Abstract
ProQ is a post-transcriptional gene regulator in bacteria known to globally interact with both messenger RNAs (mRNAs) and small regulatory RNAs (sRNAs). Here, we investigate ProQ-mRNA interactions in live bacteria using single-molecule tracking and reveal their dependence on the structural elements of ProQ. We illustrate that during exponential growth phase, stationary phase, and osmotic stress, ProQ mainly binds to mRNAs. All structural domains of ProQ, including N-terminal domain, linker region, and C-terminal domain, contribute to mRNA binding. While residue Y70 is generally required for global mRNA binding, other tested residues in the N-terminal domain have minor to moderate effects on mRNA binding, indicating that different mRNAs likely depend on different ProQ residues for binding. Finally, our data reveal that the expressed sRNA substrates have limited impacts on the fraction of mRNA-associated ProQ, likely due to the high abundance of mRNA-free ProQ inside the cell.
Similar content being viewed by others
Introduction
Bacterial cells employ several strategies to adapt to hostile environments, such as the presence of antibiotics or attacks by the host’s macrophages. RNA binding proteins (RBPs) often serve as critical regulators for gene expression during stress responses. In addition, the expression of small regulatory RNAs (sRNAs) helps to rapidly reprogram the cell’s transcriptome and proteome to optimize the metabolism against such attacks. To perform their function promptly and specifically on target mRNAs, these sRNAs also often rely on RBPs.
Hfq, as one global RBP, has been prominently investigated over the past few decades1,2,3. Hfq broadly binds sRNAs and mRNAs and plays critical roles in the sRNA-mediated gene regulation by increasing the stability of associated sRNAs and facilitating base pairing between sRNAs and target mRNAs4,5,6. Hfq exists as a homohexamer, with its proximal face, distal face, and rim all able to interact with RNAs with different sequence preferences, as revealed by genetic, biochemical, and imaging characterizations1,2,3. Specifically, using single-molecule tracking (SMT) in live Escherichia coli cells, we previously demonstrated that Hfq can dynamically interact with mRNAs and sRNAs, depending on the availability and sequence features of the sRNA substrates7. Hfq proteins mainly interact with mRNAs through the distal face. Expressed sRNAs can bind to mRNA-associated Hfq either by directly displacing mRNAs from Hfq or by co-occupying Hfq with mRNAs, depending on their binding sites on Hfq. The presence of multiple RNA binding surfaces on Hfq enables dynamic exchange among different classes of RNAs and allows potential multivalent interactions between multiple RNAs1,7,8.
More recently, additional global RBPs have been identified, including the FinO-domain-containing protein ProQ9,10. ProQ was initially characterized as a key factor in proline uptake as well as in the expression of proP for usage in osmoregulation11,12,13. Recent studies have classified ProQ as a global RBP in Gram-negative bacteria, including E. coli, Salmonella enterica, Neisseria meningitidis and Pasteurella multocida14,15,16,17,18, and have demonstrated its ability to interact with different sRNA targets and the 3’ untranslated region (UTR) of mRNAs14,16. The RNA interactome of ProQ largely differs from Hfq’s, with only a small subset of overlapping RNA targets14. However, in contrast to Hfq, for which each binding site has its own sequence preference, ProQ is suggested to preferentially bind structures in RNA partners rather than specific sequences14,16. While ProQ is functionally linked to biofilm formation, stress responses, virulence, and motility19,20,21,22, the mechanisms of ProQ-mediated gene regulation via interaction with RNA substrates remain to be fully explored. ProQ functioning as a sRNA chaperone was reported in a few cases, such as repressing translation of hupA mRNA via RaiZ sRNA19. Recent transcriptomic studies revealed that ProQ has a global impact on mRNA lifetime23. More specifically, it was shown to protect RNAs (such as mRNA cspE16, rpoS24, sRNAs RbsZ and RybB14) from degradation, likely by preventing the binding of poly(A) polymerase24.
ProQ protein consists of three structural regions: N-terminal FinO domain (NTD) that spans residues 1–119, C-terminal Tudor domain (CTD) that spans residues 176–228, and a likely unstructured extended linker region that carries positively charged residues9,25. It has been suggested that the relatively conserved NTD is the main RNA binding site for ProQ. This observation is supported by a bacterial three-hybrid (B3H) assay using specific sRNAs and RNA fragments from 3’ UTR of mRNAs26. Hereafter, we use sRNAs to collectively refer to both sRNAs and 3’UTR-containing RNA fragments. In this assay, the mutations affecting the binding of ProQ to the RNAs are mostly located within the NTD region, whereas deletion of the linker and/or the CTD does not compromise the RNA binding capabilities of ProQ26. Moreover, in vitro electrophoretic mobility shift assays (EMSA) illustrated that the dependence on the CTD for RNA-binding activity differs between different tested sRNAs. Specifically, the NTD of ProQ carries the ability to recognize sRNA substrates that possess Rho-independent terminator structure, whereas the full length ProQ can bind RNA molecules with both Rho-dependent terminator and independent terminator structures27. However, since a large fraction of ProQ RNA interactome is mRNAs, it is still unclear if ProQ follows the same structural dependence for mRNA binding as for sRNA binding.
Here, we investigated the interactions of ProQ with mRNAs and sRNAs in live E. coli cells using SMT as previously used to study Hfq7. We relied on the change in diffusivity of ProQ to distinguish the binding of cellular mRNAs to ProQ compared to free ProQ or sRNA-associated ProQ. Our results confirmed that ProQ can largely associate with cellular mRNAs. However, distinct from interactions with tested sRNAs in previous studies26,27, we found that the linker and CTD are important for mRNA binding. Mutagenesis of conserved residues in the NTD, identified as important for binding of sRNAs26, further revealed that while some residues are generally critical for mRNA binding, others have moderate to minor impact on global mRNA binding. These results indicate that the studied residues may contribute variably to distinct subsets of mRNAs. Finally, competition experiments using selected sRNAs revealed that ProQ’s linker and CTD contribute to competitive binding of sRNAs in a sRNA-dependent manner. Collectively, our findings highlight different structure requirements of ProQ for global mRNA binding and sRNA binding, suggesting that ProQ may use distinct sites to interact with different types of RNAs.
Results
Tracking ProQ proteins in live E. coli cells
To allow tracking of ProQ protein in live cells, its C-terminus was tagged with several commonly used photo-switchable fluorescent proteins (FPs) through chromosomal insertion in E. coli, including mEos3.228, mMaple329, Dendra230 or PAmCherry31. The PAmCherry tag was selected as it resulted in the best signal for SMT measurement. ProQ is known to regulate the expression of genes involved in osmotic stress resistance, among other stresses11,12,13. Therefore, to confirm the functionalities of ProQ-PAmCherry, we assessed the growth rate and imaged cell morphology of the proQ-PAmCherry strain compared to wild type (WT) and ΔproQ strains in a high salt environment (250 mM NaCl). The bacterial strain expressing ProQ-PAmCherry demonstrated a similar growth rate as the WT strain, while ΔproQ strain demonstrated a minor growth defect and reached a lower OD600 at stationary phase (Fig. 1A). In addition, there was no significant difference between the morphology of the proQ-PAmCherry strain compared to the WT strain, but ΔproQ cells were significantly smaller in size, with more cells showing a circular morphology (Fig. 1B, C), in agreement with a previous study32. Based on these observations, we concluded that tagging ProQ at the C-terminus with PAmCherry does not cause defects in E. coli cell growth and morphology, suggesting that it is not preventing ProQ from performing its normal RBP activities.
In all experiments, we performed SMT on ProQ-PAmCherry using single-molecule localization microscopy in two dimensions (2D) in live E. coli cells growing in MOPS EZ rich defined medium. Images were collected at a rate of 120 frames per second with 5 ms exposure time for each frame. We first tracked WT ProQ-PAmCherry in cells grown at exponential phase without any treatment (referred to as “no treatment”, or “NT” condition) (Fig. 2A and Supplementary Fig. 1A). Quantification of ProQ-PAmCherry localization with DNA stained by Hoechst revealed that during exponential growth, ProQ proteins have comparable localization in the nucleoid and cytoplasm regions but are depleted from the cell membrane (Fig. 2B). This localization preference differs from that of Hfq, which demonstrates uniform localization throughout the cell under NT condition7. The higher membrane localization fraction of Hfq compared to ProQ may be related to the interaction of Hfq and the inner membrane-bound RNase E during Hfq-dependent regulation events7. In addition, we calculated the one-step displacement (osd) speed of individual ProQ-PAmCherry protein at each time step and plotted osd speed as a function of the cellular coordinate in a diffusivity speed map (Fig. 2C). The speed map and quantification of the average osd speed suggest that ProQ diffuses across the cells with similar speed, similar to what was previously observed for Hfq7.
A Representative example of WT ProQ-PAmCherry in WT genetic background in a single cell during exponential growth, under no treatment (NT) condition. Panel 1: DIC image of the cell. Panel 2: Nucleoid is stained with Hoechst in live cells. Panel 3: 2D reconstruction image of ProQ-PAmCherry. Panel 4: Different diffusion trajectories from tracking algorithm are shown in different colors (“Traj”). Panel 5: One-step displacement (osd) speed map (unit: µm s−1) is shown as a scatter plot where different colors represent different speeds at each position, and the white curves represent the nucleoid regions detected by Hoechst staining. The scale bar represents 1 µm. B Enrichment of ProQ localization is calculated for cytoplasm, membrane and nucleoid regions for individual cells under NT condition. C Average osd speed of ProQ within the cytoplasm, membrane and nucleoid regions in individual cells under NT condition. Each violin plot contains 80 cells collected from 3 biological replicates. P-values calculated using two-tailed t-test are labeled on top of each bar graph.
ProQ demonstrates global mRNA binding capability during exponential growth
To investigate interactions between ProQ and cellular mRNAs with SMT, we tracked ProQ-PAmCherry in E. coli cells treated with rifampicin (RIF) (Supplementary Fig. 1A). RIF inhibits cellular transcription, leading to the loss of most cellular mRNAs and some sRNAs due to their shorter lifetimes33,34 compared to rRNAs or tRNAs35,36. We calculated the effective diffusion coefficient (Deff) by fitting the linear function to the mean squared displacement (MSD) as a function of time lag (Δt) (Fig. 3A). Deff increased under RIF treatment in comparison to NT case (Figs. 3A, 4A), indicating that a fraction of WT ProQ proteins is associated with mRNA molecules during exponential growth phase.
A Mean square displacement (MSD) is plotted against the time interval (Δt) for ProQ-PAmCherry in WT genetic background under NT and rifampicin (RIF) treated conditions. The linear fitting lines are shown. B Representative fitting of the cumulative probability density function (CDF) of one-step squared displacement (osd2) distribution for WT ProQ-PAmCherry in WT background under rifampicin treatment. CDF is better fit by a double-population model (blue) than a single-population model (red). Fitting by a triple-population model (green) does not further improve as its chi square (χ2) is larger than that of the double-population model. C CDF of osd2 distribution for WT ProQ-PAmCherry in WT background under NT and RIF treatment cases and corresponding double-population fitting (blue solid curves). D Representative posterior occupation histogram of diffusion coefficient from saSPT analysis for ProQ-PAmCherry in WT genetic background under RIF treated (top) and NT (bottom) conditions. Histogram was fit with Gaussian function. Individual Gaussian peaks are marked with red curve, and the global fit (in NT case) is marked with blue curve. The Dfast and Dslow are marked in the plots as the center of the Gaussian fit.
A Effective diffusion coefficients (Deff) are plotted for WT ProQ-PAmCherry in WT genetic background under NT and RIF treatment conditions during exponential growth, stationary phase and osmotic stress. B mRNA-associated fractions calculated using CDF fitting for WT ProQ-PAmCherry in WT genetic background under NT and RIF treatment conditions during exponential growth, stationary phase and osmotic stress. C Diffusion coefficients of the fast population (Dfast) (left) and the slow population (Dslow) (right) determined by the two-population CDF fitting for WT ProQ under three growth conditions. For exponential and stationary phase, the numbers are reported for RIF case, whereas for the osmotic stress, the numbers are reported for NT case. D Number of spots per cell for WT ProQ in the WT genetic background under NT condition. The number of spots were determined using 7500 frames consistently across all the samples. E Representative posterior occupation histogram of diffusion coefficient from saSPT analysis for WT ProQ-PAmCherry during osmotic stress. Histogram was fit with Gaussian function. Individual Gaussian peaks are marked with red curve, and the global fit is marked with blue curve. The Dfast, Dslow, Dultra-slow,1 and Dultra-slow,2 are marked in the plots as the center of each Gaussian peak. Experiments were performed for 2 to 6 biological replicates, with each data set containing ~1000 trajectories for Deff value calculation or ~2500 trajectories for mRNA-associated fraction calculation from ~50 cells from exponential growth and osmotic stress conditions; and ~200 trajectories for Deff value calculation or ~600 trajectories for mRNA-associated fraction calculation from ~50 cells for stationary phase. Error bars represent the s.d. for cases with >3 biological replicates. P-values calculated using two-tailed t-test are labeled on top of each bar graph. All fitting results from both CDF fitting and saSPT analysis are reported in Supplementary Tables 1 and 2.
To analyze the subpopulations of ProQ-PAmCherry under NT and RIF treated conditions, we fit the cumulative probability density function (CDF) of one-step squared displacement (osd2) with double populations7,37 (Fig. 3B, C). In the RIF treated case, the CDF curve can be reasonably well described by a single-population model. Double-population model improved the fitting, but triple-population model did not further improve the fitting (Fig. 3B). We therefore chose the double-population model. ~79% of ProQ population exhibited a fast diffusion in RIF treated condition with an osd2 value of 5.1 µm2 s−1 (Dfast), whereas the remaining population (~21%) was in a slow diffusion state with an osd2 value of 1.36 µm2 s−1 (Dslow) (Supplementary Table 1). Similar to our previous work on Hfq, we assigned the slow and fast diffusion states to be the mRNA-bound and mRNA-free fractions of ProQ, respectively. In the NT condition, the slow-diffusion population increased to ~68%, suggesting that a large fraction of ProQ population is associated with mRNAs during exponential growth (Figs. 3C and 4B, Supplementary Table 1). Notably, after RIF treatment, there is still a small subpopulation of ProQ with low diffusion coefficients, which may be due to ProQ interactions with other cellular components, such as ribosomes21.
We validated the population analysis using saSPT38, an alternative method using Bayesian nonparametric modeling. Consistent with the CDF fitting method described above, in the RIF treated case, the distribution of diffusion coefficient predominately showed a major peak centered around 1.2 µm2 s−1, corresponding to the mRNA-free population (Fig. 3D). In the NT condition, a second peak centered around 0.3 µm2 s−1 appeared (Fig. 3D). The slow-diffusion peak occupied ~82% of the total population (Supplementary Table 2), consistent with the mRNA-bound population estimated using the CDF fitting method. We noticed the saSPT analysis consistently estimated a ~ 4-fold slower diffusion coefficient compared to CDF fitting, mainly because saSPT estimates effective diffusion coefficient from the trajectories, whereas CDF considers one-step displacement. Nevertheless, the estimated mRNA-bound fraction of ProQ using both methods is highly consistent (Supplementary Fig. 2, Supplementary Tables 1 and 2), also as described below.
ProQ remains associated with mRNAs with a decreased abundance at the stationary phase
We next compared ProQ diffusivity under NT and RIF treatment conditions at the early stationary phase (Supplementary Fig. 1A). Both the diffusion coefficient (Fig. 4A) and the mRNA-associated fraction of ProQ (Fig. 4B) during stationary phase remain comparable to those during exponential growth. In addition, the diffusion coefficients of the mRNA-associated (Dslow) and mRNA-free population (Dfast) of ProQ at the stationary phase are comparable with the corresponding values at the exponential phase (Fig. 4C). We then estimated relative protein abundance by counting the number of spots from SMT trajectories using the same number of image frames. We found ProQ abundance to be ~10% compared to exponential phase (Fig. 4D). Collectively, these results show that ProQ remains largely associated with mRNAs at the stationary phase, however, its abundance is significantly reduced.
ProQ exhibits ultra-slow diffusion states during osmotic stress
We next tracked WT ProQ-PAmCherry in cells under osmotic stress with treatment of NaCl during exponential phase. Under osmotic shock, ProQ localization was predominantly in the cytoplasm, due to nucleoid compaction39 (Supplementary Fig. 1A). Relative ProQ abundance and ensemble Deff in osmotic stress are comparable to those in unstressed condition (Fig. 4A, D). Population analysis using both methods estimated a similar or mildly increased mRNA-bound fraction of ProQ under osmotic stress compared to unstressed conditions (Fig. 4B, Supplementary Tables 1 and 2). We observed significant cell damage upon RIF treatment together with osmotic stress (Supplementary Fig. 1B) and therefore could not perform SMT under this condition.
Interestingly, saSPT analysis revealed four diffusion states of ProQ under osmotic stress compared to two states in NT case in either exponential or stationary phase (Fig. 4E, Supplementary Table 2). The fast diffusion state, Dfast (1.07 µm2 s−1 using saSPT) under osmotic stress is comparable with Dfast under NT case (1.12 µm2 s−1 during exponential growth and 1.02 µm2 s−1 during stationary phase) (Fig. 4E, Supplementary Table 2), reflecting that the diffusion coefficient of the mRNA-free ProQ is unchanged under osmotic stress. The first slow diffusion state Dslow (0.55 µm2 s−1 using saSPT) in osmotic stress is faster compared to Dslow (0.30 µm2 s−1 during exponential growth and 0.35 µm2 s−1 during stationary phase) (Fig. 4E, Supplementary Table 2). We interpret this slight increase as due to a global decrease in translation activity of mRNAs during osmotic stress40, which reduces the average molecular weight of translated mRNAs and leads to an increase in the diffusion coefficient. In addition, two “ultra-slow” diffusion states were revealed under osmotic stress (Dultra-slow,1: 0.17 µm2 s−1, Dultra-slow,2: 0.01 µm2 s−1) (Fig. 4E, Supplementary Table 2). The exact molecular states that correspond to these ultra-slow diffusion states remain to be further investigated.
The fraction of mRNA-bound ProQ remains similar in the absence of Hfq
Since previous studies identified a partially overlapping set of RNA binding partners between Hfq and ProQ14, we next sought to identify the impact of Hfq on ProQ’s diffusivity. We knocked out the hfq gene and performed all SMT experiments on ProQ-PAmCherry in the Δhfq background. The presence or absence of Hfq had an insignificant impact on ProQ diffusivity under NT or RIF treatment conditions (Fig. 5A, Supplementary Tables 1 and 2). These results indicate that the mRNAs shared between Hfq and ProQ do not constitute a significant proportion of the total abundance of the ProQ interactome. Another possibility is that among the shared mRNA binders, ProQ and Hfq likely bind to different regions on their shared mRNA partners and do not interfere strongly with each other, consistent with Hfq and ProQ having distinct sequence or structural preferences to associate with their RNA partners14,16,27.
A Effective diffusion coefficients (Deff) of WT and six mutant ProQ-PAmCherry proteins in the Δhfq background under NT and RIF treatment conditions. B Diffusion coefficients of the fast population (Dfast) (left) and the slow population (Dslow) (right) determined by the two-population CDF fitting for WT and six mutant ProQ-PAmCherry proteins in the Δhfq background under RIF treatment condition. C mRNA-associated fractions determined by the two-population CDF fitting for WT and six mutant ProQ-PAmCherry proteins in the Δhfq background under NT and RIF treatment conditions. D Number of spots per cell for WT and six mutant ProQ-PAmCherry proteins in the Δhfq background under NT condition. The number of spots were determined using 7500 frames consistently across all the samples. Experiments were performed for 4 to 9 biological replicates, with each data set containing ~1000 trajectories for Deff value calculation or ~2500 trajectories for mRNA-associated fraction calculation from ~50 cells. Error bars in all plots represent the s.d. from all biological replicates. P-values calculated using two-tailed t-test are labeled on top of each bar graph. All fitting results from both CDF fitting and saSPT analysis are reported in Supplementary Tables 1 and 2.
The linker and CTD of ProQ significantly contribute to its endogenous mRNA binding capability in vivo
Given that mRNAs contribute to a major class of ProQ substrates under several tested conditions, we next sought to determine the structural regions of ProQ that modulate its mRNA binding capability. In a previous study, a B3H assay revealed that the ProQ NTD/FinO-domain is the primary binding site for selected sRNAs26. In vitro binding assays further illustrated that ProQ’s NTD can only bind sRNAs that possess Rho-independent terminator structures, while the full length ProQ is able to bind both Rho-dependent and independent terminators structures of sRNAs27. To explore whether similar rules affect ProQ’s interactions with mRNAs, we generated PAmCherry-tagged ProQ constructs with both the CTD and linker region deleted (referred to as ProQ NTD) or only the CTD deleted (ProQ NTD+linker). We then performed SMT of these mutant ProQ constructs under NT and RIF conditions. All experiments were conducted in a Δhfq background.
The comparison of diffusion coefficients under NT and RIF conditions between these constructs illustrates the contribution of each structural domain of ProQ to global mRNA binding. As shown in Fig. 5A, diffusion coefficients of ProQ NTD and ProQ NTD+linker constructs under NT and RIF conditions are higher than those of WT ProQ under corresponding conditions. From population analysis, the mRNA-free fraction of ProQ NTD and ProQ NTD+linker constructs have a faster diffusion coefficient compared to WT ProQ construct (Fig. 5B, Supplementary Tables 1 and 2), likely attributed to changes in the molecular weight of the protein due to truncations. Under NT conditions, the mRNA-bound fraction of ProQ NTD+linker was reduced by ~50% compared to WT ProQ, and the mRNA binding ability was nearly abolished in ProQ NTD (Fig. 5C, Supplementary Tables 1 and 2). These results suggest that ProQ NTD alone cannot interact strongly with long mRNAs in the cell, and that the presence of the linker region can partially rescue the mRNA binding.
mRNA binding activity of ProQ demonstrates differential dependence on NTD residues
Because ProQ’s NTD has been suggested to be the most critical for binding to the sRNAs, we generated constructs with single-point mutations in the NTD to assess the role of these residues in global mRNA binding. Namely, the highly conserved positively charged residues K54 and R58, aromatic residue Y70, and negatively charged residue D82 of ProQ NTD were individually mutated. These residues were shown to influence binding of ProQ to cspE 3’UTR and SibB sRNA26. SMT experiments on ProQ mutants under NT and RIF treatment showed that different mutations affect ProQ-mRNA interactions to a different extent (Fig. 5A, C, Supplementary Tables 1 and 2). Specifically, mutation Y70S nearly abolished mRNA binding activity of ProQ. This result suggests that residue Y70 is critical for mRNA binding, consistent with its universal importance for binding of sRNAs41. Mutation R58A moderately impacted mRNA binding, and mutations K54A and D82A minimally impacted mRNA binding. These results contrast with the significant roles of these residues in ProQ binding to cspE 3’UTR and SibC sRNA27, suggesting that mRNAs and sRNAs require distinct ProQ features for efficient binding. A recent study demonstrated that critical residues involved in ProQ-RNA interactions vary among sRNA substrates41. Similarly, our data show that while residue Y70 is essential to ProQ-mRNA interactions, other tested residues of NTD involved in binding of mRNAs may vary among mRNA substrates, leading to a moderate to insignificant decrease in the global mRNA-associated fraction caused by a single-point mutation.
To rule out the possibility that the decrease of the mRNA-bound fraction is due to the protein abundance change of ProQ mutants, we compared the relative abundance of different ProQ mutants. We observed a 0.5- to 2.5-fold change in the abundance of ProQ mutants compared to WT ProQ (Fig. 5D). However, the increase or decrease in the abundance of ProQ mutants does not correlate with the observed changes in the mRNA-associated fraction. Indeed, if the decrease in the mRNA-bound fraction was purely driven by the increase of the ProQ mutant abundance, leading to a sub-stoichiometry of mRNA substrate, or vice versa, one would expect that ProQ R58A (one of the highest abundances) has the largest decrease in the mRNA-bound fraction, whereas ProQ Y70S (the lowest abundance) has the smallest decrease in the mRNA-bound fraction. Our data do not demonstrate such a trend and therefore argues against the possibility that the change in the abundance of ProQ mutants is responsible for the observed changes in the mRNA-bound fraction.
CTD and linker of ProQ contribute to the competitive binding by sRNAs
Like Hfq, ProQ was revealed to interact with both mRNAs and sRNAs. We therefore next sought to investigate whether sRNAs can compete against mRNAs for ProQ binding, as in the case of Hfq. We selected three sRNA substrates of ProQ with different structural complexities from S. enterica16: (i) cspE 3’UTR, encompassing two comparably short RNA hairpins; (ii) STnc1690, containing one extended RNA hairpin, and (iii) SibC with multiple hairpins (Fig. 6A). ChiX was used as a control sRNA. Vector-based expression of these sRNAs was controlled by an inducible promoter (Plac). Using northern blots, we also verified the expression of these sRNAs and estimated their copy numbers to be in the range of 270–950 molecules per cell (Supplementary Fig. 3), consistent with previously measured sRNA copy number7,42,43.
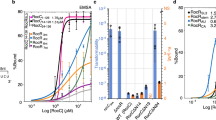
A Secondary structures of selected ProQ-binding sRNAs. B Effective diffusion coefficients (Deff) and C mRNA-associated fractions of WT ProQ, ProQ NTD and ProQ NTD+linker in the presence of an empty vector and vectors expressing STnC1690, SibC, cspE 3’UTR and ChiX determined by saSPT analysis (left) and CDF fitting (right). Experiments were performed for 2 to 8 biological replicates, with each data set containing ~1500 trajectories for Deff value calculation or ~5000 trajectories for mRNA-associated fraction calculation from ~40 cells. Error bars represent the s.d. for cases with >3 biological replicates. P-values calculated using two-tailed t-test are labeled on top of each bar graph. All fitting results are reported in Supplementary Tables 3 and 4.
To study the competitive binding to ProQ by the sRNAs, we measured the diffusion of ProQ upon expression of each sRNA. All experiments were carried in Δhfq strains expressing PAmCherry-tagged WT ProQ, ProQ NTD or ProQ NTD+linker. Due to their significantly smaller sizes, sRNAs that can bind ProQ more strongly and displace cellular mRNAs are expected to cause an increase in the diffusion coefficient of ProQ. Overexpression of all three sRNAs (cspE 3’UTR, STnc1690, SibC) mildly increased the Deff of WT ProQ compared to induction of an empty vector (Fig. 6B), suggesting that all three sRNAs displaced a small fraction of mRNAs from ProQ (~2–13% by CDF fitting and ~9–20% by saSPT) (Fig. 6C, Supplementary Fig. 2, Supplementary Tables 3 and 4). STnc1690 demonstrated the highest level of mRNA displacement from ProQ. As a control, ChiX sRNA was induced in the same way. No significant difference was observed in ProQ diffusivity in the presence of ChiX (Fig. 6C, Supplementary Tables 3 and 4), confirming that the binding of ProQ to cspE 3’UTR, STnc1690 or SibC is specific. The fractions of ProQ competed by these sRNAs were much smaller compared to ChiX competition for Hfq protein7.
ProQ has been estimated to be in the range of 2000–15,000 copies per cell21,44,45. To understand whether the low efficiency of mRNA displacement from ProQ by sRNAs is due to the much higher abundance of ProQ protein over sRNAs, we estimated the WT ProQ copy number by western blot using a standard curve generated from purified ProQ. During exponential growth in MOPS rich media, ProQ copy number was estimated to be ~17,000 copies per cell (Figure Supplementary 4). Considering that the expressed sRNAs are 270–950 copies per cell, most sRNAs may preferentially bind to mRNA-free ProQ rather than competing for the mRNA-associated ProQ.
We finally examined whether the NTD and linker region of ProQ contribute to the competitive binding by the sRNAs (Fig. 6B, C). The ability of STnc1690 to displace mRNAs from ProQ is not strongly affected by the deletion of CTD or the linker. However, SibC and cspE 3’UTR were unable to displace mRNAs to bind ProQ in the absence of the CTD or in absence of both the linker and CTD. Overall, these results demonstrate that while these sRNAs do not require CTD or the linker for binding to ProQ, their ability to compete with mRNAs for ProQ relies on these structural domains to some extent. This observation supports the previous hypothesis that the CTD of ProQ may be needed for RNA exchange on this protein12 and highlights the complex structural requirements of ProQ for various RNA substrate binding and competition.
Discussion
Using SMT, we measured the interactions of ProQ with bulk mRNAs, as well as with three sRNAs in live E. coli cells. Our results revealed that, similar to Hfq, over half of the ProQ population under exponential growth conditions is occupied by cellular mRNAs. A slightly higher mRNA-associated fraction of ProQ compared to Hfq is consistent with the previous CLIP-seq comparison between these two proteins16,46. Specifically, in Salmonella, ~450 binding sites were detected in ~167 mRNA coding genes for Hfq, and ~527 binding sites were detected in ~331 mRNA coding genes for ProQ. In contrast, the number of sRNAs that bind to ProQ (~70 peaks from CLIP-seq) is smaller than Hfq (225 peaks from CLIP-seq). Thus, compared to Hfq, these data support that ProQ interactome mainly consists of mRNAs. In addition, the number of RNA pairs that associate with ProQ is significantly smaller compared to Hfq14. This difference in RNA binding preferences between ProQ and Hfq indicates that ProQ may primarily regulate mRNAs in addition to being involved in the classic sRNA-mediated regulation as for Hfq.
By examining two stress conditions, our results show that ProQ level and diffusivity are sensitive to specific stresses. While the mRNA-associated fraction remains similar, we found ProQ level significantly decreased during stationary phase. Previous studies showed that certain ProQ mutants that are deficient in mRNA binding can be misfolded or degraded by Lon protease47. Given the global decrease in the mRNA level during stationary phase48, we speculate that ProQ level is downregulated during stationary phase to maintain a balance with the abundance of mRNA substrates. In contrast, during short osmotic stress, ProQ level remains comparable to that in the unstressed condition. We find that ProQ specifically exhibits two additional ultra-slow diffusion states compared to all other tested conditions (including different growth conditions, ProQ mutations and the presence of sRNA substrate). While the exact molecular nature and the functional relevance remain to be further investigated, several possibilities may contribute to the formation of the ultra-slow diffusion states, including ProQ interaction with even larger molecular complexes compared to translated mRNAs, interaction with spatially confined, slow-diffusing or static biomolecules, or formation of large molecular assemblies or condensates, which have been observed for other RNA binding proteins, such as TmaR and Hfq under specific stress conditions49,50.
To further understand how ProQ interacts with its mRNA substrates, we first explored the roles of its structural domains by introducing domain deletions and then examined several critical residues in NTD known for sRNA binding by mutagenesis. Our data collectively reveal that ProQ employs different structural elements to interact with mRNA and sRNA substrates.
First, we found that the CTD and the linker both contribute to the full binding activity of ProQ with cellular mRNAs, whereas NTD alone shows minimal mRNA binding activity. This observation contrasts the case of ProQ interactions with certain sRNAs, in which NTD is sufficient for interactions26, particularly for sRNAs and UTR fragments harboring Rho-independent terminator structures27. Indeed, mass spectrometry combined with hydrogen deuterium exchange experiment showed that cspE 3’UTR and SraB sRNA interact with non-identical residues on ProQ. In particular, ProQ CTD and linker region potentially contribute to the binding of larger RNA substrate, i.e., SraB25. Our findings here further support that for large mRNA substrates, linker and CTD of ProQ are necessary for efficient mRNA binding. Interestingly, a functional genetic screen revealed that mutations in the ProQ CTD that were not noted to affect binding of sRNAs can impact ProQ’s function by indirectly activating fliE transcription51. In addition, ProQ carrying mutations on the CTD also fails to protect target mRNAs from degradation51. Our data supports the importance of ProQ CTD for its regulatory functions, likely through its contributions to mRNA binding. It is worth mentioning that while we demonstrated the PAmCherry tag does not interfere with the function of WT ProQ, we cannot completely rule out the possibility that the truncated ProQ proteins are more sensitive to the tag, and therefore the impact of NTD and linker on mRNA binding might be overestimated in our assay.
Second, among the conserved residues on ProQ NTD, we found that certain residues identified to be critical for binding of tested sRNAs26 do not significantly compromise ProQ interactions with bulk mRNAs. A recent mutagenesis study on a series of selected sRNAs showed that while Y70 and R80 are universally required for all tested sRNAs, the other tested residues individually have minor to moderate effect on binding in a substrate-dependent manner41. Similarly, a mutagenesis screening of the ProQ/FinO domain-containing protein RocC of Legionella pneumophila identified Y70 as one of the most critical residues for interactions with RocR sRNA52. Reflecting these observations, our study highlights that Y70 is also generally required for global mRNA binding, while other tested residues demonstrated a minor to moderate effect on mRNA binding. However, a limitation of our method is that we cannot distinguish interactions of ProQ with different mRNA subsets. We therefore speculate that different sets of ProQ residues may be involved in interactions with distinct subclasses of mRNAs, which could explain why a single-point mutation in ProQ only leads to a moderate to negligible reduction in overall mRNA binding efficiency even if it can significantly affect the binding of a specific mRNA species. Collectively, our results indicate that while RNA binding pocket in the NTD of ProQ is defined, the dependence on specific residues for interacting with different mRNAs may vary largely. Future high-throughput sequencing experiments to globally identify RNA interactome associated with each ProQ mutant may allow establishing correlation between specific ProQ structural features and specific RNA sequence features.
Finally, we assessed the competitive binding of specific sRNAs to ProQ. Unlike sRNA binding to Hfq, we found that the tested sRNAs have limited impacts on the fraction of mRNA-associated ProQ, likely due to the high copy number of ProQ protein compared to expressed sRNAs. In addition, we observed that different sRNAs have distinct dependencies on ProQ’s structural domains during competition with mRNAs. While STnC1690 can compete for ProQ binding without the CTD or the linker, cspE 3’ UTR and SibC sRNA are not able to compete unless all three domains are present. This observation indicates that, while certain sRNAs can bind exclusively to the NTD, their ability to compete with mRNAs already bound to ProQ is likely promoted by the presence of the CTD and linker regions, supporting that the CTD may facilitate dynamic RNA exchange on ProQ12.
Methods
Plasmid construction
Genes encoding stnC1690 and sibC were cloned with a pLac-O1 promoter and a rrnB terminator into a modified pET15b vector backbone (kind gift from Perozo lab) using BglII and EcoRI restriction sites. cspE 3’UTR was cloned with a pLac-O1 promoter into a modified pET15b vector backbone using BglII and EcoRI restriction sites. First, RNA sequences were amplified from Salmonella enterica subsp. enterica serovar Typhimurium strain SL1344 using primers EM5167-EM5168 (stnC1690), EM5169-EM5170 (sibC) or EM5165-EM5195 (cspE 3’UTR). Using the resulting fragments as templates, PCR amplification using primer EM5171-EM5196 (stnC1690, sibC) or EM5171-EM5195 (cspE 3’UTR) was performed to complete the promoter sequence. Vector and fragments were digested with BglII and EcoRI, and ligated.
Bacterial strain construction
Strain KP2389 was generated by elimination of the FRT-flanked antibiotic resistance cassette from strain KP1224A (Δhfq::kan)7 after transformation with pCP20 (FLIP), as described previously53.
Strain containing chromosomal proQ-PAmCherry gene was constructed as follows. PCR1 was performed on pKD4 vector using primers EM5186-EM5187. PCR2 was performed using a vector harboring the PAmCherry gene54 as template, with primers EM5198-EM5189. PCR1 and PCR2 were annealed and amplified with primers EM5198-EM5187, and the resulting fragment was transformed into a pKD46-containing WT strain expressing the λ Red system53. The proQ-PAmCherry::kan construct was transferred to a WT strain (EM1055) or to a Δhfq strain (KP2389) through P1 transduction, yielding strain MCC1529 and KP2390, respectively. Subsequent mutants of ProQ-PAmCherry were constructed by using strain MCC1529 as a template for PCR amplification with primers EM5300-EM5191 (R80A), EM5301-EM5191 (K54A), EM5302-EM5191 (R58A), EM5303-EM5191 (Y70S), and EM5304-EM5191 (D82A). PCR products were transformed into a pKD46-containing WT strain expressing the λ Red system and transferred into strain KP2389 (Δhfq) through phage P1 transduction.
Strains containing chromosomal ProQ NTD-PAmCherry (NTD) or ProQ NTD+linker-PAmCherry (ΔCTD) were constructed by first performing PCR amplification on pKD4 using primers EM5186-EM5187 (PCR1). PCR amplification on strain MCC1529 using primers EM5305-EM5189 (NTD) or EM5306-EM5189 (NTD+linker) yielded PCR2.1 and PCR2.2. PCR 3 was achieved through annealing of PCR1 to either PCR2.1 or PCR 2.2 and subsequent amplification using primers EM5187-EM5305 (PCR2.1, NTD) or EM5187-EM5306 (PCR2.2, NTD+linker). PCR products were transformed into a pKD46-containing WT strain expressing the λ Red system and transferred into strain KP2389 (Δhfq) through P1 transduction.
Growth curve measurement
Bacterial strains WT, ΔproQ, and proQ-PAmcherry were grown overnight in LB (250 mM NaCl). The following day, samples were prepared at 3 × 106 cells/ml in triplicates in 96-well flat base polystyrene Microtest plates (Sarstedt). Growth was monitored using the Epoch 2 Microplate Spectrophotometer reader (BioTek) with the following settings: OD: 600 nm, Temperature: 37 °C, Shaking: continuous, Measurement interval: 10 min, Total time: 28 h.
Culture growth and treatment for imaging experiments
Each E. coli strain was grown in LB containing 50 µg/mL kanamycin in the cases without induction of sRNAs or LB containing 50 µg/mL kanamycin and 100 µg/mL of ampicillin in the cases with induction of sRNAs at 37 oC and 250 RPM aerobically overnight (~14 h). The following day, overnight E. coli cultures were diluted 100-fold in MOPS EZ rich defined medium (Teknova) with 0.2% fructose serving as the carbon source and were allowed to grow to the desired OD600nm of ~0.2 for measurement at exponential phase, and to OD600nm of ~1.1–1.3 for the measurement at stationary phase. 1 mL of cell culture was harvested through centrifugation at 1500 x g for 5 min. In the case of RIF treatment, RIF was added at a final concentration of 200 ug/mL when for OD600nm reached 0.2 for 15 min before harvesting. In the case of osmotic stress, 300 mM of NaCl was added to the cell culture when OD600nm reached 0.2 for 10 min before harvesting. The cells were imaged in the presence of the same NaCl concentration. Plasmid-encoded RNAs were induced by isopropyl β-D-1-thiogalactopyranoside (IPTG) for 45 min at a final concentration of 1 mM when the OD600nm reached ~0.1 before harvesting. The centrifugated cells were concentrated by pipetting out ~960 µL of the supernatant and mixing with remaining ~40 µL of solution. ~1.5 µL of concentrated cell culture was transferred to a cleaned #1.5 coverslip sealed to a custom 3D printed chamber, and the cells were laid flat and secured by placing a 5 mm × 5 mm 1% agarose gel pad made from MOPS EZ-rich defined medium, fructose and corresponding treatment reagents.
Single-molecule tracking
SMT was conducted on a Nikon Ti-E inverted microscope setup equipped with 100x NA 1.49 CFI HP TIRF oil immersion objective as previously described55. A 405 nm laser (Crstalaser) and a 561 nm laser (Coherent Obis LS) were used for photo conversion and imaging of PAmCherry respectively. Common dichroic mirror (zt405/488/561/647/752r-UF3) and emission filter (Chroma ET595/50 M) were used as excitation and emission filters. EMCCD camera (Andor iXon Ultra) was used to capture emission signals. Fast frame rates were achieved through cropping imaging region by preceding the camera with an emission image splitter (Cairn OptoSplit III). Before laser stimulation, cells were examined under Differential Interference Contrast (DIC) mode to select regions of interests. During SMT experiment, 2 × 2 binning mode and 1.5x tube lens were used resulting in a pixel size of 173 nm. After each frame of 405 nm excitation, ten frames with 561 nm excitation were recorded. The power density of the 561 nm laser was ~2750 W/cm2, and the power density of the 405 nm laser was ~7 W/cm2. Images were collected at a rate of 120 frames per second with 5 ms exposure time for each frame. Around 7500 frames were collected for each movie at room temperature. To prevent drifting in Z direction during the course of imaging, a built-in focus lock system (Nikon Perfect Focus) was engaged. All images were recorded using Nikon NIS-Element software.
Image reconstruction
SMT images were reconstructed using a custom code written in Interactive Data Language (IDL)43. Briefly, all the pixels with an intensity value above three times of the standard deviation of the individual frame pixel intensity were identified in each frame. Among those pixels, the ones having larger values than surrounding pixels in each 5 × 5 pixel region were identified as possible peak candidates, and 2D Gaussian function was fit to a 7 × 7 pixel region surrounding these candidates. Candidates with failed fitting were discarded, and precise peak positions were defined for the remaining ones. The horizontal drift was corrected by fast Fourier transformation analysis.
Calculation of effective diffusion coefficient
Regions enclosing cells in the reconstructed images composed of ~7500 frames were cropped and files containing x-y coordinates of selected peaks were extracted. A custom code written in MATLAB was used to yield diffusion trajectories56,57. To minimize artificial diffusion trajectories connecting individual peaks, we imposed a 400 nm maximum one-step displacement criteria at a time resolution of 8.27 ms. Diffusion trajectories longer than five consecutive frames were used to determine the effective diffusion coefficient. Deff values were extracted as the slopes from fitting mean squared displacement (MSD) and time lag (Δt) with linear functions (MSD = Deff x Δt) using OriginPro.
Population analysis using fitting of cumulative probability density function
A double population fitting of the cumulative probability density function (CDF) of one-step squared displacement (osd 2) was used to analyze the distribution of mRNA-associated and mRNA-free populations of ProQ:
where P1 and D1 are designated as population fraction and diffusion coefficient for fast-diffusing state interpreted as the mRNA-free fraction; P2 and D2 are designated as population fraction and diffusion coefficient for the slow-diffusing state interpreted as mRNA-associated fraction. For each ProQ construct, we fit the RIF treatment cases first and used the mean ±2x Standard deviation of D1 to constrain the fitting of D1 in the corresponding NT condition of the same ProQ construct. Two state population fittings were carried out in OriginPro with Levenberg-Marquardt iteration algorithm. We also tested the fitting with single population as well as triple population fit while double population fit gives the lowest χ2 values out of all three fits. Diffusion trajectories longer than three time steps were included for the derivation of osd2.
Population analysis using saSPT
Diffusion trajectories were fitted to a RBME (regular Brownian motion with localization error) likelihood function using saSPT 0.4.038 in Jupyter Notebook. The parameters of pixel size, frame interval, and focal depth were set to 0.173 µm, 0.00833 s (120 Hz), and 0.2 µm, respectively. Results were extracted as a posterior distribution of diffusion coefficients. The distribution of diffusion coefficient was fit with Gaussian functions to extract the average diffusion coefficient of each state using OriginPro.
Northern blot
Cells were grown until an OD600nm of 0.2 in MOPS EZ rich medium supplemented with 0.2% fructose and appropriate antibiotics, when necessary. sRNA substrate expression was induced by addition of 1 mM IPTG for 60 min, after which total RNA was extracted using the hot-phenol procedure58. Total RNA was resolved on 5% acrylamide 29:1, 8 M urea gels and RNAs were detected with the following radiolabeled DNA probes: EM5204 (stnC1690), EM5205 (SibC), EM5203 (cspE 3’UTR). Quantification was achieved by generating calibration curves using in vitro transcribed sRNAs. Templates for in vitro transcription were obtained through PCR amplification on S. enterica SL1344 using primers EM5175-EM5176 (stnC1690), EM5177-EM5178 (sibC) or EM5173-5174 (cspE 3’UTR). In vitro transcription reactions were carried as described previously59. In vitro transcribed sRNAs of various amount ranging from 1-200 fmole were migrated alongside total RNA samples. Densitometry analysis was done using Image Studio Lite software (LICOR).
ProQ protein purification
5 ml of MCC1506 (BL21-DE3 carrying pET-21b plasmids expressing ProQ-6His) overnight culture was used to inoculate 1 L of fresh LB medium and incubated at 37 °C with shaking at 200 rpm until an OD600nm of 0.7 was reached. ProQ expression was then induced with 0.5 mM IPTG for 3 hours. Cells were harvested by centrifugation at 2900 x g for 15 min and resuspended in a resuspension buffer (20 mM Tris-HCl pH 8; 200 mM NaCl; 20 mM imidazole; 5 mM MgCl2) containing 1 protease inhibitors tablet (Thermo Scientific, catalog number A32961). Cell lysis was performed using an Emulsiflex device (1000–15,000 psi, 3 passes). The lysate was centrifuged at 30,000 × g for 35 min at 4° C, and then the supernatant was collected and incubated for 1 h with 2.5 ml of Ni-NTA agarose beads resin at 4 °C. The mixture was then loaded onto a column, washed 10 times with wash buffer (20 mM Tris-HCl pH 7.5; 500 mM NaCl; 50 mM imidazole; 5 mM MgCl2; 1 mM DTT) and eluted 5 times with elution buffer (20 mM Tris-HCl pH 7.5; NaCl 500 mM; 500 mM imidazole; 5 mM MgCl2; 1 mM DTT). Fractions were analyzed by Coomassie blue staining and ProQ-containing fractions were pooled and applied to a PD-10 column with Sephadex G-25 to perform buffer exchange to the storage buffer (20 mM HEPES pH 7.5; 70 mM NH4Cl; 30 mM KCl; 7 mM MgCl2; 10% Glycerol). Protein concentration was determined by BCA assay (Thermo Scientific, catalog number 23221 for Reagent A and 23224 for Reagent B), using bovine serum albumin standard (WISENT INC, catalog number 800-095-EG).
Western blot
Strains MCC1529 (ProQ-PAmCherry) and EM1055 were cultured at 37 °C in MOPS rich medium supplemented with 0.2% fructose until reaching an OD600nm of 0.2, followed by an additional 1 h incubation in the same conditions to mimic sRNA induction time. 110 µl of ice-cold 50% TCA (trichloroacetic acid) was then added to 500 μL culture aliquots and the solutions were mixed on ice for 10 min. After precipitation by centrifugation at 13,000 rpm, 4 °C for 10 min, pellets were washed twice with 80% acetone (centrifuged at 13,000 rpm, 4 °C for 5 min each time) and resuspended in 100 μL of 1x protein buffer (125 mM Tris-HCl pH 6,8; 1% SDS; 20% glycerol; 0.02% bromophenol blue; 100 mM DTT). The total protein extraction can be temporarily stored at −20 °C. 10 µl total protein extraction was denatured at 90 °C for 3 min and separated by 12% SDS-PAGE ran at 150 V for 1 h. Proteins were then electro-transferred to a nitrocellulose membrane at 250 mA for 1 h. The membrane was blocked in TBS–5% milk and incubated overnight with a primary rabbit anti-ProQ antibody (kind gift from Dr. Janet Wood, with 1:500–1000 dilution), followed by 3 washes in TBS-T (TBS + 0.1% Tween-20). The membrane was incubated for 1 h at room temperature with an anti-rabbit fluorophore-conjugated secondary antibody (1:100,000, LICORbio, catalog number 926-32211) and washed 3 times in TBS-T. Detection was performed using the Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE, USA), and band densitometry was analyzed with the ImageJ software.
Statistics and reproducibility
Sample sizes include the number of diffusion trajectories and number of cells from each biological replicate, as well as the number of biological replicates, which are all reported in the related figure captions. The biological replicate is defined by independent bacterial cell culture used in one experiment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Code availability
Codes for analyzing diffusion coefficient, population analysis and enrichment analysis are available at GitHub: https://github.com/JingyiFeiLab/sptPALM_MSD_osd2. https://github.com/JingyiFeiLab/STORM_Diffusion-master_2023_05_05.
References
Vogel, J. & Luisi, B. F. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589 (2011).
Santiago-Frangos, A. & Woodson, S. A. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip. Rev. RNA 9, e1475 (2018).
Updegrove, T. B., Zhang, A. & Storz, G. Hfq: the flexible RNA matchmaker. Curr. Opin. Microbiol. 30, 133–138 (2016).
Wagner, E. G. H. & Romby, P. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 90, 133–208 (2015).
Storz, G., Vogel, J. & Wassarman, K. M. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43, 880–891 (2011).
Carrier, M.-C., Lalaouna, D. & Massé, E. Broadening the definition of bacterial small RNAs: characteristics and mechanisms of action. Annu. Rev. Microbiol. 72, 141–161 (2018).
Park, S. et al. Dynamic interactions between the RNA chaperone Hfq, small regulatory RNAs, and mRNAs in live bacterial cells. eLife 10, e64207 (2021).
Małecka, E. M. & Woodson, S. A. Stepwise sRNA targeting of structured bacterial mRNAs leads to abortive annealing. Mol. Cell 81, 1988–1999.e4 (2021).
Olejniczak, M. & Storz, G. ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers? Mol. Microbiol. 104, 905–915 (2017).
Mark Glover, J. N. et al. The FinO family of bacterial RNA chaperones. Plasmid 78, 79–87 (2015).
Milner, J. L. & Wood, J. M. Insertion proQ220::Tn5 alters regulation of proline porter II, a transporter of proline and glycine betaine in Escherichia coli. J. Bacteriol. 171, 947–951 (1989).
Chaulk, S. G. et al. ProQ is an RNA chaperone that controls ProP levels in Escherichia coli. Biochemistry 50, 3095–3106 (2011).
Kunte, H. J., Crane, R. A., Culham, D. E., Richmond, D. & Wood, J. M. Protein ProQ influences osmotic activation of compatible solute transporter ProP in Escherichia coli K-12. J. Bacteriol. 181, 1537–1543 (1999).
Melamed, S., Adams, P. P., Zhang, A., Zhang, H. & Storz, G. RNA-RNA interactomes of ProQ and Hfq reveal overlapping and competing roles. Mol. Cell 77, 411–425.e7 (2019).
Smirnov, A. et al. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc. Natl. Acad. Sci. USA 113, 11591–11596 (2016).
Holmqvist, E., Li, L., Bischler, T., Barquist, L. & Vogel, J. Global maps of ProQ binding in vivo reveal target recognition via RNA structure and stability control at mRNA 3’ Ends. Mol. Cell 70, 971–982.e6 (2018).
Gulliver, E. L. et al. The role and targets of the RNA-binding protein ProQ in the gram-negative bacterial pathogen Pasteurella multocida. J. Bacteriol. 204, e00592-21 (2022).
Bauriedl, S. et al. The minimal meningococcal ProQ protein has an intrinsic capacity for structure-based global RNA recognition. Nat. Commun. 11, 2823 (2020).
Smirnov, A., Wang, C., Drewry, L. L. & Vogel, J. Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA. EMBO J. 36, 1029–1045 (2017).
Westermann, A. J. et al. The major RNA-binding protein ProQ impacts virulence gene expression in salmonella enterica serovar Typhimurium. mBio 10, e02504–18 (2019).
Sheidy, D. T. & Zielke, R. A. Analysis and expansion of the role of the Escherichia coli protein ProQ. PloS ONE 8, e79656 (2013).
Avrani, S., Bolotin, E., Katz, S. & Hershberg, R. Rapid genetic adaptation during the first four months of survival under resource exhaustion. Mol. Biol. Evol. 34, 1758 (2017).
Jenniches, L. et al. Improved RNA stability estimation through Bayesian modeling reveals most Salmonella transcripts have subminute half-lives. Proc. Natl. Acad. Sci. USA 121, e2308814121 (2024).
Bergman, S., Birk, C. & Holmqvist, E. ProQ prevents mRNA degradation through inhibition of poly(A) polymerase. Nucleic Acids Res. 53, gkaf103 (2025).
Gonzalez, G. M. et al. Structure of the Escherichia coli ProQ RNA-binding protein. RNA 23, 696–711 (2017).
Pandey, S. et al. Genetic identification of the functional surface for RNA binding by Escherichia coli ProQ. Nucleic Acids Res. 48, 4507–4520 (2020).
Stein, E. M. et al. Determinants of RNA recognition by the FinO domain of the Escherichia coli ProQ protein. Nucleic Acids Res. 48, 7502–7519 (2020).
Zhang, M. et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat. Methods 9, 727–729 (2012).
Wang, S., Moffitt, J. R., Dempsey, G. T., Xie, X. S. & Zhuang, X. Characterization and development of photoactivatable fluorescent proteins for single-molecule-based superresolution imaging. Proc. Natl. Acad. Sci. USA 111, 8452–8457 (2014).
Chudakov, D. M., Lukyanov, S. & Lukyanov, K. A. Using photoactivatable fluorescent protein Dendra2 to track protein movement. BioTechniques 42, 553–563 (2007).
Subach, F. V. et al. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods 6, 153–159 (2009).
Kerr, C. H., Culham, D. E., Marom, D. & Wood, J. M. Salinity-dependent impacts of ProQ, Prc, and Spr deficiencies on Escherichia coli cell structure. J. Bacteriol. 196, 1286–1296 (2014).
Chen, H., Shiroguchi, K., Ge, H. & Xie, X. S. Genome-wide study of mRNA degradation and transcript elongation in Escherichia coli. Mol. Syst. Biol. 11, 808 (2015).
Vogel, J. et al. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 31, 6435 (2003).
Svenningsen, S. L., Kongstad, M., Stenum, T. S., Muñoz-Gómez, A. J. & Sørensen, M. A. Transfer RNA is highly unstable during early amino acid starvation in Escherichia coli. Nucleic Acids Res. 45, 793–804 (2017).
Blundell, M. R. & Wild, D. G. Altered ribosomes after inhibition of Escherichia coli by rifampicin. Biochem. J. 121, 391 (1971).
Bettridge, K., Verma, S., Weng, X., Adhya, S. & Xiao, J. Single-molecule tracking reveals that the nucleoid-associated protein HU plays a dual role in maintaining proper nucleoid volume through differential interactions with chromosomal DNA. Mol. Microbiol. 115, 12–27 (2021).
Heckert, A., Dahal, L., Tjian, R. & Darzacq, X. Recovering mixtures of fast-diffusing states from short single-particle trajectories. eLife 11, e70169 (2022).
Walker, A. M., Abbondanzieri, E. A. & Meyer, A. S. Live to fight another day: The bacterial nucleoid under stress. Mol. Microbiol. 123, 168–175 (2025).
Dai, X. et al. Slowdown of translational elongation in Escherichia coli under hyperosmotic stress. mBio 9, e02375–17 (2018).
Stein, E. M. et al. Biochemical and genetic dissection of the RNA-binding surface of the FinO domain of Escherichia coli ProQ. RNA 29, 1772–1791 (2023).
Reyer, M. A. et al. Kinetic modeling reveals additional regulation at co-transcriptional level by post-transcriptional sRNA regulators. Cell Rep. 36, 109764 (2021).
Fei, J. et al. RNA biochemistry. Determination of in vivo target search kinetics of regulatory noncoding RNA. Science 347, 1371–1374 (2015).
Dolgalev, G. V., Safonov, T. A., Arzumanian, V. A., Kiseleva, O. I. & Poverennaya, E. V. Estimating total quantitative protein content in Escherichia coli, Saccharomyces cerevisiae, and HeLa Cells. Int. J. Mol. Sci. 24, 2081 (2023).
Li, G.-W., Burkhardt, D., Gross, C. & Weissman, J. S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 (2014).
Holmqvist, E. et al. Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J. 35, 991–1011 (2016).
El Mouali, Y. et al. Scanning mutagenesis of RNA-binding protein ProQ reveals a quality control role for the Lon protease. RNA 27, 1512–1527 (2021).
Morin, M., Enjalbert, B., Ropers, D., Girbal, L. & Cocaign-Bousquet, M. Genomewide stabilization of mRNA during a “Feast-to-Famine” growth transition in Escherichia coli. mSphere 5, e00276–20 (2020).
Szoke, T. et al. Regulation of major bacterial survival strategies by transcripts sequestration in a membraneless organelle. Cell Rep. 42, 113393 (2023).
McQuail, J., Switzer, A., Burchell, L. & Wigneshweraraj, S. The RNA-binding protein Hfq assembles into foci-like structures in nitrogen starved Escherichia coli. J. Biol. Chem. 295, 12355–12367 (2020).
Rizvanovic, A., Kjellin, J., Söderbom, F. & Holmqvist, E. Saturation mutagenesis charts the functional landscape of Salmonella ProQ and reveals a gene regulatory function of its C-terminal domain. Nucleic Acids Res. 49, 9992–10006 (2021).
Kim, H. J. et al. Structural basis for recognition of transcriptional terminator structures by ProQ/FinO domain RNA chaperones. Nat. Commun. 13, 7076 (2022).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97, 6640–6645 (2000).
Buss, J. et al. A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genet. 11, e1005128 (2015).
Park, S. et al. Conducting multiple imaging modes with one fluorescence microscope. J. Vis. Exp. JoVE 28, 58320 (2018).
Crocker, J. C. & Grier, D. G. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 179, 298–310 (1996).
Sadoon, A. A. & Wang, Y. Anomalous, non-Gaussian, viscoelastic, and age-dependent dynamics of histonelike nucleoid-structuring proteins in live Escherichia coli. Phys. Rev. E 98, 042411 (2018).
Aiba, H., Adhya, S. & de Crombrugghe, B. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256, 11905–11910 (1981).
Desnoyers, G., Morissette, A., Prévost, K. & Massé, E. Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J. 28, 1551 (2009).
Acknowledgements
This work was supported by the NIH director’s new innovator Award (1DP2GM128185-01) to J.F., and by an operating grant BMB 389354 from the Canadian Institutes of Health Research (CIHR) to E.M. We thank Dr. Janet M. Wood for sharing the ProQ primary antibody.
Author information
Authors and Affiliations
Contributions
J.F. and E.M. conceptualized and supervised the project; J.F., E.M., K.P., M.C.C. and S.P. designed related experiments; Z.Z., K.P., M.C.C., S.P., N.E.A., C.D.V.V., and A.G. performed the experiments; Z.Z., K.P., M.C.C., S.P., N.E.A., J.H.M. and J.F. performed the analysis; J.F., Z.Z., and M.C.C. wrote the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Gisela Storz, Alexandre Smirnov and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Kaliya Georgieva. [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhou, Z., Prévost, K., Carrier, MC. et al. Structural dependence of ProQ-mRNA interactions in live bacterial cells. Commun Biol 8, 1780 (2025). https://doi.org/10.1038/s42003-025-09145-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09145-x