Abstract
Rotational migration is one specific form of collective cell migration when epithelial cells are confined in a spherical geometry, such as in the epithelial acini. This tissue-level rotation motion is crucial for the morphogenesis of multiple epithelial systems. Here, we introduce human pancreatic ductal organoids as a model to study rotational migration. Live imaging revealed the persistent rotation of the organoids over time. By tracking the nuclei, the three-dimensional trajectory of the cellular movement was reconstructed and the velocity of the rotation was quantified. The presence of focal adhesion clusters and prominent actin stress fibers were observed at the basal side of the organoids, suggesting the interactions between the cells and the surrounding extracellular matrix. Finally, our inhibition study showed the dependence of pancreatic ductal organoid rotational migration on myosin activity, actin polymerization, and actin branching. We hope that this model will enable future studies with human primary cells, which are more faithful to normal epithelial cells.
Similar content being viewed by others
Introduction
The collective migration of epithelial cells plays a vital role in many biological processes, including morphogenesis, wound repair, tissue homeostasis, and cancer metastasis1. One specific form of collective migration is rotational migration, which is observed when epithelial cells are confined in a spherical geometry. In rotational migration, the entire multicellular structure rotates as a result of directed collective migration. This migration mode has been observed in various systems, such as the follicle cells of Drosophila egg chambers2,3,4,5,6, canine kidney epithelial acini7,8,9, murine pancreatic organoids10,11, and human mammary acini12,13,14. This suggests that rotational migration is an inherent dynamic feature of three-dimensional multicellular structures. Moreover, the significant role of rotational migration in morphogenesis has been reported in several studies. For example, disruption of Drosophila follicle cells’ rotational migration prevented the elongation of the egg chamber15,16, a critical step in its developmental process. Recent work in human mammary gland morphogenesis has also revealed that rotational migration is needed for the budding of spherical alveoli from the mammary duct network17. Furthermore, the loss of rotational migration was associated with the malignant behavior of breast cancer cells12,13, suggesting the potential role of rotational migration in maintaining normal tissue homeostasis.
To establish the collective rotational migration, the epithelial cells need to collectively polarize towards the migration direction. Interestingly, unlike other collective cell migrations, polarizing external cues such as space or chemical gradients are not available in rotational migration. Hence, the polarization process is likely to rely on cell-to-cell or cell-to-extracellular matrix (ECM) interactions. Several studies using the invertebrate Drosophila follicle cell model have identified a number of these polarizing cell–cell interactions. The accumulation of cadherin protein Fat2 at the trailing edge of a follicle cell is essential for the polarization of the WAVE complex at the leading edge of the adjacent cell2. In another study, the authors showed the roles of formin protein DAAM and Rho GTPase Cdc42 in actin stress fibers and protrusions polarization3,4. For the mammalian systems, on the other hand, the cellular polarization mechanisms are still unknown. Nevertheless, perturbation of some key players, such as actomyosin contractility8,12,13,18, cell–cell junction protein E-cadherin18, ECM receptor integrin β112,13, signaling proteins EGFR12, and β-catenin9, has been shown to disrupt the rotational migration in mammalian models. However, most of the findings were established in immortalized cell lines. In our opinion, the study of rotational migration regulation will benefit from human primary cell models, derived from human tissues and more faithful to normal epithelial cells. This will potentially provide more accurate findings on epithelial cell behavior and biology. Recent developments in organoid culture systems open the door for in vitro human primary cell models, such as in the brain19, intestine20, pancreas21, and others22,23.
In this study, we introduce a human primary cell model for rotational migration study, the pancreatic ductal organoids, which were derived from human exocrine tissues. Upon culture in Matrigel, the pancreatic ductal epithelial cells form a cystic structure consisting of a spherical lumen encapsulated by a single layer of epithelial cells. Live imaging revealed the persistent rotation of the organoids over time. By tracking the nuclei, we reconstructed the trajectory of the cellular movement and quantified its velocity. We also observed the presence of focal adhesion clusters and prominent actin stress fibers at the basal side of the organoids, suggesting the interactions between the cells and the surrounding Matrigel. Finally, our inhibition study showed the dependence of pancreatic ductal organoid rotational migration on myosin activity, actin polymerization, and actin branching.
Results and discussion
Rotation of the human pancreatic ductal organoid
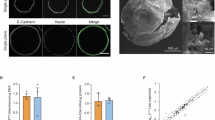
Normal human pancreatic ductal organoids were derived from the exocrine tissue provided by the Integrated Islet Distribution Program (IIDP). By culturing the exocrine tissue in Matrigel, the ductal cells grow and establish a spherical structure that consists of a lumen surrounded by a thin layer of epithelial cells (Fig. 1A)21,24. Apicobasal polarity was observed within these cells, where the immunostaining of the apical marker ZO-1 was observed at the lumen side, while the basal marker integrin α6 (ITGA6) was observed between the cells and at the outer side of the cells, which is in contact with the surrounding Matrigel. Moreover, continuous expression of the tight junction protein ZO-1 can be observed between the cells at the apical side (Fig. 1B). The cystic morphology of the pancreatic ductal organoid is also another evidence of the functional epithelial barrier. Lumen formation is initiated by the secretion of ions (such as the sodium cation and chloride anion) into the luminal space, which increases the luminal osmolality and in turn, drives the influx of water and the increase in lumen volume25,26,27. Hence, the maintenance of this cystic morphology is highly dependent on the epithelial barrier function that preserves the osmotic gradient.
A Confocal section taken at the middle of a human pancreatic ductal organoid showing the cystic morphology of the organoid, a lumen encapsulated by a layer of pancreatic ductal epithelial cells. The organoids retain their apical-basal polarity as shown by the apical marker protein ZO-1 at the lumen side (inside) and the basal marker protein ITGA6 at the organoid interface with the Matrigel (outside). Bar = 50 µm. B Maximum projection of the confocal sections taken from the pancreatic ductal organoid, showing the continuous staining of the tight junction protein ZO-1 between the ductal cells, the distribution of the integrin protein ITGA6 at the basal side of the cells, and the distribution of nuclei within the organoid. Bar = 50 µm. C Maximum projection of confocal sections taken during the live imaging of pancreatic ductal organoid nuclei, showing the rotational migration of the organoid. Pseudo-colors (red, yellow, green, and cyan) were assigned to several nuclei to illustrate the location of these nuclei over time. Bar = 50 µm.
Brightfield live imaging of the pancreatic ductal organoids revealed the rotational motion of the organoids over time (Video 1). To investigate the rotation further, the nuclei were stained with a non-toxic, cell-permeable, and highly specific live cell DNA probe, SPY650-DNA, followed by live imaging using spinning disk confocal microscopy. Indeed, we observed the directed motion of the nuclei that resulted from the rotation of the whole organoid (Fig. 1C, Video 2). We observed two key differences between the observed rotation of our pancreatic ductal organoids, and the previously reported rotation of MDCK (canine kidney) and MCF10A (human mammary) spheroids. One, rotation in MDCK and MCF10A spheroids is limited to the early, low cell-number stages (<20 cells)7,12,13, whereas rotation in the pancreatic ductal organoids can still be observed in large organoids (>300 µm in diameter, Video 1) that consist of hundreds of cells. Two, rotation in MDCK cells at later stages that involve more cells (>20 cells) is only observed in a gradient of Matrigel concentrations8,9—making it a form of durotaxis—whereas the rotation we observed in the pancreatic organoids is within a homogeneous Matrigel matrix.
As a comparison, we also looked into the pathological pancreatic ductal adenocarcinoma (PDAC) organoids. Similar to their normal counterpart, PDAC organoids also have a cystic morphology, and the apicobasal polarity is intact (Fig. S1A). However, the PDAC cells tend to have cuboid cellular morphology, resulting in a cell layer that is thicker and denser than the normal pancreatic ductal organoid. Moreover, stratified cell layers are occasionally observed within the PDAC organoids. To our surprise, the rotational migration phenotype is lost in the PDAC organoids (Fig. S1B, Video 3), suggesting the potential importance of rotational migration in maintaining normal tissue homeostasis. This observation is also in line with previous studies in breast cancer12,13.
Quantification of human pancreatic ductal organoid rotation
Tracking the movement of single cells within large organoids in three dimensions remains a significant challenge. To quantify the organoid rotational migration, live imaging data of the nucleus were processed using a deep learning segmentation method and custom tracking algorithms (see Materials and methods). This image-processing pipeline allows us to reconstruct the nucleus positions over time and derive the displacement trajectory of each nucleus within the rotating organoid (Fig. 2A, B). Next, the velocity of each nucleus at the organoid surface at a given time point was calculated by dividing the displacement by the time covered between the imaging time points (Fig. 2C). The average velocity is relatively stable over the imaging period (Fig. 2D). Looking at the velocity distribution, the velocity ranges between 0 to 35 µm/hr (Fig. 2E), with the average velocity of ~15–20 µm/hr (Fig. 2F). This rotation velocity falls within the range of velocity reported in other rotational migration models, including the Drosophila follicle cells at 12–36 µm/hr28, the murine pancreatic organoid at around 12 µm/hr11, the canine kidney epithelial line MDCK at 5–20 µm/hr9, and the human breast epithelial line MCF10A at 12.5–17.5 µm/hr13. In contrast, the trajectory reconstruction of the PDAC organoids showed that the nuclei are moving randomly within a limited region, at an average velocity of ~3 µm/hr (Fig. S1B–G).
A Overlay of maximum projections of a rotating pancreatic ductal organoid (organoid #1) taken every 30 minutes, between hours 3 and 7 of the live imaging. To illustrate the rotational migration of the cells, the nuclei were color-coded based on the time point they were imaged. The transparent green arrow indicates the general direction of the rotation. The X and Y position coordinates start from the top left of the image. Bar = 50 µm. B Three-dimensional reconstruction of the nuclei trajectories within the rotating organoid #1. C Vector plot showing the direction and the velocity of nuclei displacement between each time point. The length of the arrows is scaled to the velocity of the nucleus. The vector plot is projected to the XY axis to illustrate the rotational migration of organoid #1. D The nuclei average velocity remains relatively stable at ~12–22 µm/hr over the imaging period (mean ± SEM, N = 3 organoids from three independent experiments, n = 21–48 nuclei per organoid from 7 hours of imaging period, i.e., 15 time points). E Histogram plot showing the distribution of the recorded velocity. The velocity ranges between 0 and 35 µm/hr, with peaks at ~15–20 µm/hr (N = 3 organoids from three independent experiments, n = 21–48 nuclei per organoid from 7 hours of imaging period, i.e., 15 time points). F Average velocity of three independent pancreatic ductal organoids, showing a range between ~15 and ~18 µm/hr (mean ± SD, N = 3 organoids from three independent experiments, n = 21–48 nuclei per organoid from 7 hours of imaging period, i.e., 15 time points).
Pancreatic ductal organoid rotational migration relies on actomyosin contractility
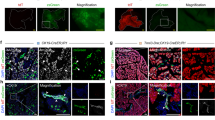
Next, we pondered how the pancreatic ductal cells migrate along the organoid rotational axis. The apical side of the organoid interacts with the fluid-filled luminal space; hence, it is less likely that the cells are migrating on the apical side. On the other hand, the basal side interacts with the Matrigel, and the cells can interact with the ECM proteins through the integrins and migrate on this interface. Indeed, we observed clusters of focal adhesion protein paxillin and prominent actin stress fibers at the basal side of the organoids (Fig. 3A), indicative of cell-ECM adhesions. Moreover, we also observed the enrichment of the phosphorylated myosin regulatory light polypeptide 9 (pMYL9) on the actin stress fibers (Fig. 3B), suggesting the activation of non-muscle myosin IIA and actomyosin contractility. This is in line with the findings in the rotating Drosophila follicle cells, where paxillin clusters and myosin-decorated actin fibers were also observed at the basal side of the follicle cells3. These observations suggest the potential involvement of actomyosin activity in regulating rotational migration.
Ai Confocal maximum projection of the basal surface of pancreatic ductal organoid stained for filamentous actin (F-actin), focal adhesion protein paxillin, integrin β1 (ITGB1), and DNA. Prominent actin stress fibers and clusters of focal adhesion protein, paxillin, were observed at the basal surface. Bar = 50 µm. ii–iii Zoomed-in images from the yellow dash boxes of Fig. 3Ai and intensity profile showing the location of paxillin clusters at the end of the stress fibers. The white arrow indicates the direction of the intensity profile and the white dash box indicates the area used to derive the intensity profile. Bar = 5 µm. Bi Confocal maximum projection of the basal surface of pancreatic ductal organoid stained for F-actin, phosphorylated myosin light chain 9 (pMYL9), non-muscle myosin heavy chain 9 (MYH9), and DNA. Colocalization of F-actin and pMYL9 was observed in some of the stress fibers. Bar = 50 µm. ii–iii Zoomed-in images from the yellow dash boxes of Fig. 3Bi and intensity profile confirm the colocalization of actin stress fibers and pMYL9. The white arrow indicates the direction of the intensity profile, and the white dashed box indicates the area used to derive the intensity profile. Bar = 15 µm.
To investigate the role of myosin contractility, the pancreatic ductal organoids were treated with a small molecule myosin II inhibitor, Blebbistatin, which has a high affinity towards the myosin-ADP-Pi complex and locks the myosin in an actin detached state29. Nuclei live imaging was performed before and after the Blebbistatin treatment on the same organoid (Fig. 4A, Video 4). From the trajectory analysis (Fig. 4B), we observed a significant slowdown of the rotation while the rotational direction was more or less maintained. Averaging the quantified velocity over the imaging period clearly showed a significant decrease in velocity after the Blebbistatin treatment (Fig. 4C, D). This suggests the reliance of rotational migration on myosin contractility. To note, myosin inhibition does not compromise the luminal structure of the organoid, where the apicobasal polarity was intact, and we still observed the continuous tight junction network at the apical side (Fig. S2A, B). Looking closer at the basal side of the Blebbistatin-treated organoids, we observed significant disruption of the actin stress fibers and the accumulation of pMYL9 at the base of the actin fibers (Fig. 4E). Instead of the long and relatively straight actin stress fibers observed in the untreated organoids (Fig. 3), the actin fibers tend to have wavy morphology (Fig. 4E) and are shorter after the Blebbistatin treatment (Fig. S2C). These observations suggest that, besides myosin activity, actin organization might also be important for organoid rotational migration.
Ai Overlay of maximum projections of a rotating pancreatic ductal organoid (organoid #1) taken before the myosin inhibitor, Blebbistatin, treatment. The images were taken every 30 minutes for 5 hours. The transparent green arrow indicates the general direction of the rotation. Bar = 50 µm. ii Overlay of maximum projections of the same organoid taken 1 hour after the 200 µM Blebbistatin treatment, showing the significant slowdown of the rotation. The images were taken every 30 minutes for 7 hours. Bar = 50 µm. The nuclei were color-coded based on the time when they were taken. The red arrow points to the nuclei of non-viable cells within the lumen of the organoid, which are excluded from the subsequent nucleus tracking analyses. B The nuclei trajectory of the corresponding pancreatic ductal organoid before i and after ii Blebbistatin treatment. C Histogram plot showing the velocity distribution of the nuclei before and after the Blebbistatin treatment. The plot shows the decrease in cell velocity after the Blebbistatin treatment on organoid #1 (n = 29–59 nuclei per organoid from 5 to 7 hours of imaging period, i.e., 11–15 time points). D Average velocity of three independent pancreatic ductal organoids before and after the Blebbistatin treatment. The decrease in velocity was observed in all three organoids (mean ± SD, *p < 0.0001, N = 3 organoids from 3 independent experiments, n = 29–87 nuclei per organoid from 3–7 hours of imaging period, i.e., 7–15 time points). Ei Confocal maximum projection of the basal surface of pancreatic ductal organoid treated with 200 µM Blebbistatin for 12 hr and stained for F-actin, pMYL9, MYH9, and DNA. Bar = 50 µm. ii, iii Zoomed-in images and intensity profile showing the disrupted actin stress fibers and the accumulation of pMYL9 at the base of the stress fibers. The white arrow indicates the direction of the intensity profile, and the white dashed box indicates the area used to derive the intensity profile. Bar = 20 µm.
Next, to confirm that rotational migration depends on myosin contractility, we inhibited myosin activity using an orthogonal approach. Organoids were treated with Y-27632, a widely used Rho-kinase (ROCK) inhibitor. ROCK enhances myosin activity by phosphorylating the myosin regulatory light chain (directly activating myosin) and the myosin-binding subunit of myosin phosphatase (suppressing phosphatase activity)30,31,32. Thus, ROCK inhibition reduces myosin phosphorylation, activation, and contractility. Consistent with this, Y-27632 treatment significantly reduced phosphorylated MYL9 levels (Fig. S3A), and quantification of rotation velocity confirmed a slowdown in migration (Fig. S3B, Video 5). Together, these results confirm that organoid rotational migration depends on myosin activity.
According to the “go-or-grow” hypothesis33,34,35, cells favor either proliferation or migration at a given time. Since pancreatic ductal organoid cells proliferate continuously (Fig. S2D), it is plausible that cell division could impede rotational migration. Indeed, we occasionally observed dividing cells during nuclear live imaging (green arrows in Video 4). However, closer inspection of nuclei before Blebbistatin treatment (Video 4, left) showed that dividing cells migrated at a speed comparable to neighboring cells, and their division did not noticeably slow rotational movement. This is intriguing because dividing cells typically round up (Fig. S2E) and detach from the substrate, suggesting that neighboring cells may help move the dividing cells to maintain rotational continuity. Although less likely, one alternative explanation is that the slowdown observed after Blebbistatin treatment could result from increased proliferation, suppressing migration. To test this, we blocked proliferation using Thymidine (Fig. S2D) prior to Blebbistatin treatment. Even in the absence of proliferation, Blebbistatin still caused a clear reduction in rotation (Video 6), indicating that its effect on rotational migration is independent of cell proliferation.
Rotational migration also depends on actin branching activity
The results above pointed towards the critical role of actin organization on pancreatic ductal organoid rotational migration. To study this, we treated the organoids with an actin polymerization inhibitor, Latrunculin A. Indeed, inhibition of actin polymerization significantly slows down the organoid rotation (Fig. S4A, Video 7). Similar to the Blebbistatin-treated organoids, Latrunculin A treatment does not disrupt the apicobasal polarity of the organoids (Fig. S4B). However, we observed wavy patterns of the tight junction protein ZO-1 (Fig. S4C), which were not observed in the untreated and Blebbistatin-treated organoids. These wavy tight junction structures might imply a decrease in cellular tension at the apical side of the Latrunculin A-treated cells. Looking closer to the basal side, Latrunculin A treatment seems to drive the actin distribution towards the perimeter of the cells, causing the enrichment of actin at the cell–cell boundaries (Fig. S4D). Additionally, the enrichment of activated myosin (pMYL9) was also observed at the periphery of some cells. Combined, these results suggest the importance of actin organization in the organoid rotational migration.
Next, we focused on actin branching, a key process in actin filament assembly and organization. Actin branching is critical for lamellipodium formation—broad, flat protrusions at the leading edge of migrating cells36. In rotating Drosophila follicle cells, lamellipodia also form at the leading edge and extend beneath neighboring cells as cryptic lamellipodia2,37. By imaging the cell–cell junction protein E-cadherin, we similarly observed cryptic lamellipodia in pancreatic ductal organoids (Fig. 5A). To test whether actin branching is required for rotational migration, we treated organoids with the Arp2/3 inhibitor CK-66638. Since actin branching nucleation depends on assembly of the Arp2/3 complex on existing actin filaments39, inhibition of this complex blocks branching. CK-666 treatment significantly slowed organoid rotation (Fig. 5B, Video 8). At the basal surface, actin stress fibers remained prominent, some decorated with pMYL9 (Fig. 5C). However, consistent with previous reports40, CK-666 disrupted both the structure (Fig. 5D) and dynamics (Video 9) of cryptic lamellipodia. Similarly, the non-rotating PDAC organoids also displayed compromised cryptic lamellipodia (Fig. S1H) when compared to the rotating pancreatic ductal organoids. Together, these results indicate that rotational migration of pancreatic ductal organoids depends on cryptic lamellipodia and actin branching.
Ai Confocal maximum projection of the basal surface of non-treated pancreatic ductal organoid stained for the cell–cell junction protein E-cadherin and DNA. Bar = 50 µm. ii An orthogonal slice was extracted from the region indicated by the white dashed line in Fig. 5Ai, showing the presence of cryptic lamellipodium at the basal side of the organoid. Bar = 20 µm. Bi Overlay of maximum projections of a rotating pancreatic ductal organoid (organoid #1) taken before the actin branching complex Arp2/3 inhibitor, CK-666, treatment. The images were taken every 30 minutes for 5 hours. The transparent green arrow indicates the general direction of the rotation. Bar = 50 µm. ii Overlay of maximum projections of the same organoid taken 1 hour after the 200 µM CK-666 treatment, showing the significant slowdown of the rotation. The images were taken every 30 minutes for 7 hours. Bar = 50 µm. The nuclei were color-coded based on the time when they were taken. The red arrow points to the nuclei of non-viable cells within the lumen of the organoid, which are excluded from the subsequent nucleus tracking analyses. iii Average velocity of three independent pancreatic ductal organoids before and after the CK-666 treatment. The decrease in velocity was observed in all three organoids (mean ± SD, *p < 0.0001, N = 3 organoids from three independent experiments, n = 14–109 nuclei per organoid from 5 to 7 hours of imaging period, i.e., 11–15 time points). C Confocal maximum projection of the basal surface of pancreatic ductal organoid treated with 200 µM CK-666 for 12 hr and stained for F-actin, pMYL9, and DNA. Actin stress fibers were still observed after actin branching inhibition by CK-666, and some of the fibers were decorated with the active pMYL9. Bar = 50 µm. Di Confocal maximum projection of the basal surface of CK-666 treated pancreatic ductal organoid stained for the cell–cell junction protein E-cadherin and DNA. Cryptic lamellipodium structures are still observed after inhibition of actin branching, but they are less prominent when compared to the non-treated organoids in Fig. 5A. Bar = 50 µm. ii An orthogonal slice was extracted from the region indicated by the white dashed line in Di, showing the presence of cryptic lamellipodia at the basal side of the organoid. Bar = 20 µm.
Conclusion
In this study, we introduce primary human pancreatic ductal organoids as a model to investigate epithelial cell rotational migration. Using this system, we quantified rotation velocity and showed that this behavior requires myosin contractility, actin polymerization, and actin branching. To enable these measurements, we developed a 3D single-cell tracking method that is transferable to other systems. The organoid is an experimentally versatile system: we can expand it at a 1:2 ratio biweekly and can genetically modify it with lentiviral or adenoviral transduction, thus enabling rapid mechanistic studies with primary human epithelial cells that better recapitulate normal physiology than immortalized lines. We hope that this model system will contribute to future studies on the unresolved regulatory mechanisms of epithelial cell rotational migration. Key questions include what initiates rotation, what sustains collective migration over time, and which downstream pathways and cellular processes depend on this behavior.
It is also important to consider the biological and experimental context of the pancreatic ductal organoid used in this study. First, the organoids were cultured in Matrigel, a hydrogel derived from mouse sarcoma that contains a complex mixture of ECM proteins, particularly laminin and collagen IV, but is known to exhibit batch-to-batch variability. Given that pancreatic ductal cells interact with the ECM (Fig. 3A), and that ECM proteins have been shown to regulate pancreatic ductal progenitors41,42 as well as phenotypes of PDAC organoids43,44,45, it is reasonable to predict that rotational migration behavior may vary when organoids are embedded in hydrogels of differing ECM composition, such as collagen I gels44,45 or polyethylene glycol matrices functionalized with defined ECM ligands43,46. Notably, however, rotational migration was consistently observed across multiple batches of Matrigel employed in this study, indicating that this behavior is not contingent upon batch-to-batch variability of Matrigel. Second, the organoid culture media contain growth factors, hormones, and supplements that may influence migration—for example, A83-01 and mNoggin inhibit TGF-β signaling. Because TGF-β is known to regulate cell migration47, its inhibition in culture could confound our results. However, organoid rotation persisted even after removing the TGF-β inhibitors (Video 10), suggesting that this behavior is not solely an artifact of TGF-β suppression. In addition, organoid architecture adds complexity: epithelial cells migrate within a spherical tissue enclosing a pressurized lumen, exposing the apical surface to hydrostatic pressure. Both hydrostatic pressure and surface curvature are known to influence cell behavior48,49,50, suggesting that these mechanical cues may modulate rotational migration. Taken together, these considerations position pancreatic ductal organoids as a physiologically relevant and versatile platform for dissecting the biochemical and mechanical regulation of epithelial rotational migration.
Methods
Pancreatic ductal organoid culture
Normal human pancreatic ductal organoids were derived from pancreatic exocrine tissues isolated from cadaver donors (RRID: SAMN17277513 and SAMN38518868), which were acquired through the IIDP. The tissue was obtained from deidentified cadaver donors; therefore, the study was exempt from review by the Florida State University Institutional Review Board, Tallahassee, Florida, USA. IIDP provided 0.5 mL exocrine tissue, which is equal to ~20 million cells. The exocrine cells were seeded into Matrigel (Corning) to allow for the growth and expansion of pancreatic ductal organoids. The Matrigel culture and expansion of pancreatic ductal organoids were performed following the Tuveson Laboratory Murine and Human Organoid Protocols (http://tuvesonlab.labsites.cshl.edu/wp-content/uploads/sites/49/2018/06/20170523_OrganoidProtocols.pdf), which were kindly compiled based on the lab’s previous studies21,24. The human wash media and complete feeding media were used, as suggested by the Tuveson Lab protocol. The wash media consists of Advanced DMEM/F-12 (ThermoFisher), supplemented with 10 mM HEPES (ThermoFisher), 1× GlutaMAX supplement (ThermoFisher), 100 µg/mL Primocin (Invivogen), and 0.1% BSA (ThermoFisher). The complete feeding media consists of Advanced DMEM/F-12, supplemented with 10 mM HEPES, 1× GlutaMAX supplement, 100 µg/mL Primocin, 1× Wnt3a conditioned media (refer to Tuveson lab protocol), 1× R-spondin1 conditioned media (refer to Tuveson lab protocol), 1× B27 supplement (ThermoFisher), 10 mM Nicotinamide (Sigma), 1.25mM N-acetylcysteine (Sigma), 100 ng/mL mNoggin (Peprotech), 50 ng/mL hEGF (Peprotech), 100 ng/mL hFGF (Peprotech), 10 nM hGastrin I (Tocris), 500 nM A83-01 (Tocris), and 1 µM PGE2 (Tocris). Considering the cells' doubling time, the organoids were passaged 1:2 every 10–14 days. For the passage procedure, the organoid containing Matrigel domes was mechanically disrupted by pipetting the complete media up and down within the well plate. The cell suspension was transferred to a 5 mL protein LoBind tube (Eppendorf) and centrifuged at 200 × g, 4 °C for 5 minutes. The supernatant was aspirated and the cell pellet was resuspended in ice-cold wash media, followed by centrifugation at 200 × g, 4 °C for 5 minutes. The supernatant was aspirated, and the cell pellet was resuspended in ice-cold Matrigel. The Matrigel cell suspension was pipetted onto a pre-warmed well plate (~37 °C) to form the Matrigel domes containing the pancreatic ductal cells, e.g., 50 µL Matrigel dome per well of a 24-well plate. Due to the growth limitation of the human pancreatic ductal organoid, the experiments were done on organoids with passage numbers less than 10. The primary tumor PDAC organoids were acquired from ATCC, as part of the Human Cancer Models Initiative, with the organoid name HCM-CSHL-0092-C25. The culture procedure of the PDAC organoids was very similar to the normal pancreatic ductal organoid culture described above, except for the exclusion of PGE2 in the complete feeding media.
Inhibitor treatment
Pancreatic ductal organoids were treated with 200 µM Blebbistatin (Sigma), 80 µM Y-27632 (Tocris), 1 µM Latrunculin A (VWR), or 200 µM CK-666 (MedChemExpress). Treatment concentrations were selected based on titration experiments to determine the lowest dose of each drug that slows down organoid rotation (Videos 11–14). For live imaging, organoids were pre-treated for 1 hour, followed by overnight imaging with the inhibitor within the culture media. Of the tested inhibitors, 80 µM Y-27632 and 1 µM Latrunculin A disrupted organoid morphology after the overnight treatment (Fig. S5). However, washout restored rotational migration for all treatments, including the recovery of organoid morphology after Y-27632 and Latrunculin A exposure (Video 15). This suggests that the chosen concentrations caused only limited toxicity within the overnight treatment window and that they are reversible. For immunofluorescence analysis, organoids were treated for 12 hours before fixation. To block DNA replication and cell proliferation, the organoids were treated with 2 µM Thymidine (Sigma) for 4 days before subsequent treatments.
Immunofluorescence imaging and immunoblot
Pancreatic ductal organoids within the Matrigel domes were fixed in 4% formaldehyde (EMS) for 15 min, permeabilized by 0.5% Triton-X (Sigma) for 10 min, blocked by 5% BSA (VWR), and incubated overnight in the primary antibodies against ZO-1 (1:250, ThermoFisher #61-7300), ITGA6 (1:250, EMD Millipore #MAB1378), phosphorylated MYL9 (pSer19) (1:250, ThermoFisher #MA5-15163), MYH9 (1:250, Proteintech #11128-1-AP), Paxillin (1:500, Abcam #ab32084), ITGB1 (1:250, DSHB #AIIB2), E-cadherin (1:250, Cell Signaling #3195S), or CDK5RAP2 (1:500, gift from Timothy Megraw51). The samples were incubated in the corresponding Alexa Fluor secondary antibodies (1:500, ThermoFisher) for 1.5 hours. DNA was stained with 2 µg/mL DAPI (Sigma) for 15 min. Actin filaments were stained with 1 µg/mL Phalloidin-TRITC (Sigma) for 1 hour. To monitor DNA replication, 20 µM EdU (Abcam) was introduced to the organoid culture for 2 hours prior to fixation, followed by the staining of EdU-labeled cells with 100 mM Tris (pH 8.5, Sigma), 1 mM CuSO4 (Sigma), 100 µM Cy5 azide dye (Lumiprobe), and 100 mM ascorbic acid (Sigma) for 30 min at room temperature. Samples were thoroughly washed, and the DNA was co-stained with DAPI as described above. Confocal imaging was done using a Zeiss LSM 880 system with a ×63/1.4 NA oil immersion objective.
For the immunoblot, organoids from at least two Matrigel domes were harvested and lysed in Pierce RIPA buffer (ThermoFisher) with the presence of phosphatase inhibitors (Sigma). Proteins extracted were denatured in the presence of NuPAGE LDS buffer (ThermoFisher) at 80 °C for 10 minutes, followed by protein separation using the NuPAGE Bis-Tris 4–12% gels (ThermoFisher). Membrane transfer was performed using the iBlot2 system (ThermoFisher). The membrane was incubated overnight at 4 °C in 1 µg/mL primary antibodies against phosphorylated MYL9 and β-actin (Santa Cruz, #sc-47778), followed by the corresponding IRDye secondary antibodies (1:2500, LiCor). The membrane was imaged using the Odyssey Dlx Imager (LiCor).
Live imaging
For the live imaging of the nuclei, the pancreatic ductal organoids within the Matrigel domes were stained with 1× SPY650-DNA (Cytoskeleton) for 16 hr prior to the live imaging. For live imaging of the cell membrane, the organoids were stained with 5 µg/mL CellMask Orange (ThermoFisher) for 1 hr prior to the live imaging. The corresponding stains were present throughout the live imaging. Live imaging was performed using an Andor Revolution Spinning Disk Confocal system (Oxford Instruments) with either a ×20/0.75 NA or ×40/0.8 NA objective, within an incubator chamber maintained at 37 °C and 5% CO2. Z-stack images were taken every 10 or 30 minutes with a z-distance of 1 or 2 µm between each stack. Meanwhile, the brightfield live imaging was performed using an Olympus IX71 with a digital sCMOS camera (Prime 95B, Photometrics) and a ×10/0.3 NA objective, within an incubator chamber maintained at 37 °C and 5% CO2.
Image-processing and velocity quantification
To acquire the position of the nuclei, the confocal z-stack images from each time point were subjected to the tracking and segmentation algorithms TrackMate v.7.14.052 and StarDist v.0.9.153 within Fiji v.2.15.154. The TrackMate-StarDist analysis provides the x, y, and z locations of the detected nuclei segments, and represents each continuous nucleus segment along the z-stack as a single track. Hence, a track of nucleus segments detected here represents one individual nucleus. Next, by using an in-house MATLAB (MathWorks) algorithm, the mid-z location of each nucleus was determined by recording the location of the nucleus segment with the highest mean intensity along each track. At this point, each nucleus of a given confocal z-stack image was represented by an x-y-z coordinate. Finally, the trajectory tracking of the nuclei over time was done by using the particle tracking algorithm55, which was adapted into MATLAB. The velocity of each nucleus was calculated from the displacement along the trajectory divided by the time needed to cover the distance. To note, occasionally there are nuclei of non-viable cells within the lumen of the organoids (e.g., Figs. 4A and 5B, S3B, and S4A). As the aim of this analysis is to measure the displacement and velocity of the migrating epithelial cells, the nuclei of these non-viable cells were excluded from the analysis. The 3D plot of nuclei position was done using scatter3sph v.1.5.0.0 in MATLAB.
Statistics and reproducibility
Statistical analyses were conducted using Welch’s t test between sample pairs. The analyses were performed using GraphPad Prism v.10. All statistical tests were considered significant if p < 0.05. The plots show mean ± SEM or mean ± SD as indicated in the figure caption.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The numerical source data for the plots presented in this manuscript can be obtained in Supplementary Data 1. All other data are available from the corresponding authors upon request.
Code availability
The MATLAB algorithms can be accessed through https://github.com/IriantoLab/Rotational-migration-in-human-pancreatic-ductal-organoids-depends-on-actin-and-myosin-activity.git.
References
Friedl, P. & Gilmour, D. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10, 445–457 (2009).
Williams, A. M., Donoughe, S., Munro, E. & Horne-Badovinac, S. Fat2 polarizes the WAVE complex in trans to align cell protrusions for collective migration. Elife 11, e78343 (2022).
Sherrard, K. M., Cetera, M. & Horne-Badovinac, S. DAAM mediates the assembly of long-lived, treadmilling stress fibers in collectively migrating epithelial cells in Drosophila. Elife 10, e72881 (2021).
Popkova, A. et al. A Cdc42-mediated supracellular network drives polarized forces and Drosophila egg chamber extension. Nat. Commun. 11, 1921 (2020).
Williams, A. M. & Horne-Badovinac, S. Fat2 polarizes Lar and Sema5c to coordinate the motility of collectively migrating epithelial cells. J. Cell Sci. 137, jcs261173 (2024).
Barlan, K., Cetera, M. & Horne-Badovinac, S. Fat2 and Lar define a basally localized planar signaling system controlling collective cell migration. Dev. Cell 40, 467–477 e465 (2017).
Marmaras, A. et al. A mathematical method for the 3D analysis of rotating deformable systems applied on lumen-forming MDCK cell aggregates. Cytoskeleton 67, 224–240 (2010).
Chin, A. S. et al. Epithelial cell chirality revealed by three-dimensional spontaneous rotation. Proc. Natl. Acad. Sci. USA 115, 12188–12193 (2018).
Hirata, E., Ichikawa, T., Horike, S. I. & Kiyokawa, E. Active K-RAS induces the coherent rotation of epithelial cells: a model for collective cell invasion in vitro. Cancer Sci. 109, 4045–4055 (2018).
Hof, L. et al. Long-term live imaging and multiscale analysis identify heterogeneity and core principles of epithelial organoid morphogenesis. BMC Biol. 19, 37 (2021).
Tan, T. H. et al. Emergent chirality in active solid rotation of pancreas spheres. PRX Life 2, 033006 (2024).
Tanner, K., Mori, H., Mroue, R., Bruni-Cardoso, A. & Bissell, M. J. Coherent angular motion in the establishment of multicellular architecture of glandular tissues. Proc. Natl. Acad. Sci. USA 109, 1973–1978 (2012).
Wang, H., Lacoche, S., Huang, L., Xue, B. & Muthuswamy, S. K. Rotational motion during three-dimensional morphogenesis of mammary epithelial acini relates to laminin matrix assembly. Proc. Natl. Acad. Sci. USA 110, 163–168 (2013).
Brandstatter, T. et al. Curvature induces active velocity waves in rotating spherical tissues. Nat. Commun. 14, 1643 (2023).
Haigo, S. L. & Bilder, D. Global tissue revolutions in a morphogenetic movement controlling elongation. Science 331, 1071–1074 (2011).
Crest, J., Diz-Munoz, A., Chen, D. Y., Fletcher, D. A. & Bilder, D. Organ sculpting by patterned extracellular matrix stiffness. Elife 6, e24958 (2017).
Fernandez, P. A. et al. Surface-tension-induced budding drives alveologenesis in human mammary gland organoids. Nat. Phys. 17, 1130–1136 (2021).
Lu, L. et al. Polarity-driven three-dimensional spontaneous rotation of a cell doublet. Nat. Phys. 20, 1194–1203 (2024).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Boj, S. F. et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015).
Kim, J., Koo, B. K. & Knoblich, J. A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584 (2020).
Corsini, N. S. & Knoblich, J. A. Human organoids: new strategies and methods for analyzing human development and disease. Cell 185, 2756–2769 (2022).
Huch, M. et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32, 2708–2721 (2013).
Dasgupta, S., Gupta, K., Zhang, Y., Viasnoff, V. & Prost, J. Physics of lumen growth. Proc. Natl. Acad. Sci. USA 115, E4751–E4757 (2018).
Narayanan, V. et al. Osmotic gradients in epithelial acini increase mechanical tension across E-cadherin, drive morphogenesis, and maintain homeostasis. Curr. Biol. 30, 624–633 e624 (2020).
Indana, D. et al. Lumen expansion is initially driven by apical actin polymerization followed by osmotic pressure in a human epiblast model. Cell Stem Cell 31, 640–656 e648 (2024).
Chen, D. Y., Lipari, K. R., Dehghan, Y., Streichan, S. J. & Bilder, D. Symmetry breaking in an edgeless epithelium by Fat2-regulated microtubule polarity. Cell Rep. 15, 1125–1133 (2016).
Kovacs, M., Toth, J., Hetenyi, C., Malnasi-Csizmadia, A. & Sellers, J. R. Mechanism of blebbistatin inhibition of myosin II. J. Biol. Chem. 279, 35557–35563 (2004).
Kimura, K. et al. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273, 245–248 (1996).
Kawano, Y. et al. Phosphorylation of myosin-binding subunit (MBS) of myosin phosphatase by Rho-kinase in vivo. J. Cell Biol. 147, 1023–1038 (1999).
Totsukawa, G. et al. Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J. Cell Biol. 150, 797–806 (2000).
Hoek, K. S. et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 68, 650–656 (2008).
Schultz, C., Lemke, N., Ge, S., Golembieski, W. A. & Rempel, S. A. Secreted protein acidic and rich in cysteine promotes glioma invasion and delays tumor growth in vivo. Cancer Res. 62, 6270–6277 (2002).
Giese, A. et al. Dichotomy of astrocytoma migration and proliferation. Int. J. Cancer 67, 275–282 (1996).
Vinzenz, M. et al. Actin branching in the initiation and maintenance of lamellipodia. J. Cell Sci. 125, 2775–2785 (2012).
Squarr, A. J. et al. Fat2 acts through the WAVE regulatory complex to drive collective cell migration during tissue rotation. J. Cell Biol. 212, 591–603 (2016).
Nolen, B. J. et al. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460, 1031–1034 (2009).
Mullins, R. D., Heuser, J. A. & Pollard, T. D. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. USA 95, 6181–6186 (1998).
Ozawa, M. et al. Adherens junction regulates cryptic lamellipodia formation for epithelial cell migration. J. Cell Biol. 219, e202006196 (2020).
Zook, H. N. et al. Activation of ductal progenitor-like cells from adult human pancreas requires extracellular matrix protein signaling. iScience 27, 109237 (2024).
Quijano, J. C. et al. Methylcellulose colony assay and single-cell micro-manipulation reveal progenitor-like cells in adult human pancreatic ducts. Stem Cell Rep. 18, 618–635 (2023).
Below, C. R. et al. A microenvironment-inspired synthetic three-dimensional model for pancreatic ductal adenocarcinoma organoids. Nat. Mater. 21, 110–119 (2022).
Randriamanantsoa, S. et al. Spatiotemporal dynamics of self-organized branching in pancreas-derived organoids. Nat. Commun. 13, 5219 (2022).
Papargyriou, A. et al. Heterogeneity-driven phenotypic plasticity and treatment response in branched-organoid models of pancreatic ductal adenocarcinoma. Nat. Biomed. Eng. 9, 836–864 (2025).
Usman, O. H. et al. Differential modulation of cellular phenotype and drug sensitivity by extracellular matrix proteins in primary and metastatic pancreatic cancer cells. Mol. Biol. Cell 34, ar130 (2023).
Deng, Z. et al. TGF-beta signaling in health, disease, and therapeutics. Sig. Transduct. Target Ther. 9, 61 (2024).
Chan, C. J. et al. Hydraulic control of mammalian embryo size and cell fate. Nature 571, 112–116 (2019).
Yoshino, D. et al. Hydrostatic pressure promotes endothelial tube formation through aquaporin 1 and Ras-ERK signaling. Commun. Biol. 3, 152 (2020).
Schamberger, B. et al. Curvature in biological systems: its quantification, emergence, and implications across the scales. Adv. Mater. 35, e2206110 (2023).
Barrera, J. A. et al. CDK5RAP2 regulates centriole engagement and cohesion in mice. Dev. Cell 18, 913–926 (2010).
Ershov, D. et al. TrackMate 7: integrating state-of-the-art segmentation algorithms into tracking pipelines. Nat. Methods 19, 829–832 (2022).
Schmidt, U., Weigert, M., Broaddus, C. & Myers, G. In Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. (eds. A. F. Frangi, J. A. Schnabel, C. Davatzikos, C. Alberola-López & G. Fichtinger) 265–273 (Springer International Publishing, Cham; 2018).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Crocker, J. C. & Grier, D. G. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 179, 298–310 (1996).
Acknowledgements
The authors would like to thank the Confocal Microscopy Laboratory at FSU College of Medicine for access to the confocal microscopes. The authors would like to thank Dr. Terra Bradley for the careful editing of the manuscript and Prof. Timothy L Megraw for the antibody against CDK5RAP2. J.I. and G.X. were supported by startup funds from Florida State University, awards from the Florida Department of Health’s Bankhead-Coley Cancer Research Program (award number 21B11) and Live Like Bella Pediatric Cancer Research Initiative (award number 23L06). Y.J.W. was supported by the American Diabetes Association grant #11-22-JDFPM-03 and the Islet Award Initiative (IIDP-IAI; https://iidp.coh.org/Investigators/Islet-Award-Initiative). Human pancreatic exocrine tissues were provided by the NIDDK-funded IIDP (RRID:SCR_014387) at City of Hope, NIH Grant # U24DK098085. The PDAC organoid line is from the Human Cancer Models Initiative (HCMI), https://ocg.cancer.gov/programs/HCMI. D.T. and X.L. were supported by the National Science Foundation (grant no. PHY 2310639), the Collie-Welch Chair through the Welch Foundation (F-0019).
Author information
Authors and Affiliations
Contributions
G.X., C.M. and J.I. wrote the manuscript. G.X., C.M., and O.H.U. designed and performed experiments, and data analysis. R.W.F.T., N.C., and G.D.J. performed experiments and data analysis. Y.J.W. and D.T. provide insights into data interpretation and manuscript preparation. X.L. established the image-processing pipeline. X.L. and J.I. provide supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Mostafa Bakhti and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors. Simona Chera and Ophelia Bu.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xie, G., Modak, C., Usman, O.H. et al. Rotational migration in human pancreatic ductal organoids depends on actin and myosin activity. Commun Biol 8, 1749 (2025). https://doi.org/10.1038/s42003-025-09136-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09136-y