of some species associated with obesity and metabolism, such as Lachnospiraceae_bacterium_28_4, Blautia, and Lachnospiraceae, increased after NMN treatment (Supplementary Fig. 3K).
Taken together, these findings suggest that NMN treatment alters the gut microbiota in obese mice, but not to the same extent as that seen in obese mice compared with control mice.
NMN treatment alters the abundance of gut microbiota components that interact functionally with lipid metabolites to impair spermatogenesis
Given the subtle changes that we observed in the gut microbiota of obese mice treated with NMN, next, we predicted the function of the gut microbiota components in the three groups using PICRUSt to determine the potential impact of these changes. Functional abundance at each classification level was analyzed using the KEGG database, and a Venn diagram was constructed to depict the number of unique and shared genes among the different groups (Supplementary Fig. 4A). At level 1, the most abundant gut microbiota components among the three groups were enriched in metabolism, human diseases, and cellular processes (Supplementary Fig. 4B). Single-gene annotation showed that the major genes were involved in metabolic pathways, mainly those related to carbohydrate, metabolic diseases, energy, and lipid metabolism (Supplementary Fig. 4C). A functional abundance clustering heatmap showed that an increase in metabolic diseases, cell growth, and cell death among microbial functions in the Obe group that decreased after NMN treatment (Supplementary Fig. 4D). Next, we conducted spearman correlation analysis among differential gut microbiota, differential serum metabolites, lipid metabolism-related indicators, and semen quality-related indicators. The results showed significant positive and negative correlations between gut microbes and differentially expressed metabolites. Specifically, Colidextribacter, Helicobacter, Bacteroides, and Escherichia-Shigella correlated positively with N-(2-cyanoethyl)-N’-(2,6-dimethylphenyl)-N-isopropylthiourea, N-Arachidonoyl-L-serine, and CAR-related metabolites; Bacteroides, Helicobacter, and Colidextribacter were positively correlated with LDL, TC, TG, and abnormal sperm count (Supplementary Fig. 4E); and Enterorhabdus, Akkermansia, and Dubosiella were positively correlated with HDL, sperm count, sperm viability, and testicular and epididymal indices (Supplementary Fig. 4F). These results imply that gut microorganisms and metabolites can interact with each other, and that NMN can reduce the negative effects of obesity on spermatogenesis by altering the abundance of gut microorganisms that impair sperm development.
Treatment with NMN promotes glycolysis in the testes of obese mice
The metabonomics and gut microbiome analyses suggested that NMN improves spermatogenesis in obese mice by affecting energy metabolism. Therefore, we performed transcriptome sequencing of testicular tissue to determine whether NMN treatment alters the expression of genes related to energy metabolism in the testes. PCA clearly separated the Obe group from the Ctrl group, and the Obe+NMN group was similarly distinct from the Ctrl group (Fig. 3A). Compared with the control group, 1426 genes were upregulated and 1487 genes were downregulated in the Obe group. In addition, 426 genes were upregulated and 370 genes were downregulated in the Obe+NMN group compared with the Obe group (Fig. 3B). A total of 278 genes were differentially expressed among the three groups, 273 of which exhibited opposite expression trends in Obe mice versus Obe+NMN mice compared with Obe mice versus Ctrl mice (Fig. 3C). Trend clustering analysis of the differentially expressed genes showed that the gene expression levels in the Obe group were significantly different from those in the Ctrl and Obe+NMN groups, and the gene expression levels in the Obe+NMN group were similar to those in the Ctrl group (Fig. 3D). Functional enrichment analysis using the KEGG and GO databases showed that the differentially expressed genes were significantly enriched in metabolic pathways such as the energy metabolism pathway, the lipid metabolism pathway, and the insulin metabolism pathway (Fig. 3E). Further cluster analysis of the expression levels of differentially expressed genes enriched in these pathways showed that expression of the testis-specific glycolysis rate-limiting enzyme LDHC was downregulated in the Obe group and upregulated after NMN treatment (Fig. 3F). Moreover, qRT-PCR analysis confirmed that the expression of genes encoding Occludin, CX43, the anti-apoptotic factor Bcl2, the key glycolysis rate-limiting enzyme Hk2, Pkm2, Ldha, and Ldhc was downregulated, and that the expression of genes encoding the pro-apoptotic factors Bax and Caspase-3 was upregulated in the Obe group compared with the Ctrl group, while these effects were partially reversed by NMN treatment (Fig. 3G–I). Immunohistochemistry (IHC) and western blotting confirmed the qRT-PCR results (Supplementary Fig. 5C–H).
A Principal component analysis of the Ctrl, Obe, and Obe+NMN groups. B Volcano plot of testicular DEGs between the Obe and Ctrl groups (1) and the Obe+NMN and Obe groups (2). C Venn diagram of DEGs. D KEGG functional enrichment analysis. E Heatmap of differentially expressed genes. F Cluster analysis of differentially expressed genes in the mouse testes. G Occludin (1) and Cx43 (2) mRNA expression levels, as detected by qRT-PCR (n = 3). H Bcl2 (1), Bax (2), and Caspase-3 (3) mRNA expression levels, as detected by qRT-PCR (n = 3). I Hk2 (1), Pkm2 (2), Ldha (3), and Ldhc (3) mRNA expression levels, as detected by qRT-PCR (n = 3). All data are presented as median with interquartile range or mean ± SEM. n = 4 *p < 0.05; **p < 0.01.
IHC analysis of Vimentin, an SC marker, showed that obese mice exhibited a significantly lower number of Vimentin-positive cells in the testes than the Ctrl group, and this number increased significantly after NMN treatment (Supplementary Fig. 5A, B). Furthermore, lactate and NAD+ expression in the testes were significantly lower in Obe mice compared with NC mice and significantly higher in Obe+NMN mice compared with Obe mice (Supplementary Fig. 5I), indicating that, in addition to improving energy metabolism in obese mice, NMN also improves SCs lactate production.
These results suggest that NMN-induced improvement of sperm cell morphology and function in obese mice may be mediated by the glycolytic pathway.
NMN treatment inhibits LDHC acetylation in obese mice
Many enzymes involved in glycolysis are modified by lysine acetylation, which may affect enzyme activity and regulate metabolic responses in the testes. Given that our transcriptome analysis suggested a role for glycolysis in NMN-mediated improvement of spermatogenesis in obese mice, we next examined whether lysine acetylation level of key enzymes involved in glycolysis influenced glucose metabolism homeostasis in the testes. Panacetylation levels in testicular tissue were elevated in the Obe group compared with the Ctrl group and decreased after NMN treatment (Fig. 4A). Furthermore, LC-MS/MS analysis of protein acetylation in testicular tissue (Supplementary Figs. 6–8) showed that principal components of the Ctrl, Obe, and Obe+NMN groups intersected (Fig. 4B). In total, 7406 proteins were identified as being differentially acetylated, with increased acetylation levels in the Obe group compared with the NC group and decreased acetylation levels in the Obe+NMN group compared with the Obe group (Fig. 4C, D). GO functional classification showed that the proteins that exhibited significant changes in acetylation levels in the Obe group compared with the NC group were mainly involved in metabolic processes (Fig. 4E). KEGG analysis showed that the differentially acetylated proteins were significantly enriched in metabolic pathways such as the fatty acid degradation, citric acid cycle, pyruvate metabolism, and glycolysis pathways (Fig. 4F).
A Panacetylation levels in the testes of Ctrl, Obe, and Obe+NMN groups. B Principal component analysis of the testicular proteome in the Ctrl, Obe, and Obe+NMN groups. C Venn diagram showed the overlap among acetylated peptides in the Ctrl, Obe, and Obe+NMN groups. D Major peptides in the Ctrl, Obe, and Obe+NMN groups, as detected by WGCNA analysis. E Top 20 enriched GO terms in the Ctrl, Obe, and Obe+NMN mice. F Bubble diagrams based on the top 20 enriched KEGG pathways in the Ctrl, Obe, and Obe+NMN groups. G Heatmap of differentially acetylated peptides in the glycolysis/gluconeogenesis pathway. Clustering results were derived from four independent replicates of data from the Ctrl, Obe, and Obe+NMN groups. H Enzymes in the glycolysis pathway that were differentially acetylated in the Ctrl, Obe, and Obe+NMN groups. I Spearman correlation analysis of acetylation levels at the three acetylation sites in LDHC, fat weight, lipid metabolism index, and spermatogenesis index. J Correlation network between the three acetylation sites in LDHC and differentially expressed metabolites (1), and the three acetylation sites in LDHC and the genus of the differentially abundant gut microbiota (2).
Construction of a heatmap showed that differentially acetylated proteins associated with glycolysis exhibited increased acetylation in the testes of obese mice compared with NC mice that decreased after NMN treatment (Fig. 4G). During glycolysis, enzymes involved in the conversion of glucose-6-phosphate to pyruvate, enzymes that catalyze the conversion of pyruvate to acetyl-CoA, and enzymes that catalyze the conversion of pyruvate to lactate were hyperacetylated in the testis of obese mice (Fig. 4H).
One of the differentially acetylated proteins that was identified as being hyperacetylated in the Obe+NMN group was LDHC, a testis-specific glycolysis rate-limiting enzyme that is involved in the production a lactate, a key energy source during spermatogenesis. Therefore, we asked whether LDHC acetylation mediates the changes in glycolysis induced by NMN in obese mice to promote spermatogenesis. Spearman correlation analysis showed that the level of LDHC acetylation was positively correlated with LDL, TC, TG, fat mass, and abnormal spermatogenesis, and was negatively correlated with HDL, sperm count, spermatozoa viability, epididymal indices, and testicular indices (Fig. 4I). Furthermore, network analysis showed that LDHC acetylation correlated negatively with 3-methylindole, LPC 40:7, PC 38:5, methyl isoquinoline-3-carboxylate, 11-deoxy prostaglandin F1α, Desulfovibrio, Lachnospiraceae NK4A136 group, Dubosiella, Lactobacillus, Akkermansia, Enterorhabdus, Methanosarcina, and Ligilactobacillus (Fig. 4J).
Taken together, these findings suggest that NMN treatment decreases protein acetylation in the testes, and that LDHC acetylation in particular may promote spermatogenesis in obese mice.
LDHC acetylation at lysine residues 5, 17, and 90 regulates Sertoli cell lactate and NAD+ production in vitro
LDHC reduces pyruvate to lactate, an intermediate metabolite in the glycolysis pathway that is the main source of energy generated by SCs to support spermatogenesis. Therefore, we next tested the effect of NMN on Ldhc expression and acetylation, as well as lactate production, in vitro to verify the results from our animal experiments at a cellular level. First, we treated the TM4 cell with a range of NMN concentrations and confirmed that NMN did not affect cell viability (Supplementary Fig. 9A, B). Next, we established a hyperlipid cell model by exposing TM4 cells to 0.4 mM palmitic acid (PA)31 and then treating them with 50 μM NMN or leaving them untreated. qRT-PCR and western blot analysis showed that the expression levels of CX43, Occludin, and androgen-binding protein, markers of SCs function, were significantly decreased in the PA group compared with untreated cells and increased after NMN treatment (Supplementary Fig. 10A–C). Furthermore, Ldhc expression was decreased in the PA group compared with untreated cells and increased in the NMN group (Supplementary Fig. 10D–F).
Next, we examined global acetylation in TM4 cells to verify the results obtained from the earlier animal experiments. Global protein acetylation levels in TM4 cells increased after PA exposure and decreased after NMN treatment. Next, to verify which of the three acetylation sites in LDHC are affected by obesity, constructed a vector to express LDHC in 293 cells and found that the exogenous protein was acetylated at all three sites, and the acetylation level increased after PA exposure (Fig. 5A, B, Supplementary Fig. 11). Mutating K5, K17, or K90, the major acetylation sites in LDHC (Fig. 4G), to Arg (K5R, K17R, or K90R) did not significantly alter LDHC acetylation levels, whereas mutation of all three sites (3KR) abrogated LDHC acetylation (Fig. 5C), suggesting that triple acetylation of this protein is functionally significant. Importantly, LDHC-3KR exhibited significantly less acetylation than wild-type LDHC (LDHC-WT) in untreated cells, and treatment with PA did not further decrease LDHC-3KR acetylation in vivo. In addition, lactate production and LDH activity in cells expressing LDHC-3KR were higher than those in cells expressing LDHC-WT (Fig. 5D, E).
A Panacetylation levels were determined by Western blot. B 293T cells were treated with PA or left untreated. Cytoplasmic proteins were extracted and subjected to co-immunoprecipitation. C (1) The LDHC-K5R mutant plasmid was transfected into 293T cells, and the mutant protein was immunoprecipitated and subjected to Western blot. (2) The LDHC-K17R mutant plasmid was transfected into 293T cells, and the mutant protein was immunoprecipitated and subjected to Western blot. (3) The LDHC-K90R mutant plasmid was transfected into 293T cells, and the mutant protein was immunoprecipitated and subjected to Western blot. (4) The LDHC-3KR mutant plasmid was transfected into 293T cells, and the mutant protein was immunoprecipitated and subjected to Western blot. D Lactate levels in the 293T cells, as detected by ELISA. E NAD+ levels in the 293T cells, as detected by ELISA. All data are presented as median with interquartile range or mean ± SEM. *p < 0.05; **p < 0.01.
Because LDHC-mediated lactate production is accompanied by redox of NADH to NAD+, we next assessed lactate and NAD+ levels in TM4 cells treated with PA and/or NMN to determine the effect on energy metabolism. Both lactate and NAD+ levels were clearly lower in the PA group compared with untreated cells and increased in response to treatment with NMN (Supplementary Fig. 7G). Moreover, inhibiting LDH by treating the cells with galloflavin (GF) markedly inhibited Ldhc and LDH activities, as well as lactate levels, while treatment with NMN reversed these effects (Supplementary Fig. 10H).
Together, these findings suggest that NMN treatment decreases Ldhc expression and acetylation at lysine residues 5, 17, and 90, thereby promoting SC lactate and NAD+ production.
Discussion
The aim of this study was to determine the effect of supplementation with NMN, an essential precursor molecule of NAD+, on spermatogenesis in an HFD-induced mouse model of obesity. We found that NMN treatment reduced body weight, fat mass, and liver fat content, partially reversed TG accumulation, and improved sperm parameters including sperm count, morphology, and motility in obese mice. NMN exerted these effects by improving SC function and NAD+ production, which inhibited acetylation of LDHC, a testis-specific LDH, increasing its activity and promoting glycolysis and lactate production.
Similar to previous studies, we found that obesity decreases sperm quality, and our findings suggest that NAD+ is responsible, at least in part, for this effect. Indeed, a number of cross-sectional and longitudinal studies, as well as systematic reviews and meta-analyses, have demonstrated a marked negative correlation between semen parameters and obesity, and one study including 13,077 men showed that overweight and obesity are associated with a higher prevalence of azoospermia and oligozoospermia32,33,34,35. Additionally, studies performed in animal models have reported that obesity decreases sperm production, increases sperm DNA fragmentation, and impairs spermatogenesis36,37. Our previous meta-analysis concluded that supplementation with NAD+ precursors improved TG, TC, LDL, and HDL levels in humans, and that NMN regulates fat cell metabolism8. Consistent with this, a recent study showed that NAD+ levels decrease with obesity in multiple murine tissues, including adipose tissue, skeletal muscles, liver, and the hypothalamus38. These findings suggested that treatment with NAD+ could indirectly improve sperm parameters by ameliorating the effects of obesity, and this hypothesis was confirmed in the present study. To our knowledge, this is the first study to show that spermatogenesis can be effectively improved by increasing NAD+ levels.
In addition to improving sperm parameters, we found that NMN treatment improves SCs glycolysis, which previous studies have shown to be essential for spermatogenesis and impaired in obesity. Importantly, the immunofluorescence co-staining of SCs marker genes and key glycolysis pathway genes further supported this conclusion (Supplementary Fig. 12C). The testis is a naturally oxygen-deprived organ, and lactate produced by SCs glycolysis is an important source of adenosine triphosphate for the rapidly proliferating spermatogenic cells39,40. Regeneration of intracellular NAD+ by LDH is necessary for continued glycolysis38, as well as hypoxia/anoxia tolerance, and low NAD+ levels have been shown to impair glycolysis, leading to cell death41. Furthermore, reduced glucose consumption, LDH activity, and lactate production in SCs promote apoptosis, leading to impaired spermatogenesis and male infertility42. A recent study showed that LDH activity is reduced in testicular SCs from morbidly obese individuals compared individuals of normal weight13. The same study showed that metabolic alteration of SCs increased with the degree of obesity. In line with this, LDHA expression in the testes of the obese mice is inversely correlated with the degree of obesity43. Moreover, obesity is linked to dysregulation of glycolysis; for example, HFD-induced obesity impairs glycolysis in mice during early pregnancy, and expression of the key glycolytic enzymes Hk2, Pkm2, and Ldha is significantly decreased in mice fed an HFD compared with mice fed a normal diet44. Here, we showed that impaired glycolysis in obesity is an important reason for SCs-mediated impairment of spermatogenesis, and that treatment with the NAD+ intermediate NMN increased NAD+ levels by regulating glycolytic pathways. Previous studies have shown that an HFD increases SC lactate levels and leads to decreased sperm quality12,45, which contradicts our finding that HFD-induced obesity in mice reduces SC lactate production. This could be due to differences in animal models, given that the two earlier studies were performed in rats, rather than in mice like the current study. Importantly, we previously found that treating obese mice with icariin promotes normal SCs glycolysis, thus enhancing spermatogenesis, supporting our conclusion that NMN-mediated regulation of SCs glycolysis accounts for the improvement in spermatogenesis seen in obese mice31. In addition, two studies performed in diabetic mice showed that NMN not only improves impaired glucose tolerance by restoring normal NAD+ levels, but also regulates SCs glycolysis, reduces spermatogenic cell apoptosis, and improves spermatogenesis24,46. Further research is needed to determine whether the link between lactate production and spermatogenesis varies among species.
In addition to increasing SCs glycolysis, we found that NMN treatment improves SCs-mediated maintenance of the BTB, which is in line with previous studies showing that BTB integrity is important for spermatogenesis. SCs play a key role in maintaining the BTB during spermatogenesis, in part by generating and preserving tight junctions with developing spermatozoa47. Obesity disrupts BTB integrity, resulting in impaired spermatogenesis and infertility48,49. Furthermore, treating SCs with PA to create an in vitro model of obesity resulted in a decrease in the expression of tight junction–related proteins such as CX43 and Occludin, which are crucial for maintenance of the BTB50. In the present study, we found that NMN treatment restored the expression levels of junction proteins in obese mice to levels comparable to those seen in normal-weight mice, thereby reversing the impairment in BTB integrity caused by obesity. Thus, our findings show that NMN treatment improves multiple aspects of SC function in obesity that promote proper spermatogenesis.
Protein acetylation is closely related to energy metabolism, and our findings confirmed that fat mass and lipid metabolism indicators were positively correlated with lysine acetylation in obese mice. A large number of non-histone proteins are acetylated in the adipose tissue of obese individuals, and lysine acetylation decreases with weight loss51. In addition, spermatogenesis indicators are negatively correlated with lysine acetylation52. Importantly, all enzymes involved in glycolysis are acetylated at key lysines in a variety of mammalian tissues and cell types, and this modification plays a significant role in controlling energy metabolism53,54. In the current study, we found that acetylation of metabolism-related proteins within the testes was significantly altered in an HFD-induced mouse model of obesity, as confirmed by label-free quantitative acetylomics. In particular, LDHC was found to by acetylated at multiple specific sites, and acetylation at all three sites correlated with decreased SC lactate production and impaired spermatogenesis. This is consistent with an earlier study showing that acetylation-mediated negative regulation of LDHA activity in pancreatic cancer cells reduces lactate production and accumulation55. Our study provides evidence that increasing NAD+ levels via NMN treatment reduces LDHC acetylation and improves the physiological function of SCs in vivo and in vitro.
However, the present study is not without limitations. As an NAD+ precursor, NMN may affect cellular metabolic networks through multiple pathways rather than targeting a single pathway56. Although this study successfully demonstrated that SIRT2 is the deacetylase of LDHC, NMN may also exert its effects through other deacetylases or acetylases such as PARP1(Supplementary Fig. 12A). Furthermore, mitochondria have a bidirectional regulatory role, and NMN can improve mitochondrial function through an ATF4-dependent pathway. However, long-term intervention may interfere with the normal threshold of cellular stress signals and increase sensitivity to oxidative stress57. There are also potential risks associated with the metabolic byproducts of NMN. Animal studies have demonstrated that although no significant toxicity is observed with continuous oral administration of 500 mg/kg NMN for 12 weeks, increased serum bilirubin levels are present, indicating an increased burden on liver metabolism. Additionally, due to species differences between mice and humans, where the basal metabolic rate of mice is seven times that of humans, the half-life of NAD+ can affect pharmacokinetics, and the current findings cannot be fully applied to clinical settings. Furthermore, population heterogeneity was not considered, and factors such as age, sex, and basal metabolic status can potentially influence the effects of NMN. Therefore, in future studies, we will consider constructing transgenic mice expressing specific human metabolic enzymes (such as NAMPT) to improve the accuracy of pathological modeling. Alternatively, we will evaluate the impact of NMN on the local microenvironment using human intestinal/testicular organoids to reduce interference from animal models.
In summary, our findings showed that NMN treatment increases NAD+ levels in the testes of obese mice, which inhibits LDHC acetylation at residues K5, K17, and K90, thereby promoting glycolysis and lactate production and improving obesity-induced impaired spermatogenesis. These findings suggest that LDHC acetylation is a potential therapeutic target for improving spermatogenesis in obese individuals.
Methods
Chemicals and reagents
NMN was purchased from Shenzhen Hygieia Huining Biotechnology Co., Ltd (batch number: 1094-61-7). The TransScript® One-Step135 gDNA Removal and cDNA Synthesis Super Mix (AT311-02) kit was purchased from TransGen Biotech (Beijing, China). The 20× Metal Enhanced DAB Substrate Kit (DA1015) was purchased from Beijing Solarbio Science & Technology Co., Ltd (Beijing, China). The diaminobenzidine (DAB) chromogenic kit was purchased from Beijing Zhongshan Jinqiao Biotechnology Co., Ltd. HiScript III RT SuperMix for qRT-PCR (+gDNA wiper) was purchased from Nanjing Vazyme Biotech Co., Ltd (batch number: R323-01). The BCA Protein Assay Kit (CW0014S), eECL Western Blot Kit (CW0049M), and SDS-PAGE Gel Kit (CW0022S) were purchased from Beijing ComWin Biotech Co., Ltd (Beijing, China). Trizol reagent (15596026 and 15596018) was purchased from Thermo Fisher Scientific (Waltham, MA, USA). All primers were designed and synthesized by Shanghai Sangon Biotechnology Co., Ltd (Shanghai, China). FK866 (HY-50876) and Galloflavin (HY-W040118) was purchased from MedChemExpress (MCE, China).
Animals and treatments
Three-week-old male C57BL/6 mice (n = 48) were purchased from the Laboratory Animal Center of the University of South China (Hengyang, China; permit number: SYXK (Xiang) 2020-0002). The animals were maintained in a facility with a room temperature of 23 ± 2 °C, a 12/12-h light/dark cycle (7 a.m./7 p.m.), and 50% to 60% relative humidity. After 1 week of adaption, the mice were randomly divided into two groups. One group (normal control, NC, n = 18) received a normal diet, while the other group (obese group, HFD, n = 30) was fed an HFD. After 12 weeks, the mice in the HFD group exhibited body weights 20% higher than those seen in the control group. The obese mice were further divided into two subgroups: the obesity model group (Obe, n = 18), which was fed the HFD and subjected to oral gavage once a day with normal saline, and the NMN intervention group (Obe+NMN, n = 12), which was fed the HFD and subjected to oral gavage once a day with 500 mg/kg/d NMN. The mice were sacrificed after 19 weeks.
Histological analysis
At the end of the 19-week experimental period, the animals were anesthetized by intraperitoneal administration of 0.6 mg/kg urethane, weighed, and sacrificed. Samples of liver, adipose, testis, and epididymis tissue were fixed in 4% paraformaldehyde for 24 h and then rinsed with water for 24 h. The fixed tissues were then embedded in paraffin and sectioned into 5-μm-thick slices. The slices were placed on slides, washed in methanol for 24 h and running water for another 24 h, and stained with hematoxylin and eosin. The morphology of the liver, adipose, testis, and epididymis tissues was evaluated under a light microscope (BX43; Olympus, PA, USA). The remaining liver, testis, and epididymis tissues were snap-frozen in liquid nitrogen and stored at −80 °C.
Serum lipid parameter analysis
After anesthesia, blood was collected from the mice via the abdominal aorta and allowed to rest for 30 min at room temperature. Then, the samples were centrifuged at 1200 × g for 15 min at 4 °C, and the supernatants containing the serum were frozen at −20 °C. To determine the serum lipid levels, the serum samples were thawed at room temperature, the reagents were prepared according to the TC, TG, LDL-C, and HDL-C kit instructions, and the optical density at a wavelength of 510 nm was determined. TC, TG, HDL, and LDL contents were then calculated.
Semen quality analysis
Sperm function parameters, including sperm motility, sperm count, and morphological abnormalities, were assessed following WHO guidelines (World Health Organization, 2021). Sperm samples from all groups were diluted to evaluate motility under a light microscope (E400; Nikon, Tokyo, Japan). Another portion of each sperm sample was incubated at 37 °C for 15 min, and the sperm count was determined using an optical microscope (TC20; Bio-Rad, WA, USA). The same portion of the sperm sample was then placed on a slide, air-dried at room temperature, fixed with methanol (Honeywell, WA, USA) for 5 min, stained with a mixture of Eosin Y (E4009; Sigma-Aldrich, MO, USA) and ethanol (Bioman), rinsed with 75% ethanol (Bioman), and dried. The slides were evaluated using a light microscope (DM1000; Leica) to estimate the percentage of sperm exhibiting normal morphology out of a minimum of 100 spermatozoa.
Oil red O staining of frozen liver tissue sections
Frozen liver tissue was sliced into 15-μm-thick sections, placed on slides, and fixed with 10% formalin for 10 min. The sections were then soaked in distilled water for 2 s, immersed in 60% isopropyl alcohol for 25 s, placed in oil red O staining solution for 15 min, immersed in 60% isopropyl alcohol for 2 s to remove the dye, soaked in distilled water for 2 s, restained with Mayer hematoxylin solution for 2 min, rinsed with running water for 10 min, and rinsed with distilled water for 2 s. The remaining water was wicked away with filter paper, and the slides were covered with coverslips, which were sealed with neutral gum.
UPLC-MS/MS analysis of serum metabolites
UPLC-MS/MS serum analysis was carried out by Novogene (Beijing, China). Briefly, the serum samples (100 μL) were mixed with prechilled 80% methanol by vortexing, incubated on ice for 5 min, and centrifuged at 15,000 × g at 4 °C for 20 min. The supernatant was diluted with LC-MS–grade water to final concentration of 53% methanol. The diluted samples were subsequently transferred to a fresh Eppendorf tube and centrifuged at 15,000 × g at 4 °C for 20 min. Finally, the supernatant was injected into the LC-MS/MS system for analysis.
The UHPLC-MS/MS analyses were performed using a Vanquish UHPLC system (ThermoFisher, Germany) coupled with an Orbitrap Q ExactiveTM HF mass spectrometer or Orbitrap Q ExactiveTM HF-X mass spectrometer (Thermo Fisher, Germany) by Novogene Co., Ltd (Beijing, China). Samples were injected onto a Hypersil Gold column (100 × 2.1 mm, 1.9 μm) using a 12-min linear gradient at a flow rate of 0.2 mL/min. The eluents for the positive and negative polarity modes were eluent A (0.1% FA in water) and eluent B (methanol). The solvent gradient was set as follows: 2% B, 1.5 min; 2% to 85% B, 3 min; 85% to 100% B, 10 min; 100% to 2% B, 10.1 min; 2% B, 12 min. The Q ExactiveTM HF mass spectrometer was operated in positive/negative polarity mode with a spray voltage of 3.5 kV, capillary temperature of 320 °C, sheath gas flow rate of 35 psi, aux gas flow rate of 10 L/min, S-lens RF level of 60, and aux gas heater temperature of 350 °C.
Gut microbiota analysis
Fecal samples were collected from the duodenum of the mice after NMN treatment. Total DNA was isolated and sequenced using a HiSeq platform (Illumina, San Diego, CA, USA) at the Novogene Bioinformatics Institute (Beijing, China). Briefly, the DNA samples were amplified, the PCR products were mixed and purified, and the library was constructed by performing end repair, adding poly-A tails, adding adapter sequences, and purifying. The raw reads were spliced and filtered to obtain the clean reads. Operational taxonomic units (OTU) clustering was performed using Uparse software (Uparse v7.0.1001, Robert C. Edgar, Tiburon, CA, USA), and sequences with ≥97% similarity were clustered to the same OTU. Taxonomic annotation was conducted using the ribosomal database project classifier (Version 2.2, Qiong Wang, East Lansing, MI, USA). The data were normalized to the sample with the lowest bacterial abundance to obtain the relative abundances, which were used for further analysis.
Alpha diversity analysis, beta diversity analysis, principal component analysis (PCA), and NMDS were performed using Past3 software. Alpha and beta diversity analyses were performed to determine the species richness and evenness of the gut microbiota.
The NMDS analysis was performed to explore differences in microbial community structure among the different groups. A heatmap was generated using Heml software to show the distribution of microbial species in the different samples. LEfSe analysis was conducted online to identify biomarkers with statistically significant differences in expression among the different groups. The correlation coefficient between relative microbial abundance and mouse body weight was determined by Spearman’s correlation analysis (SPSS Statistic 17.0, IBM, Armonk, NY, USA). Based on 16S rRNA sequencing data and the Greengene database (http://greengenes.secondgenome.com), bacterial gene functions were predicted using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt).
Acetylated peptide enrichment
The testis samples were reconstituted in 1.4 mL of precooled IAP Buffer, and pretreated anti-Ac-K antibody beads (PTM Scan Acetyl-Lysine Motif (Ac-K) Kit, Cell Signaling Technology) were added. The mixture was incubated at 4 °C for 1.5 h and centrifuged at 2000 × g for 30 s, and the supernatant was discarded. The pelleted anti-Ac-K antibody beads were washed with 1 mL precooled IAP buffer three times, then washed with precooled water three times. Next, 40 μL of 0.15% TFA was added to the washed beads, and the mixture was incubated for 10 min at room temperature. Then, 0.15% TFA was added again, the mixture was centrifuged at 2000 × g for 30 s, and the supernatant was desalted using C18 Stage Tips.
LC-MS/MS analysis of protein acetylation
Testicular protein acetylation analysis was performed by Shanghai Applied Protein Technology (Shanghai, China) using LC-MS/MS. Briefly, 100 mg frozen testicular tissue, 200 μL ultrapure water, and 800 μL methanol/acetonitrile (1:1, v/v) were added to a microcentrifuge tube. Then, the sample was homogenized with an MP homogenizer and ultrasonic instrument, incubated at −20 °C for 60 min, and centrifuged at 14,000 rpm at 4 °C for 15 min. The supernatant was collected and subjected to mass spectrometry analysis using a 5500 QTRAP mass spectrometer (AB-SCIEX). Multiquanta software was used to extract the peak area and retention time, which were normalized to the standard energy metabolism substance for metabolite identification. For clustering analysis, the quantitative information of the target metabolite set was first normalized to (−1, 1) intervals, and then cluster 3.0 software and Java Tree view software were used to analyze the samples and metabolites and to generate a hierarchical clustering heatmap.
Transcriptome sequencing and bioinformatics analysis
Testicular RNA sequencing was performed by Novogene (Beijing, China). First, total RNA was extracted, and the purity and concentration were assessed using a NanoPhotometer® spectrophotometer (Implen Inc., CA, USA) and the Qubit® RNA Assay Kit with a Qubit® 2.0 Fluorometer (Life Technologies, CA, USA). Next, transcriptome sequencing libraries were generated from 3g RNA per sample using the NEBNext® UltraTM RNA Library Preparation Kit for Illumina® (California, USA) according to the manufacturer’s instructions, and indexing codes were added to determine the sequence attributes of each sample. Index-coded samples were clustered on the cBot Cluster Generation System using the TruSeq PE Cluster Kit v3cBot-HS (Illumina, CA, USA). The prepared libraries were then sequenced on an Illumina Hiseq platform to obtain 150-bp paired-end reads. The DESeq2 R package (1.10.1) was used to analyze differential expression among the groups by adjusting the p-values of the genes. GO terms with corrected p-values less than 0.05 were considered significantly enriched. KEGG pathways (http://www.genome.jp/kegg/) significantly enriched in DEGs were identified using the cluster Profiler R package.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from mouse testicular tissue using TRIzol reagent (Invitrogen, Carlsbad, USA). The RNA was reverse-transcribed to cDNA using a TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TRAN, Beijing, China). Real-time PCR was performed using SYBR Green Real-Time PCR Master Mix (ThermoFisher Scientific NO: 4309155) and an ABI7900 PCR system (Applied Biosystems, Foster City, USA). Gapdh was included as an internal control. The relative mRNA expression levels were calculated using the 2−ΔΔCt method. The primer sequences are listed in Supplementary Table 1.
Western blotting analysis
Frozen testicular tissue, 300 μL RIPA lysis buffer (P0013K, Beyotime Biotechnology, Nanjing, China) mixed with PMSF (Solarbio) at 94:6, and three small steel balls (3.2 mm in diameter) were placed in a 1.5 mL tube, and a multi-tissue grinder (Tissuelyser-24; Shanghai Jingxin Experimental Technology, Shanghai, China) was used to homogenize the mix at 65 Hz for 60 min. TM4 cells were lysed on ice with the RIPA lysis buffer mixed with PMSF at 100:1 for 30 min, and the lysate was centrifuged at 4 °C for 20 min to collect the supernatant. Protein concentration was measured using a BCA Protein Assay Kit (CWBIO), and proteins were denatured by boiling at 100 °C for 10 min. The denatured proteins were separated on a 10% SDS-PAGE gel and electrophoretically transferred onto polyvinylidene fluoride membranes, which were then blocked in phosphate buffered saline with 0.1% Tween-20 (PBST) containing 5% skim milk powder for 2 h. After washing with PBST, the membranes were incubated with antibodies against tubulin, Bax, Bcl2, caspase-3, Hk2, Pkm2, Ldhc, Ldha, CX43, Occludin, acetylmethionine, and Tubulin overnight at 4 °C. After the membranes were washed with PBST, they were incubated with Horseradish peroxidase (HRP)-conjugated Affinipure goat anti-mouse IgG (H+L) or goat anti-rabbit IgG (H+L) for 2 h at room temperature. Finally, ECL (CW0049M, CWBIO) was added, and a T anon-5500 Chemiluminescence Imaging System was used to detect the chemiluminescent protein bands. The antibodies used in this experiment are listed in Supplementary Table 2.
Immunohistochemical analysis
Testicular tissues were fixed in 4% paraformaldehyde, embedded in paraffin, and sliced into 4-μM-thick sections. The sections were washed with 3% hydrogen peroxide for 30 min, boiled in 0.1 M sodium citrate three times for antigen retrieval, permeabilized with 1% Triton X-100 and PBST for 30 min, and blocked with 5% bovine serum albumin for 45 min. Next, the sections were incubated with rabbit primary antibodies to Bax, Bcl2, PCNA, Vimentin, Hk2, Pkm2, Ldha, and Ldhc overnight at 4 °C. After washing with PBST, the sections were incubated with a goat anti-rabbit HRP-conjugated antibody at room temperature for 45 min. The Bax, Bcl2, PCNA, Vimentin, Hk2, Pkm2, Ldha, and Ldhc signals were visualized using DAB. PBS was used as a negative control. Finally, the stained sections were observed and photographed under a light microscope (BX43, Olympus). The antibodies used in this experiment are listed in Supplementary Table 3.
Cell culture and transfection
Both cell lines (293T and TM4) used in this study were cultured in Dulbecco’s Modified Eagle’s Medium (Gibco) supplemented with 10% fetal bovine serum (FBS) at 37 °C in a 5% CO2 atmosphere. Two hours before transfection, the medium was replaced with serum-free medium. Plasmid DNA was diluted in Opti-MEM, then PEI transfection reagent was added, and the mixture was incubated for 20 min at room temperature. After incubation, the PEI-DNA complexes were added to the culture dish. After 18 h of transfection, the medium was replaced with complete medium (containing serum).
Co-immunoprecipitation (Co-IP)
Co-IP was performed using Anti-DYKDDDDK-Tag mAb magnetic beads (M20118M, Abmart) or IgG (#3900; Cell Signaling Technology) according to the manufacturer’s instructions. In brief, cell lysates were incubated with the beads at 4 °C for 2 h. Then, the beads were washed extensively and boiled in SDS loading buffer. The immunoprecipitated proteins were analyzed by Western blot.
Lactate measurement
Culture medium was collected from treated TM4 cells, and lactate levels were determined by an enzymatic method using reagents prepared according to the Lactic Acid Assay Kit instructions (A019-2-1, Nanjing Jiancheng). The OD of each sample at a wavelength of 510 nm was measured using a microplate reader, and the lactate concentration was calculated using the formula provided in the kit manual.
NAD+ measurement
Frozen mouse testicular tissues and fresh TM4 cells (1 × 106 cells/sample) were collected, and intracellular NAD+ levels were determined using a Coenzyme I NAD (H) Content Test Kit according to the manufacturer’s instructions. In brief, cells and tissues were first lysed with 200 μL of cold lysis buffer. To measure total NAD+/NADH, 20 μL of lysates from each sample was added to a 96-well plate. To measure NADH, the cell lysates and tissue lysates were incubated at 60 °C for 30 min, and 20 μL from each sample was added to a 96-well plate. Subsequently, 90 μL of alcohol dehydrogenase was added to each well, and the plate was incubated at 37 °C for 10 min. Finally, 10 μL of the chromogenic solution was added to each well, and the mixture was incubated at 37 °C for 30 min. A standard curve was generated and measured at the same time as the samples. The absorbance values were measured at 450 nm and analyzed on a plate reader. The amount of NAD+ was derived by subtracting the NADH content from the total NAD+/NADH content.
Assessment of lactate dehydrogenase (LDH) activity
LDH activity was assessed using a LDH assay kit according to the manufacturer’s instructions (A020, Nanjing Jiancheng). In brief, 1 × 106 cells were homogenized in cold assay buffer and then centrifuged at 10,000 × g at 4 °C for 15 min. Next, 50 mL of the supernatant from each sample was added to a 96-well plate. Then, 50 μL of the reaction mix was added. OD450 was measured at time 0 and then measured again after incubation at 37 °C for 30 min, and the LDH activity was calculated according to the kit protocol.
Site-directed mutagenesis
A plasmid encoding mouse Ldhc was purchased from VectorBuilder (Guangzhou, China). LDHC acetylation mutants were generated using a site-directed mutagenesis kit (GeneAll, Seoul, Korea) to convert each lysine residue (K5, K17, and K90) to glutamic acid (codon change from AAG or AAA to GAG or GAA). The mutations were confirmed by DNA sequencing (conducted by Shanghai Sangon Biotechnology Co., Ltd, Shanghai, China).
Statistics and reproducibility
Data were analyzed using GraphPad Prism 8.0 (GraphPad Software, CA, USA). Data normality was assessed using the Shapiro–Wilk test. For normally distributed data, one-way ANOVA was performed, followed by Tukey’s post-hoc test, and results were expressed as mean ± standard deviation. When two independent groups were compared, an unpaired t-test was applied. In cases where significant differences were found among multiple groups, Bonferroni’s correction was used for post-hoc comparisons. For non-normally distributed data, the Mann-Whitney U test (two-tailed) was employed, and results were reported as median values with interquartile ranges. A p-value less than 0.05 was considered statistically significant. The results from the western blotting, qRT-PCR, and cell culture experiments were obtained from three independent replicates, with representative data shown. The sample size was not predetermined using statistical methods.
Metabolomics data were analyzed and processed using Compound Discoverer 3.1 (CD 3.1) software (Thermo Fisher Scientific Inc., Massachusetts, America). The preprocessed data were analyzed using the Majorbio Cloud Platform (https://www.majorbio.com). PCA and orthogonal least partial binary discriminant analysis (OPLS-DA) were carried out using the R software package ropls (version 1.6.2). Differentially expressed metabolites were defined as those with p < 0.05, VIP > 1, and 1.5-fold change. Metabolic enrichment and pathway analyses based on a KEGG database search (https://www.genome.jp/kegg/) were used to compile and map differentially expressed metabolites between the two groups onto their respective biochemical pathways.
Gut microbiota data were preprocessed and analyzed using the NovoMagic platform (https://magic.novogene.com). Venn diagrams, column plots of relative species abundance, ternary phase diagrams, phylogenetic trees, alpha diversity, beta diversity, PCoA, and NMDS were generated and performed using QIIME2 and R (Version 3.5.3) software. Alpha and beta diversity analyses were performed to determine the species richness and evenness of the gut microbiota. NMDS analysis was used to explore the differences in microbial community structure among different groups. LEfSe analysis was used to identify biomarkers whose expression differed significantly among different groups. Spearman correlation analysis was used to evaluate the correlation between the relative abundance of microorganisms and differential metabolites, lipid metabolism indicators, spermatogenic function indicators, and LDHC acetylation levels in obese mice. Based on 16S rRNA sequencing data and the Greengene database, bacterial gene function was predicted using PICRUSt software.
Spearman’s rank correlation was used to model the nonlinear microbiota–metabolite associations, accommodating complex biological relationships.
Ethical approval
We have complied with all relevant ethical regulations for animal use. All experimental procedures were approved by the Animal Ethics Committee of the University of South China (permit number: USC2020031602).
Data availability
Metabolomics data is available at the NIH Common Fund’s National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org where it has been assigned Study ID ST004149. Transcriptome and gut microbiota data will be available from the Sequence Read Archive under accession number PRJNA1314453 and PRJNA1308262. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org) with the dataset identifier PXD068628. Any additional information required to reanalyze the data reported in this paper is available from the lead author upon request. The uncropped/unedited blots are provided in the Supplementary Fig. 13 are available in Supplementary information. The raw data are available in Supplementary Data 1 and Supplementary Data 2.
References
Bellastella, G. et al. How much does obesity affect the male reproductive function?. Int. J. Obes. Suppl. 9, 50–64 (2019).
Gadde, K. M., Martin, C. K., Berthoud, H.-R. & Heymsfield, S. B. Obesity: pathophysiology and management. J. Am. Coll. Cardiol. 71, 69–84 (2018).
Jaacks, L. M. et al. The obesity transition: stages of the global epidemic. Lancet Diabetes Endocrinol. 7, 231–240 (2019).
Bieniek, J. M. et al. Influence of increasing body mass index on semen and reproductive hormonal parameters in a multi-institutional cohort of subfertile men. Fertil. Steril. 106, 1070–1075 (2016).
Qi, X. et al. Restoring impaired fertility through diet: observations of switching from high-fat diet during puberty to normal diet in adulthood among obese male mice. Front. Endocrinol. 13, 839034 (2022).
Zhao, L. et al. Single-cell analysis of developing and azoospermia human testicles reveals central role of Sertoli cells. Nat. Commun. 11, 5683 (2020).
Yan, W. et al. Protective effects of metformin on reproductive function in obese male rats induced by high-fat diet. J. Assist. Reprod. Genet. 32, 1097–1104 (2015).
Wang, J. et al. Effects of antioxidant supplementation on metabolic disorders in obese patients from randomized clinical controls: a meta-analysis and systematic review. Oxid. Med. Cell. Longev. 2022, 7255413 (2022).
Tomar, A. et al. Epigenetic inheritance of diet-induced and sperm-borne mitochondrial RNAs. Nature 630, 720–727 (2024).
Crisóstomo, L. et al. Molecular mechanisms and signaling pathways involved in the nutritional support of spermatogenesis by Sertoli cells. Methods Mol. Biol. 1748, 129–155 (2018).
Alves, M. G. et al. Hormonal control of Sertoli cell metabolism regulates spermatogenesis. Cell. Mol. Life Sci. 70, 777–793 (2013).
Luo, D. et al. High fat diet impairs spermatogenesis by regulating glucose and lipid metabolism in Sertoli cells. Life Sci. 257, 118028 (2020).
Martins, A. D. et al. Leptin modulates human Sertoli cells acetate production and glycolytic profile: a novel mechanism of obesity-induced male infertility?. Biochim. Biophys. Acta 1852, 1824–1832 (2015).
Pan, D., Fujimoto, M., Lopes, A. & Wang, Y.-X. Twist-1 is a PPARdelta-inducible, negative-feedback regulator of PGC-1alpha in brown fat metabolism. Cell 137, 73–86 (2009).
Nunnari, J. & Suomalainen, A. Mitochondria: in sickness and in health. Cell 148, 1145–1159 (2012).
Zhang, Y. et al. The pivotal role of protein acetylation in linking glucose and fatty acid metabolism to β-cell function. Cell Death Dis. 10, 66 (2019).
Choi, S.-E. et al. Elevated microRNA-34a in obesity reduces NAD+ levels and SIRT1 activity by directly targeting NAMPT. Aging Cell 12, 1062–1072 (2013).
Romanick, S. S. et al. Obesity-mediated regulation of cardiac protein acetylation: parallel analysis of total and acetylated proteins via TMT-tagged mass spectrometry. Biosci. Rep. 38, BSR20180721 (2018).
Limami, A. M., Glévarec, G., Ricoult, C., Cliquet, J.-B. & Planchet, E. Concerted modulation of alanine and glutamate metabolism in young Medicago truncatula seedlings under hypoxic stress. J. Exp. Bot. 59, 2325–2335 (2008).
Cantó, C. et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 15, 838–847 (2012).
Mouchiroud, L. et al. The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 154, 430–441 (2013).
Yoshino, J., Mills, K. F., Yoon, M. J. & Imai, S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 14, 528–536 (2011).
Dall, M., Hassing, A. S. & Treebak, J. T. NAD+ and NAFLD - caution, causality and careful optimism. J. Physiol. 600, 1135–1154 (2022).
Yoshino, J., Baur, J. A. & Imai, S.-I. NAD+ intermediates: the biology and therapeutic potential of NMN and NR. Cell Metab. 27, 513–528 (2018).
Lian, C.-Y., Zhai, Z.-Z., Li, Z.-F. & Wang, L. High fat diet-triggered non-alcoholic fatty liver disease: a review of proposed mechanisms. Chem. Biol. Interact. 330, 109199 (2020).
Ollero, M. et al. Plasma lipidomics reveals potential prognostic signatures within a cohort of cystic fibrosis patients. J. Lipid Res. 52, 1011–1022 (2011).
Syromyatnikov, M. et al. Characteristics of the gut bacterial composition in people of different nationalities and religions. Microorganisms 10, 1866 (2022).
Behary, J. et al. Gut microbiota impact on the peripheral immune response in non-alcoholic fatty liver disease related hepatocellular carcinoma. Nat. Commun. 12, 187 (2021).
Li, Z. et al. Dietary butyrate ameliorates metabolic health associated with selective proliferation of gut Lachnospiraceae bacterium 28-4. JCI Insight 8, e166655 (2023).
Xia, X. et al. Electroacupuncture promoted intestinal defensins and rescued the dysbiotic cecal microbiota of high-fat diet-induced obese mice. Life Sci. 309, 120961 (2022).
Luo, M. et al. Icariin ameliorates spermatogenesis disorder in obese mice induced by high-fat diet through regulating the glycolytic pathway. Mol. Nutr. Food Res. 67, e2200524 (2023).
Sermondade, N. et al. BMI in relation to sperm count: an updated systematic review and collaborative meta-analysis. Hum. Reprod. Update 19, 221–231 (2013).
Macdonald, A. A., Stewart, A. W. & Farquhar, C. M. Body mass index in relation to semen quality and reproductive hormones in New Zealand men: a cross-sectional study in fertility clinics. Hum. Reprod. 28, 3178–3187 (2013).
Andersen, J. M. et al. Body mass index is associated with impaired semen characteristics and reduced levels of anti-Müllerian hormone across a wide weight range. PLoS ONE 10, e0130210 (2015).
Eisenberg, M. L. et al. The relationship between male BMI and waist circumference on semen quality: data from the LIFE study. Hum. Reprod. 30, 493–494 (2015).
Barbagallo, F. et al. Molecular mechanisms underlying the relationship between obesity and male infertility. Metabolites 11, 840 (2021).
Lainez, N. M. & Coss, D. Obesity, neuroinflammation, and reproductive function. Endocrinology 160, 2719–2736 (2019).
Okabe, K., Yaku, K., Tobe, K. & Nakagawa, T. Implications of altered NAD metabolism in metabolic disorders. J. Biomed. Sci. 26, 34 (2019).
Boussouar, F. & Benahmed, M. Lactate and energy metabolism in male germ cells. Trends Endocrinol. Metab. TEM 15, 345–350 (2004).
Yi, X.-D., Zhanf, Y.-N., Xiao, S. & Lei, X.-C. [Role and regulatory mechanism of glycometabolism of Sertoli cells in spermatogenesis]. Zhonghua Nan Ke Xue Natl J. Androl. 25, 923–927 (2019).
Ismond, K. P., Dolferus, R., de Pauw, M., Dennis, E. S. & Good, A. G. Enhanced low oxygen survival in Arabidopsis through increased metabolic flux in the fermentative pathway. Plant Physiol. 132, 1292–1302 (2003).
Alves, M. G., Martins, A. D., Cavaco, J. E., Socorro, S. & Oliveira, P. F. Diabetes, insulin-mediated glucose metabolism and Sertoli/blood-testis barrier function. Tissue Barriers 1, e23992 (2013).
Han, J. et al. Obesity induces male mice infertility via oxidative stress, apoptosis, and glycolysis. Reproduction 166, 27–36 (2023).
Chen, Z. et al. Dysregulated glycolysis underpins high-fat-associated endometrial decidualization impairment during early pregnancy in mice. Biochim. Biophys. Acta Mol. Basis Dis. 1869, 166659 (2023).
Suleiman, J. B. et al. Orlistat reverses intratesticular lactate transport decline and infertility in male obese rats. Reproduction 160, 863–872 (2020).
Kane, A. E. & Sinclair, D. A. Sirtuins and NAD+ in the development and treatment of metabolic and cardiovascular diseases. Circ. Res. 123, 868–885 (2018).
Jiang, Y. et al. Identification and verification of potential biomarkers in sertoli cell-only syndrome via bioinformatics analysis. Sci. Rep. 13, 12164 (2023).
Wang, S. et al. LncRNA Tug1 maintains blood-testis barrier integrity by modulating Ccl2 expression in high-fat diet mice. Cell. Mol. Life Sci. 79, 114 (2022).
Fan, Y. et al. Diet-induced obesity in male C57BL/6 mice decreases fertility as a consequence of disrupted blood-testis barrier. PLoS ONE 10, e0120775 (2015).
Pelletier, R.-M., Layeghkhavidaki, H., Kumar, N. M. & Vitale, M. L. Cx30.2 deletion causes imbalances in testicular Cx43, Cx46, and Cx50 and insulin receptors. Reciprocally, diabetes/obesity alters Cx30.2 in mouse testis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 318, R1078–R1090 (2020).
Gao, W., Liu, J.-L., Lu, X. & Yang, Q. Epigenetic regulation of energy metabolism in obesity. J. Mol. Cell Biol. 13, 480–499 (2021).
Lund, P. J. et al. Stable isotope tracing in vivo reveals a metabolic bridge linking the microbiota to host histone acetylation. Cell Rep. 41, 111809 (2022).
Liu, Y. et al. Protein acetylation: a novel modus of obesity regulation. J. Mol. Med. Berl. Ger. 99, 1221–1235 (2021).
Li, J., Wang, T., Xia, J., Yao, W. & Huang, F. Enzymatic and nonenzymatic protein acetylations control glycolysis process in liver diseases. FASEB J. 33, 11640–11654 (2019).
Zhao, D. et al. Lysine-5 acetylation negatively regulates lactate dehydrogenase A and is decreased in pancreatic cancer. Cancer Cell 23, 464–476 (2013).
Yu, B. et al. The versatile multi-functional substance NMN: Its unique characteristics, metabolic properties, pharmacodynamic effects, clinical trials, and diverse applications. Front. Pharmacol. 15, 1436597 (2024).
Xiong, X. et al. NAD+-boosting agent nicotinamide mononucleotide potently improves mitochondria stress response in alzheimer’s disease via ATF4-dependent mitochondrial UPR. Cell Death Dis. 15, 744 (2024).
Acknowledgements
This study was supported by funding from the Guangxi Natural Science Foundation (2023GXNSFAA026043, 2024GXNSFAA999479), the National Natural Science Foundation of China (82360292), the Students Innovation and Entrepreneurship Training Program of National College (202210601043, 202311102563), and the horizontal scientific research project of university of South China-Hengyang Maternal and Child Health Hospital Joint Training Base for Top Innovative Graduate Talents.
Author information
Authors and Affiliations
Contributions
K.L., J.Y.W., H.H., and X.C.L. designed the study. K.L., W.H.N., Y.H.W., and Y.Z.L. performed the experiments and drafted the manuscript. G.Q.H., J.J.Y., and Z.Y.T. were responsible for the data acquisition and analysis. H.P., X.C., and X.C.L. provided technical support. X.D.W. and L.L.H. revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Rupinder Kaur and Dario Ummarino.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, K., Nong, W., Huang, H. et al. Nicotinamide mononucleotide treatment improves spermatogenesis in obese mice by reducing lysine acetylation of lactate dehydrogenase C. Commun Biol 8, 1761 (2025). https://doi.org/10.1038/s42003-025-09094-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09094-5