Abstract
Cancer is increasingly recognized as a systemic disease, extending beyond local alterations to systemic alterations in distant organs through the release of various factors that promote tumor progression and metastasis. Here, we applied hyperspectral stimulated Raman scattering (hSRS) microscopy to study metabolic alterations in the liver driven by distant tumors, revealing unprecedented accumulation of vitamin A-enriched lipid droplets. Quantitative spectral analysis uncovered increased unsaturation levels and abnormal vitamin A ester. Notably, inhibition of secretory pathways in remote tumors effectively abrogated these metabolic alterations, with FABP5 in tumor-derived extracellular vesicles identified as a key mediator. These findings uncover a unique aspect of cancer progression mechanisms, implicating tumor-driven systemic lipid metabolic remodeling and vitamin A dysregulation in metastatic progression and therapeutic response.

Similar content being viewed by others
Introduction
Although cancer originates in specific tissues or organs, it is now recognized as a systemic disease1. Recent studies have identified metabolic reprogramming within cancer cells and their microenvironment as a hallmark of cancer progression and metastasis2,3. Cancer-induced metabolic reprogramming not only promotes tumor growth and metastasis4 but also facilitates immune evasion5 and therapy resistance6. Beyond local reprogramming, cancer cells release metabolites, secretory proteins, and extracellular vesicles (EVs) that induce metabolic alterations in distant organs7. This systemic effect supports the formation of pre-metastatic niches, creating a favorable environment and providing nutritional support for the development of metastasis6,8,9,10,11. For example, EVs are key regulators of this process, transporting specific signaling molecules and mRNA to distant sites12 and driving metabolic alterations, in pro-metastatic tissues13,14. Given the complexity and importance of these systematic interactions of cancer and organs, identifying tumor-induced metabolic signatures in distant organs could offer valuable insights into the mechanisms of systematic alterations and open new avenues for early diagnosis and targeted treatment.
Recent studies employing various -omics approaches, including transcriptomics, proteomics, and metabolomics, have advanced our understanding of cancer-organ communication, revealing distinct metabolic profiles in multiple tumor-affected organs10,12,15. For example, in the lungs of tumor-bearing mice, enhanced glycolysis and lactate production have been observed, with lactate promoting PD-L1 expression in macrophages, leading to an immunosuppressive phenotype9. Additionally, tumor-derived EVs have been shown to alter fatty acid metabolism and promote lipid accumulation in liver tissues, facilitating the formation of pre-metastasis niches conducive to cancer metastasis6. These findings highlight the significance of cancer-induced systematic alterations; however, the substantial heterogeneity of tissues and the unique spatial distribution of metabolites - closely tied to metabolic status and function – emphasize the need to characterize tumor-induced metabolic profiles at single-cell or even subcellular resolution.
For in situ detection and imaging of metabolites, vibrational spectroscopy provides direct measurement of chemical bond vibrations, enabling high-resolution chemical analysis in cells and tissues16. Raman-based techniques, including spontaneous Raman and stimulated Raman scattering (SRS) microscopy, allow label-free chemical imaging by detecting intrinsic vibrational signatures of different metabolites17,18,19,20,21,22. SRS offers enhanced signal levels and faster acquisition, enabling imaging of large tissue regions with high spatial resolution and minimal background interference from autofluorescence. Here, we visualize cancer-induced metabolic alterations in the liver, a primary metastatic site of cancer23 and hub for systematic metabolism in an organism, using hyperspectral stimulated Raman scattering (hSRS) imaging in both C-H stretching and fingerprint regions. Through SRS imaging in the high-wavenumber C-H stretching window, we observed a significant increase in lipid droplets (LDs), a major lipid storage organelle, in the livers of mice with aggressive breast tumors. Spectroscopic analysis of these LDs further revealed an enrichment of unsaturated lipids. Notably, fingerprint hSRS imaging identified previously unrecognized accumulation of vitamin A in the liver, primarily stored in esterified form within LDs. Mechanistically, these metabolic alterations in the liver were abrogated by inhibiting the EV secretion from the primary tumor, where FABP5 was identified as a key cargo protein within EVs. Collectively, these results demonstrate tumor-induced metabolic alterations in distant organs and reveal molecular signatures with potential applications in diagnosis and targeted therapy.
Results
hSRS imaging reveals altered LD content and composition in the liver of TB mice
Previous studies have shown that tumor-secreted factors promote liver metastasis24. More recently, tumor-secreted factors were also reported to induce liver metabolic dysfunctions prior to metastasis4, suggesting a potential link between tumor-induced metabolic alterations and the preferential colonization of certain cancers in the liver. To investigate such tumor-induced metabolic abnormalities, we focused on triple-negative breast cancer (TNBC), which shows a strong tendency for liver metastasis25,26,27.
To investigate the impact of distant tumors on liver metabolism, MDA-MB-231 breast cancer cells were orthotopically implanted in NSG mice to establish the tumor-bearing (TB) mice, while PBS-implanted mice served as the tumor-free (TF) control group. 50 days post tumor implantation, liver tissues were collected and frozen-sectioned for hSRS imaging (Fig. 1A, B). hSRS imaging was performed in both the high wavenumber C-H stretching region, which is used to quantify lipid and protein contents, and the fingerprint region, which provides more detailed compositional information. From the high wavenumber images, a distinct distribution pattern of CH-rich biomolecules was observed in the liver tissues of TB mice compared to TF mice (Fig. 1C, Supplementary Movie 1 and 2). Phasor analysis28 of the C-H spectra allowed segmentation of lipid-rich and protein-rich regions (Fig. 1D), revealing a significant accumulation of LDs in the liver of TB mice. Quantification of LD size and number further confirmed an increase in LDs in the TB liver compared to the TF liver (Fig. 1E), especially for LDs larger than 2 μm2, which were scarcely present in the TF liver. The overall lipid content was measured from each LD by taking the sum SRS intensity at 2855 cm−1, showing significantly higher lipid levels in the TB liver (Fig. 1F).
A Schematic illustration of tumor-induced metabolic reprogramming in the liver. B Schematic illustration of animal model. C SRS images of liver tissues from TF and TB mice in the C-H stretching window. D Segmentation of lipid-rich (green) and protein-rich (blue) regions by phasor analysis of the C-H stretching spectra. E Quantification of LD size measured from the lipid-rich region. F Quantification of lipid intensity at 2855 cm−1 for each LD. G Mean SRS spectra of lipid-rich and protein-rich regions in the C-H stretching window. H Intensity ratio of 3015 cm−1 and 2855 cm−1 peaks, representing relative unsaturated lipid content for each LD. I Mean SRS spectra of lipid-rich and protein-rich regions in the fingerprint region. *p < 0.05; ****p < 0.0001. Error bars represent SD. Scale bars: 20 μm. Part of the figure is created in BioRender. Lee, H. (2025) https://BioRender.com/jozm8oh.
To further investigate the composition of these LDs, SRS spectra were extracted from lipid-rich and protein-rich regions (Fig. 1G). Both lipid and protein spectra exhibited distinct profiles between TB and TF livers, indicating significant molecular alterations of the tissue. Specifically, the lipid spectrum from TB liver showed a peak at 3015 cm−1, corresponding to the =C–H bond in unsaturated lipids (Supplementary Fig. 1). The ratio of unsaturated lipids to total lipids in each LD was quantified by calculating the intensity ratio of 3015 cm−1 and 2855 cm−1 peaks (Fig. 1H), showing a significantly increased unsaturation level in TB liver compared to TF liver. Interestingly, the 3015 cm−1 peak was also observed in the protein spectrum from TB liver, suggesting the presence of unsaturated lipid species associated with proteins in the cytoplasm.
Several unsaturated fatty acids and esterified metabolites, such as cholesterol and fat-soluble vitamins, may accumulate in LDs, particularly within the liver29. Gene Set Enrichment Analysis (GSEA) of bulk RNA-seq data from TB and TF livers further indicated significant enrichment of genes associated with the metabolism of fat-soluble vitamins (Supplementary Fig. 2). To explore the composition of these unsaturated lipid species, we performed hSRS imaging in the fingerprint region. Using masks derived from the high wavenumber segmentation, we obtained fingerprint SRS spectra from lipid-rich and protein-rich regions (Fig. 1I, Supplementary Movie 3 and 4). In the lipid-rich region of TB liver, a sharp peak at 1655 cm−1 was observed, corresponding to the acyl C = C bond in lipids (Supplementary Fig. 3), confirming the high unsaturation level indicated by the 3015 cm−1 peak. Notably, a prominent peak at 1595 cm−1, representing conjugated C = C bonds30,31, was observed exclusively in TB liver. In the protein-rich region, a broad peak at 1655 cm−1, corresponding to the amide I band of proteins (Supplementary Fig. 3)32, was present in both TF and TB livers. Interestingly, the 1595 cm−1 peak was also detected in the protein-rich regions of TB liver. These results collectively suggest that the presence of tumors significantly alters lipid metabolism in distant livers.
Fingerprint imaging identifies abnormal accumulation of vitamin A metabolites in LDs in TB liver
Liver is a central organ in lipid, cholesterol, and vitamin metabolism, serving as a hub for related metabolites33. To explore the origin of the 1595 cm−1 peak observed in TB liver, we investigated several metabolites containing conjugated C = C bonds, including cholesterol, vitamin A, and vitamin D (Fig. 2A). In contrast to the C-H stretching region, where the spectra of these biomolecules overlap significantly (Fig. 2B), the fingerprint region reveals well-separated peaks between 1600 and 1670 cm−1 (Fig. 2C). Specifically, cholesterol exhibits a peak at 1668 cm−1, vitamin D2 shows peaks at 1650 cm−1 and 1603 cm−1, and vitamin D3 shows a peak at 1650 cm−1 with shoulder peaks at 1603 cm−1 and 1631 cm−1 (Fig. 2C). In contrast, vitamin A and its derivatives, including retinoic acid, retinal, retinol (vitamin A), and retinyl ester (vitamin A ester), exhibit distinct peaks at 1573 cm−1, 1583 cm−1, 1595 cm−1, and 1595 cm−1, respectively (Fig. 2D). Comparison of these spectra with those from lipid-rich regions of the TB liver suggests that retinol and retinyl ester are likely the metabolites contributing to the 1595 cm−1 peak in TB liver.
A Chemical structures of cholesterol, ergosterol, and fat-soluble vitamins. B Normalized SRS spectra of cholesterol, vitamins D2, D3, and A in the C–H stretching region. C Normalized SRS spectra of cholesterol, vitamins D2, D3, and A in the fingerprint region. D Normalized SRS spectra of vitamin A and its derivatives in the fingerprint region. E Normalized SRS spectra of olein and vitamin A ester mixtures with varying molar ratios at the fingerprint region. F SRS images of MDA-MB-231 cells with or without 10 μM retinol treatment at 1595 cm−1 and its corresponding spectra at fingerprint region (G). H SRS images of liver tissues from TF and TB mice at 1595 cm−1. I Quantification of vitamin A levels in each LD within liver tissues from TF and TB mice, measured by the ratio of SRS intensity at 1595 cm−1 to 1455 cm−1. ****p < 0.0001. Error bars represent SD. Scale bar: 20 μm.
Approximately 80% of vitamin A in the body is stored in LDs of hepatic stellate cells as retinyl ester34. To verify that hSRS imaging can detect vitamin A in LD, we measured SRS spectra of triacylglycerol (TAG), vitamin A ester, and a TAG/vitamin A ester mixture (Supplementary Fig. 4). In the C–H stretching region, SRS spectra of TAG, vitamin A ester, and the mixture showed only subtle differences (Supplementary Fig. 4A). In contrast, in the fingerprint region, vitamin A ester exhibited a highly distinctive Raman peak at 1595 cm−1, with minimal overlap with the C = C peak of TAG at 1655 cm−1 (Supplementary Fig. 4B). The SRS spectrum of the TAG/vitamin A ester mixture was dominated by the 1595 cm−1 peak, with only a small shoulder at 1655 cm−1 (Supplementary Fig. 4C). This is mainly due to the significantly enhanced Raman cross-section of the conjugated polyene structure in vitamin A.
To further validate the extremely strong Raman signature of vitamin A ester, we prepared mixtures of TAG and vitamin A ester at different molar ratios and measured their SRS spectra in the fingerprint region (Fig. 2E). The 1595 cm−1 peak first became visible at a concentration of 0.4%, with peak intensity increasing as the concentration of retinyl ester increased. When the concentration of retinyl ester exceeded 60%, the 1655 cm−1 peak from TAG was nearly undetectable due to the overwhelming signal from retinyl ester. At 1.1% concentration of retinyl ester, the intensities of the 1595 cm−1 and 1655 cm−1 peaks were comparable, and the conjugated C = C bond signal from retinyl ester was about two orders of magnitude stronger than the acyl C = C bond in TAG, indicating that the 1595 cm⁻1 peak can serve as a sensitive and isolated marker for detecting vitamin A in biological samples. To validate these in vitro measurements in a cellular context, we supplemented HepG2 cells with retinol (the predominant vitamin A species for uptake) and performed SRS imaging at 1595 cm−1. The retinol-supplemented cells displayed increased signals corresponding to vitamin A ester (Fig. 2F, G), consistent with its esterification and storage in LDs. Notably, the C-H stretching spectra showed no significant increase in the 3015 cm⁻1 peak after retinol supplementation (Supplementary Fig. 5), as no resonance enhancement occurs in this region. Together, these results demonstrate that SRS imaging at 1595 cm−1 provides a highly sensitive approach for visualizing vitamin A species in biological systems.
To quantitatively assess the altered vitamin A metabolism in the TB liver, we selected 1595 cm−1 peak for SRS imaging and quantified the vitamin A levels in LDs (Fig. 2H). The SRS images showed a significant enrichment of vitamin A in TB liver compared to TF liver. Vitamin A levels in individual LDs were calculated by the ratio of SRS intensity at 1595 cm−1 to intensity at 1455 cm−1 (C–H bending mode), revealing significantly higher Vitamin A levels in LDs from TB liver compared to TF liver (Fig. 2I). Moreover, differential gene expression (DEG) analysis of genes associated with vitamin A metabolism (Supplementary Fig. 4) identified increased expression of SCARB1, which facilitates plasma lipid uptake35, supporting the accumulation of LDs and vitamin A storage. Meanwhile, elevated expression of RDH14, responsible for the conversion of retinal to retinol36, was found in TB liver, further supporting the accumulation of vitamin A (retinol and retinyl ester) in the tissue. The expression of FABP5, a key retinoic acid and fatty acid binding protein37, also showed a trend toward higher expression in TB liver, suggesting enhanced vitamin A-related metabolic activity. To assess the presence of liver metastasis at this stage, we performed Ki-67 immunostaining on the same liver tissues. Several small potential metastatic nodules were detected in TB livers (Supplementary Fig. 6). Together, the combination of fingerprint and high-wavenumber SRS imaging revealed the accumulation of vitamin A-rich, highly unsaturated LDs in the liver of TB mice, suggesting that tumors may regulate lipid metabolism, particularly vitamin A metabolism, in distant organs.
Suppressing tumor-derived EV secretion impairs the accumulation of vitamin A metabolites in the liver
Tumor-derived EVs play a crucial role in reprogramming the metabolism of distant organs prior to metastasis9,10. We hypothesized that breast cancer-derived EVs mediate lipid and vitamin A metabolic reprogramming in the liver. To test this, Rab27a, a key regulator of exocytosis38, was stably knocked down in MDA-MB-231 breast cancer cells using shRNA to inhibit EV secretion (MDA-MB-231/Rab27a knockdown, KD). These cells were then implanted into mice to assess their effect on liver metabolism (Fig. 3A). Rab27a protein levels were significantly reduced in MDA-MB-231 cells expressing Rab27a shRNA, with MDA-MB-231/Rab27a KD #2 showing the greatest knockdown efficiency (Fig. 3B). Suppression of EV secretion was further confirmed by measuring the particle size and quantity, demonstrating a significant reduction in EV particle number from MDA-MB-231/Rab27a KD #2 cells (Fig. 3C, Supplementary Fig. 7). Therefore, MDA-MB-231/Rab27a KD #2 cells were selected for implantation into mice to investigate the role of cancer-derived EVs in modulating hepatic lipid and vitamin A metabolism.
A Schematic of the animal model. B Western blot analysis of Rab27a expression in MDA-MB-231 and MDA-MB-231/shRab27a #1 and #2 cells. C Nano-flow cytometer analysis of EV number from MDA-MB-231 and MDA-MB-231/shRab27a #2 cells. D SRS images of liver tissues from TB and Rab27a KD TB mice in the C–H stretching region. E Quantification of LD size in lipid-rich regions. F Quantification of lipid intensity at 2855 cm−1 for each LD. G Intensity ratio of 3015 cm−1 and 2855 cm−1 peaks, representing relative unsaturated lipid content in each LD. H Mean SRS spectra of lipid-rich and protein-rich regions in the fingerprint region. I SRS images of liver tissues from TB and Rab27a KD TB mice at 1595 cm−1. J Quantification of vitamin A levels in each LD within liver tissues from TB and Rab27a KD TB mice, measured by the ratio of SRS intensity at 1595 cm−1 to 1455 cm−1. Error bars represent SD. ****p < 0.0001. Scale bars: 20 μm. Part of the figure is created in BioRender. Lee, H. (2025) https://BioRender.com/jozm8oh.
High-wavenumber SRS images showed a significant reduction in the number of LDs in the liver of Rab27a KD TB mice compared to wild-type TB mice (Fig. 3D, Supplementary Movie 5 and 6). To quantify LDs, phasor analysis was performed to segment lipid-rich and protein-rich regions based on their C–H stretching spectral profiles (Supplementary Fig. 8A). While the total number of LDs was significantly reduced in the Rab27a KD TB liver (Fig. 3E), the lipid contents of each LD remained similar (Fig. 3F). Interestingly, the composition of the LDs differed significantly between Rab27a KD and wild-type TB livers. Specifically, the peak intensity at 3015 cm−1 was reduced in Rab27a KD TB liver compared to the wild-type (Supplementary Fig. 8B), suggesting a decrease in lipid unsaturation. Quantification of the ratio of SRS intensities at 3015 cm−1 and 2855 cm−1 further confirmed the reduced unsaturated lipid content in Rab27a KD liver (Fig. 3G).
In the fingerprint region, SRS spectra exhibited the absence of the 1595 cm−1 peak associated with vitamin A metabolites, in both lipid-rich and protein-rich regions of the Rab27a KD TB liver (Fig. 3H, Supplementary Movie 7 and 8). SRS imaging at 1595 cm−1 further showed a marked depletion of vitamin A metabolites in the Rab27a KD TB liver compared to the wild-type TB liver (Fig. 3I, J). Additionally, DEG analysis of bulk RNA-seq data indicated significant downregulation of RDH9, RDH11, and FABP5 in the Rab27a KD TB liver, suggesting suppressed retinal-to-retinol conversion and retinol-related metabolic activity after Rab27a KD in the tumor (Supplementary Fig. 6). Together, these results indicate that impairing EV secretion via Rab27a KD specifically suppresses the accumulation of vitamin A metabolites in the liver, providing evidence for a potential regulatory mechanism by which tumors induce metabolic alterations through EV-mediated communication.
TNBC cells promote vitamin A accumulation in hepatocytes
To further investigate the mechanism by which tumors alter lipid and vitamin A metabolism in liver, we collected conditioned medium (CM) from MDA-MB-231, MDA-MB-231/Rab27a KD, and non-cancerous MCF10A cells, and cultured HepG2 cells with each CM (Fig. 4A). HepG2 cells served as a hepatic model, while MCF10A CM as a non-cancerous control to ensure that observed metabolic changes were tumor-specific. SRS images at 2850 cm−1 showed that only HepG2 cells treated with MDA-MB-231 CM exhibited increased lipid accumulation relative to untreated controls, whereas cells exposed to MDA-MB-231/Rab27a KD CM or MCF10A CM showed no increase, with lipid levels comparable to untreated HepG2 cells (Fig. 4B). SRS spectra of LDs in the C-H stretching region revealed no significant spectral differences among groups (Fig. 4C), and quantitative analysis confirmed significantly higher lipid levels in HepG2 cells treated with MDA-MB-231 CM compared to the MCF10A CM (Fig. 4D).
A Schematic of the cell model. B SRS images at 2850 cm−1 of HepG2 cells treated with 10 μM retinol and different CM. C SRS spectra at C–H stretching region in LDs of HepG2 cells treated with 10 μM retinol and different CM. D Quantification of total lipid level in HepG2 cells treated with 10 μM retinol and different CM. and 1595 cm−1, calculated as the intensity of LD area at 2850 cm−1 divided by the area of the entire cells at 2930 cm−1. E SRS images at 1595 cm−1 of HepG2 cells treated with 10 μM retinol and different CM. F SRS spectra at fingerprint region in LDs of HepG2 cells treated with 10 μM retinol and different CM. G Quantification of vitamin A level in HepG2 cells treated with 10 μM retinol and different CM, calculated as the ratio of SRS intensities at 1595 cm−1 to 2850 cm−1 within LD area. *p < 0.05; **p < 0.01. Error bars represent SD. Scale bar: 30 μm. Part of the figure is created in BioRender. Lee, H. (2025) https://BioRender.com/jozm8oh.
In the fingerprint region, SRS images at 1595 cm−1 revealed elevated vitamin A content in LDs of HepG2 cells treated with MDA-MB-231 CM, whereas cells exposed to MDA-MB-231/Rab27a KD CM or MCF10A CM showed vitamin A levels similar to untreated controls (Fig. 4E). SRS spectra likewise demonstrated the highest 1595 cm−1 peak intensity in the MDA-MB-231 CM group (Fig. 4F). Furthermore, quantitative analysis confirmed a significant increase in vitamin A content by MDA-MB-231 CM compared with all other groups (Fig. 4G). Together, these results indicate that tumor-secreted factors from TNBC cells promote vitamin A accumulation in hepatocytes, further supporting a cancer-specific effect.
FABP5, a key cargo in TNBC-derived EVs, induces vitamin A accumulation in hepatocytes
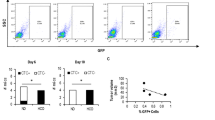
To identify potential cargoes in tumor-derived EVs, we analyzed EV protein profiles from TNBC patient serum and healthy controls using ProteomeXchange Datasets39. We focused on proteins related to lipid and vitamin A metabolism. Among the enriched cargoes in TNBC EVs, FABP5, APOF, and RBP4 were significantly upregulated (Fig. 5A, Supplementary Data 2). Using GEPIA2 and TCGA data, we examined the expression of these genes across breast cancer subtypes. FABP5 expression was notably high in basal-like (TNBC) breast cancer (Fig. 5B), whereas RBP4 expression was comparable across subtypes (Supplementary Fig. 9), and APOF expression was low or absent in TNBC (Supplementary Fig. 10), suggesting that APOF in serum EVs may not originate from TNBC cells. Importantly, high FABP5 expression correlated with poor overall survival in basal-like breast cancer patients (Fig. 5D), but showed no negative association in HER2+ non-luminal, luminal A, and luminal B subtypes (Supplementary Fig. 11). Based on these observations, we hypothesized that FABP5 may be a key cargo in tumor-derived EVs driving vitamin A metabolic alterations in the liver.
A Heatmap of different protein levels in serum EVs between healthy individuals and TNBC patients from database. B Transcript profiling of FABP5 in different breast cancer subtypes. C The association of FABP5 expression level with TNBC patient overall survival rate. D FABP5 protein expression in plasma EVs from healthy individuals and TNBC patients. E FABP5 protein levels in EVs from MCF10A and MDA-MB-231 cells. F FABP5 mRNA expression in MDA-MB-231 cells after FABP5 knockdown. G FABP5 protein expression in cells and EVs after FABP5 knockdown. H SRS images at 2850 cm−1 and 1595 cm−1 of HepG2 cells treated with 10 μM retinol and different CM, and (I) its corresponding SRS spectra in LD area. J Quantification of total lipid and vitamin A levels in HepG2 cells treated with 10 μM retinol and different CM, calculated as the intensity of LD area at 2850 cm−1 divided by the area of the entire cells at 2930 cm−1 and the ratio of SRS intensities at 1595 cm−1 to 2850 cm−1 within LD area, respectively. *p < 0.05; **p < 0.01. Error bars represent SD. Scale bar: 30 μm.
We further validated higher FABP5 levels in TNBC EVs using a small independent serum cohort, which showed significantly higher FABP5 in TNBC EVs compared to healthy controls (Fig. 5B). Similarly, EVs from MDA-MB-231 cells contained higher FABP5 protein levels than EVs from MCF10A cells (Fig. 5E). To examine whether FABP5 contributes to tumor-induced liver metabolic alterations across cancer types, we analyzed livers from KPC pancreatic cancer mouse models. TB liver tissues from KPC-bearing mice showed significantly increased lipid and vitamin A content (Supplementary Figs. 12, 13). Importantly, all cancer cells associated with increased hepatic vitamin A consistently displayed higher FABP5 levels in both cells and EVs compared to non-cancerous MCF10A cells (Supplementary Fig. 14), supporting a general role for FABP5 in mediating these metabolic alterations. Together, these results identify FABP5 as a critical cargo in tumor-derived EVs that drives liver metabolic alterations.
To directly test whether FABP5 is required for tumor-derived EV-induced lipid and vitamin A accumulation in hepatocytes, we generated stable FABP5 knockdown in MDA-MB-231 cells (Fig. 5F), which significantly reduced FABP5 protein levels in both cells and EVs (Fig. 5G). HepG2 cells treated with MDA-MB-231/FABP5 KD CM showed markedly reduced LD content and vitamin A level compared with MDA-MB-231 CM (Fig. 5H). SRS spectra of LDs from HepG2 cells treated with MDA-MB-231/FABP5 KD CM showed a significant reduction in the 1595 cm−1 peak intensity (Fig. 5I). Quantitative analysis confirmed that both lipid and vitamin A levels in the LDs were significantly lower than in cells treated with MDA-MB-231 CM, returning to levels comparable to untreated HepG2 cells (Fig. 5J). Together, these results indicate that FABP5 is a key molecule in tumor-derived EVs responsible for inducing lipid and vitamin A metabolic alterations in hepatocytes.
Discussion
In summary, this study reveals a novel mechanism by which tumors induce aberrant accumulation of lipids and vitamin A metabolites in the liver by in situ measurement of LDs in liver tissues using hSRS imaging in the fingerprint region. We also identified tumor-derived EVs as potential mediators of this metabolic alteration, with FABP5 serving as a key cargo protein involved in the process. Elevated FABP5 levels were strongly correlated with poor prognosis, specifically in TNBC, but not in other breast cancer subtypes, highlighting its critical role in TNBC progression. These findings suggest that FABP5, delivered via tumor-derived EVs, drives lipid and vitamin A metabolic alterations in the liver. These changes represent a form of metabolic remodeling in the liver niche, which is distinct from metabolic reprogramming centered on energy metabolism (Fig. 6).
FABP5, a key cargo in tumor-derived EVs, induces lipid and vitamin A (retinol) uptake and accumulation in the liver. Created in BioRender. Lee, H. (2025) https://BioRender.com/v2pcn4t.
The functions of vitamin A and lipids in cancer and its systemic environment are complex: while lipids have been widely recognized for their structural and energetic roles, they also act as signaling molecules that influence immune response and tumor progression40,41,42. Neutral lipid accumulation in pre-metastatic niches supports tumor growth43 and has been shown to impair immune responses, particularly by bioenergetically “poisoning” natural killer cells and reducing their cytotoxic function44. Tumor-derived EVs may drive lipid accumulation by disrupting normal fatty acid oxidation and oxidative phosphorylation, which leads to reduced fatty acid consumption and increased lipid storage6,43. Together with these observations, our findings support a model in which tumor-derived EVs contribute to the accumulation of unsaturated lipids in the liver, thereby promoting immune suppression and creating a nutrition-rich environment conducive to metastasis.
Meanwhile, vitamin A and its derivatives play essential roles in cellular functions45,46 and have demonstrated anticancer properties across various cancers47. For instance, vitamin A deficiency increases the incidence of spontaneous and chemically induced tumors in animal models48, suggesting that adequate vitamin A levels are protective. However, recent studies indicate that hepatic vitamin A storage may favor hepatic carcinogenesis, as seen in models where knockout of LRAT, a protein essential for vitamin A storage, in mice reduced liver cancer rates49. Additionally, retinoic acid, the active form of vitamin A, has been implicated in fostering an immunosuppressive microenvironment by promoting the differentiation of monocytes into tumor-associated macrophages rather than immunostimulatory dendritic cells50. This immunosuppression is further exacerbated by retinoic acid-induced upregulation of CD38, which promotes resistance to PD-1/PD-L1 immunotherapy51. In our study, the tumor-induced accumulation of vitamin A-rich LDs in the liver may facilitate immune evasion and possibly facilitate metastatic cancer progression, suggesting a novel regulatory role for vitamin A metabolism in the formation of metastatic niches in the liver.
Traditionally, the study of cancer-induced metabolic reprogramming has relied on omics approaches, such as genomics, transcriptomics, proteomics, and metabolomics52,53. While metabolomics based on mass spectrometry provides comprehensive chemical information, it often lacks the subcellular spatial resolution needed to capture the metabolic and cellular heterogeneity inherent to tissues such as the liver. High spatial resolution is therefore valuable to map metabolic changes within complex tissues. In this study, we utilize hSRS imaging to visualize the spatial distribution of lipids and vitamin A in liver tissues with submicron resolution. Our results demonstrate that hSRS imaging enables in situ measurement of submicron-sized organelles, allowing us to identify vitamin A accumulation in LDs as a tumor-induced metabolic signature.
Multiphoton imaging has been widely used for in vivo studies of lipid and vitamin A deposition in the liver, combining various contrast mechanisms such as second harmonic generation, third harmonic generation, and multiphoton-excited fluorescence (two-photon or three-photon excitation fluorescence)54,55. SHG highlights ordered structures such as fibrillar collagen; THG is sensitive to refractive-index interfaces (for example, at lipid–aqueous boundaries); and two-/three-photon excited fluorescence detects endogenous fluorophores, including NAD(P)H, flavins, and retinoids56. In contrast, SRS is also a nonlinear, multiphoton process but probes molecular vibrations directly, providing chemical bond-selective contrast without labeling. For histopathological analysis, SRS enables quantitative mapping of molecular content and composition in frozen sections, allowing detection of early molecular changes that are not apparent from structural alterations alone. Thus, while complementary to other multiphoton modalities, SRS adds a distinct molecular dimension that enhances histopathological evaluation and demonstrates its technical uniqueness for studying liver metabolic alterations.
While SRS provides unique advantages in chemical specificity and spatial resolution, certain technical limitations should be recognized. Spontaneous Raman spectroscopy offers more complete spectral information but at slower speeds; imaging mass spectrometry provides comprehensive molecular identification but requires sample destruction and limited spatial resolution57; and single-cell RNA sequencing offers gene-level regulatory insights but not spatial molecular compositional context58. SRS enables rapid, label-free mapping of specific chemical bonds within intact tissue structures. However, its spectral window is often limited, making it less suitable for comprehensive molecular identification; the imaging depth is constrained for in vivo applications; and the instrumentation remains technically demanding. Future integration of SRS with complementary molecular and genetic techniques will further enhance its potential for early diagnosis, disease subtyping, and development of targeted therapy.
In conclusion, this study demonstrates that tumor-derived EVs drive the accumulation of lipid- and vitamin A-rich LDs in the liver and demonstrates hSRS imaging as an enabling label-free approach to visualize these metabolic alterations in situ. These findings reveal a novel mechanism of tumor-induced metabolic alteration in distant organs and highlight hSRS as a promising platform for future studies of systemic metabolic regulation in cancer.
Methods
Chemicals. Chemicals in this study are shown in Supplementary Table 1
Cell lines and cell culture
HepG2, and MDA-MB-231 (HTB-26) cell line was purchased from American Type Culture Collection (ATCC). Both of HepG2 and MDA-MB-231 cells were cultured in DMEM (Gibco, C11995500BT) supplemented with 10% fetal bovine serum (FBS; Procell, 164210) and 1% penicillin/streptomycin. Cells were incubated in a humidified 37 °C incubator with 5% CO2. All cell lines were treated by mycoplasma removal agent (Beyotime Biotechnology, C0288S) and BeyoColor™ (Beyotime Biotechnology, C0305S) was used to detect mycoplasma to permit no mycoplasma contamination, regularly.
For in vitro vitamin A uptake, HepG2 cells were cultured in conditioned medium (supernatant collected from MDA-MB-231, MDA-MB-231/Rab27a KD, MCF10A, and MDA-MB-231/FABP5 KD) mixed with DMEM at a 1:1 ratio. Vitamin A (retinol, 10 μM) was then added to induce the absorption of vitamin A.
Immunofluorescence staining
The liver tissue was cryo-sectioned at 15 μm thickness and washed with PBS. The tissue sections were fixed with 4% PFA, followed by 0.05% Triton X-100 (Solarbio, #T8200). Then, 4% normal Donkey serum (Solarbio, #A410D051) was added onto the slides and incubated with anti-rabbit Ki-67 primary antibody (GeneTex, #GTX20833, 1:500) for overnight at 4 °C. After that, the slides were incubated with the secondary antibody (Alexa Fluor 546 donkey anti-rabbit, Invitrogen, #A10040) for 2 h at room temperature. DAPI was used for nuclear staining for 30 min at room temperature.
Stable knockdown cell lines
To generate a Rab27a knockdown MDA-MB-231 cell line, short hairpin RNA (shRNA) targeting Rab27a sequence at 5´-GCTGCCAATGGGACAAACATA-3´ and 5´-CCAGTGTACTTTACCAATATA-3´ was designed using BLOCK-iT RNAi Designer (Thermo Fisher Scientific), and cloned into pLKO.1 puro vector (Addgene, 8453). To generate a FABP5 knockdown MDA-MB-231 cell line, shRNA, 5´-CCGGCACCTGTACTCGGATCTAT CTCGAGATAGATCCGAGTACAGGTGTTTTTG-3´ and 5´-AATTCAAAAACACCTGTACT CGGATCTATCTCGAGATAGATCCGAGTACAGGTGCCGG-3´, was cloned into pLKO.1 puro vector to perform FABP5 knockdown. Lentiviruses were generated by transfecting 293 T cells with shRNA and the packaging plasmids psPAX2 (Addgene, 12260) and pMD2.G (Addgene, 12259) using Lipofectamine 2000 (Life Technologies). 48 h after transfection, Viral supernatants were collected and filtered through a 0.45 μm filter. MDA-MB-231 cells were infected by the lentivirus carrying this shRNA and selected using puromycin.
Orthotopic animal models
We have complied with all relevant ethical regulations for animal use. All animal procedures are approved by the institutional animal care and use committee (IACUC) at the College of Life Science, Wuhan University (WDSKY0202008) and the First Affiliated Hospital Zhejiang University School of Medicine (2025-034). There were no exclusions in the analysis.
Female NSG mice aged 6-8 weeks were obtained from Shanghai Model Organisms Center and used for implantation of human breast cancer cell lines (MDA-MB-231 and Rab27a-KD MDA-MB-231), and 5 mice per group (determined based on previous studies59). Mice were maintained under specific pathogen-free conditions and housed at a 12 h light/dark cycle at a relative humidity of 30-70% and room temperature of 22.2 ± 1.1 °C (four to five mice per cage) and were allowed free access to food and water. For breast cancer cell implantation, 2 × 105 of MDA-MB-231 or Rab27a-KD MDA-MD-231 cells suspended in 100 μL of PBS were subcutaneously inoculated into the left fourth pair of mammary glands in NSG mice. Mice were monitored at least once daily, and tumor growth was monitored twice a week. Mice were euthanized by cervical dislocation following deep anesthesia induced by isoflurane inhalation for liver tissue collection 50 days post-implantation. At 50 days post-implantation, primary tumors reached volumes of 700 - 1000 mm3 while body weights remained stable, consistent with previous reports59. As a control, mice were injected with the same volume of PBS following the same procedure.
The mouse KPC (KrasG12D; Trp53R172H; Pdx1-Cre) cell line was a kind gift from the laboratory of Professor Tingbo Liang60. KPC cells suspended in a 1:1 mixture of culture medium and Matrigel (1 × 108/mL). Then, six-week-old male mice (two C57BL/6 and two nude mice) were inoculated orthotopically with a volume of 25 μL of the cell suspension into the pancreas under general anesthesia to establish the pancreatic tumor model. As a control, mice were injected with the same volume of PBS following the same procedure (two C57BL/6 and two nude mice). Tissue collection procedures were performed as described for the breast cancer model.
Tissue preparation
Liver tissues were collected and snap-frozen in OCT (Tissue-Tek, 4583). The OCT-embedded tissues were sectioned at a 20-μm thickness using a cryotome (Leica, CM3050 S). For SRS imaging, tissue sections were washed in PBS for 5 min to remove OCT, then fixed in 10% neutral buffered formalin (Sigma-Aldrich, HT501128) for 15 min at room temperature. After washing with PBS 3 times, tissue sections were immersed in PBS, sandwiched with a coverslip, and sealed for SRS imaging.
hSRS imaging
Hyperspectral SRS imaging was performed on a home-built picosecond SRS microscope61, where the Raman shift was tuned by controlling the pump beam wavelength. An ultrafast laser system with dual output at 80 MHz (picoEmerald, Applied Physics & Electronics) provided both pump (tunable wavelength 700-990 nm) and Stokes beams (fixed wavelength 1031 nm). The Stokes beam was modulated at 20 MHz by an electro-optic modulator. A 60× water objective (1.2 numerical aperture, Olympus) was used to focus the light into the sample. Then, an oil condenser (1.4 numerical aperture, Olympus) was used to collect the forward-going pump and Stokes beams through the sample. A short-pass filter (1000-nm short-pass, Thorlabs) was used to block the Stokes beam and transmit only the pump beam onto a large area photodiode (10 × 10 mm) to detect the stimulated Raman loss signal. The powers of the pump and the Stokes beams at the specimen were maintained at 45 mW and 45 mW, respectively. The images were acquired with 10 μs pixel dwell time. For hSRS imaging of the C–H stretching vibration modes, the pump beam was tuned from 784.4 to 800.0 nm while the Stokes beam was fixed at 1031 nm to obtain 50-image stacks from 3050 to 2800 cm−1. For hSRS imaging of the fingerprint region, the pump beam was tuned between 877.3 to 901.0 nm while the Stokes beam was fixed at 1031 nm to obtain 50 image stacks from 1700 to 1400 cm−1.
Image and spectral analysis
hSRS images were processed and analyzed using ImageJ (v1.53e). The Phasor Analysis plug-in (https://imagejdocu.list.lu/plugin/segmentation/spectral_phasor/)28 was applied to segment lipid-rich and protein-rich regions within tissue images, based on their SRS spectral profiles in the high-wavenumber C-H stretching window62. Segmentation results were used to generate masks for lipid-rich and protein-rich areas, and used for extraction of their respective SRS spectra in the fingerprint window.
Western blotting
The cells were lysed using NP40/RIPA buffer supplemented with protease and phosphatase inhibitor cocktail (Sigma, 11836153001). The extracted proteins were separated by 10% SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore, SLHVR33RB). Then, membranes were blocked with 5% milk for 60 min, followed by incubation with primary antibodies (Santa Cruz, sc-74586, 1:1000) overnight at 4 °C. After washing 3 × 10 min with TBST, the membranes were incubated with HRP-conjugated secondary antibodies (Thermo Fisher Scientific, 31430 and 31460) for 1 h at room temperature. Following the same washing procedures, bands were visualized in ECL substrate (Thermo Fisher Scientific, 34578) and photographed by a chemiluminescence imaging analyzer (Tanon 5200). Primary antibodies list as follows: Rab27a (E-8) mouse monoclonal antibodies (Santa Cruz, sc-74586, 1:1000), FABP5 Rabbit pAb (ABclonal, A0947, 1:10000), Beta Actin Monoclonal antibody (Proteintech, 66009-1-Ig, 1:20000), and Alix Polyclonal antibody (Proteintech, 12422-1-AP, 1:5000).
qPCR assay
BeyoFast™ SYBR Green One-Step qRT-PCR Kit (Beyotime Biotechnology, D7268) was adopted to perform reverse transcription and qPCR to detect target gene expression. The qPCR primers were listed as follows:
ACTB-H forward: TGGCACCACACCTTCTACAA;
ACTB-H reverse: CCAGAGGCGTACAGGGATAG;
FABP5-H forward: TGAAGGAGCTAGGAGTGGGAA;
FABP5-H reverse: TGCACCATCTGTAAAGTTGCAG;
Actb-M forward: GGCTGTATTCCCCTCCATCG;
Actb-M reverse: CCAGTTGGTAACAATGCCATGT;
Fabp5-M forward: TGAAAGAGCTAGGAGTAGGACTG;
Fabp5-M reverse: CTCTCGGTTTTGACCGTGATG.
EV isolation
This study follows ISEV2018 guideline. To harvest EVs from cells, the conditioned medium was collected from cells grown in a medium containing EV-depleted serum for 48 h. Then, EVs were isolated from conditioned medium via ultracentrifugation. Briefly, conditioned medium was collected from cells cultured in medium supplemented with EV-depleted serum for 48 h. The medium was sequentially centrifuged at 500 × g for 15 min to remove floating cells, followed by centrifugation at 12,500 × g for 20 min to eliminate cellular debris. The resulting supernatant was then subjected to ultracentrifugation at 110,000 × g for 70 min to pellet EVs. The EV pellet was washed in PBS under the same ultracentrifugation conditions and finally resuspended in PBS for subsequent experimental applications and characterizations using Flow NanoAnalyzer U30 (NanoFCM, Xiamen, China) following the manufacturer’s instructions.
RNA sequencing
The RNA sequencing data is analyzed from the previous RNA sequencing dataset we constructed59. Specifically, RNA sequencing was performed by Majorbio (Shanghai, China) and the transcriptome library was prepared following the TruSeq RNA sample preparation Kit from Illumina (San Diego, CA) using 1 μg of total RNA. The detailed steps have been described in previous work63. Libraries were size-selected for cDNA target fragments of 300 bp through 2% Low Range Ultra Agarose followed by PCR amplified using Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific) for 15 PCR cycles. After quantified by TBS380, the paired-end RNA library was sequenced with the Illumina HiSeqxten/NovaSeq 6000 sequencer (2 × 150 bp read length). The raw paired-end reads were trimmed and quality controlled by SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) with default parameters. Then clean reads were separately aligned to reference genome with orientation mode using HISAT2 software. The mapped reads of each sample were assembled by StringTie (https://ccb.jhu.edu/software/stringtie/index.shtml) in a reference-based approach. The expression level of each transcript was calculated according to the transcripts per million reads (TPM) method. RSEM was used to quantify gene abundance. Eventually, differential expression analysis was performed using DESeq2. The normalized gene expression values were utilized for gene set enrichment analysis performed using the standalone GSEA software (v4.2.2).
Database analysis
Proteomic data of serum EVs were obtained from the ProteomeXchange database. Differential expression analysis was performed using the limma package in R (version 4.3.2). Survival analysis and FABP5 expression analysis across different breast cancer subtypes were conducted using The Gene Expression Profiling Interactive Analysis (GEPIA2) platform (http://gepia.cancer-pku.cn/), with data obtained from The Cancer Genome Atlas (TCGA).
Patients and specimen collection
All ethical regulations relevant to human research participants were followed. All human specimens were obtained from Renmin Hospital of Wuhan University (Wuhan, China) and approved by the Clinical Research Ethical Committee of Renmin Hospital of Wuhan University in accordance with the principle of the Helsinki Declaration. Informed consent was obtained from the patients. Peripheral blood samples were collected from three healthy individuals and three patients with triple-negative breast cancer using 6 mL K2EDTA tubes. The freshly drawn blood was centrifuged at 1550 × g for 30 min to obtain cell-free plasma. The supernatant was aliquoted and stored at –80 °C until EV isolation.
EV protein cargo analysis by mass spectrometry
Total protein was extracted from the EVs using urea lysis buffer (6 M urea, 100 mM NH₄HCO₃, 0.2% SDS) supplemented with protease inhibitor. Protein concentration was determined with a BCA Protein Assay Kit (Thermo Fisher Scientific, #23225). The EV protein cargo was analyzed by mass spectrometry on a Q Exactive HF mass spectrometer (Thermo Scientific) coupled to an EASY-nLC 1000 nanoflow LC system (Thermo Scientific). Detailed analytical procedures were described in a previous study64.
Statistics and reproducibility
SRS spectra were plotted by Origin 2019b software. Statistical significance was determined by two-tailed unpaired Student’s t tests using GraphPad Prism 7.0 software. P-value < 0.05 was considered statistically significant. Data are presented as mean ± SD unless otherwise noted. Details on the number of biological replicates and the sample sizes are provided in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data required to evaluate the conclusions of this study are included within the paper. The RNA-seq data generated in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) with accession code GSE222707 (liver tissue from mice xenografted with MDA-MB-231 cells and the tumor-free control group). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org) with the dataset identifier PXD069130 (proteome of 231 EV, https://www.iprox.cn/page/PSV023.html;?url=1733464844096CNk4, password: nGs3) and the dataset identifier PXD069253 (proteome of 10 A EV, https://www.iprox.cn/page/PSV023.html;?url=1733464899910oQzg, password: KUWQ). One of the paired measurements for MDA-MB-231 and MCF10A EV in Fig. 5E is supplemented in the Supplementary Data 1 file. The raw mass spectrometry-based proteomic data for Fig. 5A are from the dataset identifier PXD047470. Source data for graphs and charts are supplied as Supplementary Data 1 with this paper. Uncropped and unedited blot/gel images are included in the Supplementary Information file.
References
Swanton, C. et al. Embracing cancer complexity: Hallmarks of systemic disease. Cell 187, 1589–1616 (2024).
Suhail, Y. et al. Systems biology of cancer metastasis. Cell Syst. 9, 109–127 (2019).
Faubert, B., Solmonson, A. & DeBerardinis, R. J. Metabolic reprogramming and cancer progression. Science 368, eaaw5473 (2020).
Cao, M. et al. Cancer-cell-secreted extracellular vesicles suppress insulin secretion through miR-122 to impair systemic glucose homeostasis and contribute to tumour growth. Nat. Cell Biol. 24, 954–967 (2022).
Gong, Z. et al. Lipid-laden lung mesenchymal cells foster breast cancer metastasis via metabolic reprogramming of tumor cells and natural killer cells. Cell Metab. 34, 1960–1976.e1969 (2022).
Wang, G. et al. Tumour extracellular vesicles and particles induce liver metabolic dysfunction. Nature 618, 374–382 (2023).
Psaila, B., Kaplan, R. N., Port, E. R. & Lyden, D. Priming the ‘soil’ for breast cancer metastasis: the pre-metastatic niche. Breast Dis. 26, 65–74 (2006).
Lucotti, S., Kenific, C. M., Zhang, H. & Lyden, D. Extracellular vesicles and particles impact the systemic landscape of cancer. EMBO J. 41, e109288 (2022).
Morrissey, S. M. et al. Tumor-derived exosomes drive immunosuppressive macrophages in a pre-metastatic niche through glycolytic dominant metabolic reprogramming. Cell Metab. 33, 2040–2058.e2010 (2021).
Yang, E. et al. Exosome-mediated metabolic reprogramming: the emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal. Transduct. Target Ther. 5, 242 (2020).
Rabas, N., Ferreira, R. M. M., Di Blasio, S. & Malanchi, I. Cancer-induced systemic pre-conditioning of distant organs: building a niche for metastatic cells. Nat. Rev. Cancer. https://doi.org/10.1038/s41568-024-00752-0 (2024).
Wortzel, I., Dror, S., Kenific, C. M. & Lyden, D. Exosome-mediated metastasis: Communication from a distance. Dev. Cell 49, 347–360 (2019).
Liu, Y. & Cao, X. Characteristics and Significance of the Pre-metastatic Niche. Cancer Cell 30, 668–681 (2016).
Jiang, C., Jiang, Z., Sha, G., Wang, D. & Tang, D. Small extracellular vesicle-mediated metabolic reprogramming: from tumors to pre-metastatic niche formation. Cell Commun. Signal 21, 116 (2023).
Twelkmeyer, B., Tardif, N. & Rooyackers, O. Omics and cachexia. Curr. Opin. Clin. Nutr. 20, 181–185 (2017).
Cheng, J. X. & Xie, X. S. Vibrational spectroscopic imaging of living systems: An emerging platform for biology and medicine. Science 350, aaa8870 (2015).
Li, Y. et al. Review of stimulated raman scattering microscopy techniques and applications in the biosciences. Adv. Biol. (Weinh.) 5, e2000184 (2021).
Huang, X., Xue, Z., Zhang, D. & Lee, H. J. Pinpointing fat molecules: Advances in coherent Raman scattering microscopy for lipid metabolism. Anal. Chem. 96, 7945–7958 (2024).
Huang, J. & Ji, M. Stimulated Raman scattering microscopy on biological cellular machinery. J. Innovative Optical Health Sci. 16, 2230010 (2022).
Xu, J. et al. Unveiling Cancer Metabolism through Spontaneous and Coherent Raman Spectroscopy and Stable Isotope Probing. Cancers 13, 1718 (2021).
Fujita, K. Raman imaging as a window into cellular complexity: a future perspective. Nat. Methods 22, 890–892 (2025).
Min, W., Cheng, J.-X. & Ozeki, Y. Theory, innovations and applications of stimulated Raman scattering microscopy. Nat. Photonics 19, 803–816 (2025).
Hess, K. R. et al. Metastatic patterns in adenocarcinoma. Cancer 106, 1624–1633 (2006).
Costa-Silva, B. et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 17, 816–826 (2015).
Xia, L. et al. The cancer metabolic reprogramming and immune response. Mol. Cancer 20, 28 (2021).
Gao, Y. et al. Metastasis organotropism: redefining the congenial soil. Dev. Cell 49, 375–391 (2019).
Yang, K. et al. The role of lipid metabolic reprogramming in tumor microenvironment. Theranostics 13, 1774–1808 (2023).
Fereidouni, F., Bader,. A. N. & Gerritsen, H. C. Spectral phasor analysis allows rapid and reliable unmixing of fluorescence microscopy spectral images. Opt. Express 20, 12729–12741 (2012).
Welte, M. A. & Gould, A. P. Lipid droplet functions beyond energy storage. Biochim. Biophys. Acta-Mol. Cell Biol Lipids 1862, 1260–1272 (2017).
Schulz, H., Baranska, M. & Baranski, R. Potential of NIR-FT-Raman spectroscopy in natural carotenoid analysis. Biopolymers 77, 212–221 (2005).
Surmacki, J. M. & Abramczyk, H. Confocal Raman imaging reveals the impact of retinoids on human breast cancer via monitoring the redox status of cytochrome c. Sci. Rep.-UK 13, 15049 (2023).
Zhang, M. et al. Fingerprint stimulated Raman scattering imaging unveils Ergosteryl ester as a metabolic signature of azole-resistant Candida albicans. Anal. Chem. 95, 9901–9913 (2023).
Trefts, E., Gannon, M. & Wasserman, D. H. The liver. Curr. BIOL 27, R1147–r1151 (2017).
Senoo, H., Kojima, N. & Sato, M. Vitamin A-storing cells (stellate cells). Vitam. Horm. 75, 131–159 (2007).
Lamon-Fava, S., Diffenderfer, M. R. & Marcovina, S. M. Lipoprotein(a) metabolism. Curr. Opin. Lipidol. 25, 189–193 (2014).
Kumar, S., Sandell, L. L., Trainor, P. A., Koentgen, F. & Duester, G. Alcohol and aldehyde dehydrogenases: retinoid metabolic effects in mouse knockout models. Biochim. Biophys. Acta 1821, 198–205 (2012).
Napoli, J. L. Cellular retinoid binding-proteins, CRBP, CRABP, FABP5: Effects on retinoid metabolism, function and related diseases. Pharmacol. Therapeut. 173, 19–33 (2017).
Ostrowski, M. et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19–30 (2010).
Hyon, J. Y. et al. Extracellular vesicle proteome analysis improves diagnosis of recurrence in triple-negative breast cancer. J. Extracell. Vesicles 14, e70089 (2025).
Zechner, R. et al. FAT SIGNALS–lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 15, 279–291 (2012).
Lim, S. A., Su, W., Chapman, N. M. & Chi, H. Lipid metabolism in T cell signaling and function. Nat. Chem. Biol. 18, 470–481 (2022).
Wang, N. et al. Blockade of Arf1-mediated lipid metabolism in cancers promotes tumor infiltration of cytotoxic T cells via the LPE-PPARγ-NF-κB-CCL5 pathway. Life Metabolism 2, load036 (2023).
Li, P. et al. Lung mesenchymal cells elicit lipid storage in neutrophils that fuel breast cancer lung metastasis. Nat. Immunol. 21, 1444–1455 (2020).
Kobayashi, T. et al. Increased lipid metabolism impairs NK cell function and mediates adaptation to the lymphoma environment. Blood 136, 3004–3017 (2020).
O’Connor, C., Varshosaz, P. & Moise, A. R. Mechanisms of feedback regulation of vitamin A metabolism. Nutrients 14, 1312 (2022).
Yuan, Y. & Chen, L. Transporters in vitamin uptake and cellular metabolism: impacts on health and disease. Life Metabolism 4, loaf008(2025).
Vitamin A and cancer Lancet (Lond., Engl.) 1, 575–576 (1980).
Niles, R. M. Vitamin A and cancer. Nutr. (Burbank, Los Angeles Cty., Calif.) 16, 573–576 (2000).
Kluwe, J. et al. Absence of hepatic stellate cell retinoid lipid droplets does not enhance hepatic fibrosis but decreases hepatic carcinogenesis. Gut 60, 1260–1268 (2011).
Devalaraja, S. et al. Tumor-derived retinoic acid regulates intratumoral monocyte differentiation to promote immune suppression. Cell 180, 1098–1114.e1016 (2020).
Chen, L. et al. CD38-mediated immunosuppression as a mechanism of tumor cell escape from PD-1/PD-L1 blockade. Cancer Discov. 8, 1156–1175 (2018).
Peinado, H. et al. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer 17, 302–317 (2017).
Xi, Y. et al. Exosome-mediated metabolic reprogramming: Implications in esophageal carcinoma progression and tumor microenvironment remodeling. Cytokine Growth Factor Rev. 73, 78–92 (2023).
Xu, C., Nedergaard, M., Fowell, D. J., Friedl, P. & Ji, N. Multiphoton fluorescence microscopy for in vivo imaging. Cell 187, 4458–4487 (2024).
Wang, H. et al. Real-time histology in liver disease using multiphoton microscopy with fluorescence lifetime imaging. Biomed. Opt. Express 6, 780–792 (2015).
Débarre, D. et al. Imaging lipid bodies in cells and tissues using third-harmonic generation microscopy. Nat. Methods 3, 47–53 (2006).
Körber, A., Anthony, I. G. M. & Heeren, R. M. A. Mass spectrometry imaging. Anal. Chem. 97, 15517–15549 (2025).
Kolodziejczyk, A. A., Kim, J. K., Svensson, V., Marioni, J. C. & Teichmann, S. A. The technology and biology of single-cell RNA sequencing. Mol. Cell 58, 610–620 (2015).
Li, M. X. et al. Tumor-derived miR-9-5p-loaded EVs regulate cholesterol homeostasis to promote breast cancer liver metastasis in mice. Nat. Commun. 15, 10539 (2024).
Sun, K. et al. Elevated protein lactylation promotes immunosuppressive microenvironment and therapeutic resistance in pancreatic ductal adenocarcinoma. J. Clin. Investig. 135, e187024 (2025).
Huang, J. et al. Lipid metabolic heterogeneity during early embryogenesis revealed by hyper-3d stimulated raman imaging. Chem. Biomed. Imaging 3, 15–24 (2025).
Fu, D. & Xie, X. S. Reliable cell segmentation based on spectral phasor analysis of hyperspectral stimulated Raman scattering imaging data. Anal. Chem. 86, 4115–4119 (2014).
Hu, Y. et al. Cancer-cell-secreted miR-204-5p induces leptin signalling pathway in white adipose tissue to promote cancer-associated cachexia. Nat. Commun. 14, 5179 (2023).
Zhang, X. et al. Arachidonic acid triggers spermidine synthase secretion from primary tumor to induce skeletal muscle weakness upon irradiation. Cell Metab. 37, 1766–1782.e1712 (2025).
Acknowledgements
The authors thank Dr. Yongqing Zhang for his expert assistance with the SRS imaging system, and Drs. Young-Ho Chung, Hyo-Il Jung, and Seung Il Kim for advising on their previous database. This work was supported by the National Natural Science Foundation of China (82372011), Zhejiang Provincial Natural Science Foundation of China (LZ25H180001), Noncommunicable Chronic Diseases-National Science and Technology Major Project (2024ZD0525100/2024ZD0525103), and the Fundamental Research Funds for the Central Universities (226-2025-00034, 2025ZFJH01-01).
Author information
Authors and Affiliations
Contributions
X.H. and H.J.L. designed the study. X.H. performed SRS imaging and analysis. M.L., W.H., and W.Y. designed and performed the animal and RNA-seq experiments. W.H. performed Western blotting and EV characterizations. Y.Z. performed SRS spectral measurement of fatty acids. D.Z. developed and supervised SRS imaging experiments. W.Z. performed cell experiments. X.H. and W.Z. performed bioinformatics analysis. Q.Z. and Y.Z. performed pancreatic cancer animal experiments. X.H. and H.J.L. wrote the manuscript with inputs from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Yusuke Oshima and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Kaliya Georgieva. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huang, X., Zhou, W., Hou, W. et al. Tumor-secreted factors induce aberrant accumulation of vitamin A–enriched lipid droplets in the liver. Commun Biol 9, 125 (2026). https://doi.org/10.1038/s42003-025-09404-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-09404-x