Abstract
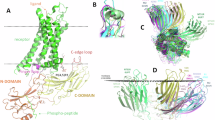
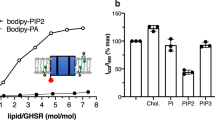
The class B1 G protein-coupled receptor (GPCR) subfamily is a class of receptors known for their regulatory roles in metabolism and neuronal activity and as important drug targets. Lipids play key functional roles in modulation of GPCR signalling, yet our understanding of the molecular level detail of specific lipid interactions with class B1 GPCRs remains limited. Here we present coarse-grained molecular dynamics (MD) simulations of the active and inactive states of 15 human class B1 family members and use aiida-gromacs to capture full provenance for the set-up of simulations in complex plasma membranes. Receptors exhibit state-dependent lipid interactions with the regulatory sterol cholesterol and phospholipid phosphatidylinositiol-3,4-bisphosphate (PIP2) at defined locations on the receptor surface. Global analysis of trends across the subfamily reveals conserved patterns of lipid interaction dynamics. The glycosphingolipid GM3 exerts a modulatory influence on the dynamics of class B1 extracellular domains in both simulations and in vitro time-resolved FRET assays.
Similar content being viewed by others
Data availability
All data are available in the main text or the supplementary materials. Additional data including raw data for plots presented has been deposited on Zenodo: https://doi.org/10.5281/zenodo.1435905691; https://doi.org/10.5281/zenodo.1472841492.
References
Cong, Z. et al. Structural perspective of class B1 GPCR signaling. Trends Pharmacol. Sci. 43, 321–334 (2022).
Karageorgos, V. et al. Current understanding of the structure and function of family B GPCRs to design novel drugs. Hormones 17, 45–59 (2018).
Cary, B. P. et al. New insights into the structure and function of class B1 GPCRs. Endocrine Rev. 44, 492–517 (2023).
Latorraca, N. R., Venkatakrishnan, A. J. & Dror, R. O. GPCR dynamics: Structures in motion. Chem. Rev.117, 139–155 (2017).
Hauser, A. S. et al. GPCR activation mechanisms across classes and macro/microscales. Nat. Struct. Mol. Biol. 28, 879–888 (2021).
Laganowsky, A. et al. Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510, 172–175 (2014).
Yeagle, P. L. Non-covalent binding of membrane lipids to membrane proteins. Biochimica et Biophysica Acta - Biomembranes 1838, 1548–59 (2014).
Byrne, E. F. X. et al. Structural basis of Smoothened regulation by its extracellular domains. Nature 535, 517-522 (2016).
Oates, J. & Watts, A. Uncovering the intimate relationship between lipids, cholesterol and GPCR activation. Curr. Opin. Struct. Biol. 21, 802–7 (2011).
Kiriakidi, S. et al. Effects of cholesterol on GPCR function: Insights from computational and experimental studies. Adv. Exp. Med. Biol. 1135, 89–103 (2019).
Janetzko, J. et al. Membrane phosphoinositides regulate GPCR-β-arrestin complex assembly and dynamics. Cell 185, 4560–4573 (2022).
Ansell, T. B., Song, W. & Sansom, M. S. P. The glycosphingolipid GM3 modulates conformational dynamics of the glucagon receptor. Biophys. J. 119, 300–313 (2020).
Hedger, G. & Yen, H.-Y. The influence of phosphoinositide lipids in the molecular biology of membrane proteins: recent insights from simulations. J. Mol. Biol. 437, 168937 (2025).
Pluhackova, K., Gahbauer, S., Kranz, F., Wassenaar, T. A. & Böckmann, R. A. Dynamic cholesterol-conditioned dimerization of the G protein coupled chemokine receptor type 4. PLoS Comput. Biol. 12, e1005169 (2016).
Guixà-González, R. et al. Membrane omega-3 fatty acids modulate the oligomerisation kinetics of adenosine A2A and dopamine D2 receptors. Sci. Rep. 6, 19839 (2016).
Dawaliby, R. et al. Allosteric regulation of G protein-coupled receptor activity by phospholipids. Nat. Chem. Biol. 12, 35–9 (2016).
Yen, H. Y. et al. PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature 559, 423–427 (2018).
Oqua, A. I. et al. Molecular mapping and functional validation of GLP-1R cholesterol binding sites in pancreatic beta cells. Elife 13, RP101011 (2024).
Buenaventura, T. et al. Agonist-induced membrane nanodomain clustering drives GLP-1 receptor responses in pancreatic beta cells. PLoS Biol. 17, e3000097 (2019).
Cheng, Y. Membrane protein structural biology in the era of single particle cryo-EM. Curr. Opin. Struct. Biol. 52, 58–63 (2018).
Zhang, M. et al. G protein-coupled receptors (GPCRs): Advances in structures, mechanisms and drug discovery. Signal Transduct. Target Ther. 9, 88 (2024).
Souza, P. C. T. et al. Martini 3: A general purpose force field for coarse-grained molecular dynamics. Nat. Methods 18, 382–388 (2021).
Han, X. & Gross, R. W. The foundations and development of lipidomics. J. Lipid Res. 63, 100164 (2022).
Páll, S. et al. Heterogeneous parallelization and acceleration of molecular dynamics simulations in GROMACS. J. Chem. Phys. 153, 134110 (2020).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Heo, L. & Feig, M. Multi-state modeling of G-protein coupled receptors at experimental accuracy. Proteins: Struct. Funct. Bioinf. 90, 1873–1885 (2022).
Discovery, C. et al. Chai-1: Decoding the molecular interactions of life. bioRxiv https://doi.org/10.1101/2024.10.10.615955 (2024).
Bereau, T., Walter, L. J. & Rudzinski, J. F. Martignac: Computational workflows for reproducible, traceable, and composable coarse-grained martini simulations. J. Chem. Inf. Model 64, 9413–9423 (2024).
Huber, S. P. et al. AiiDA 1.0, a scalable computational infrastructure for automated reproducible workflows and data provenance. Sci. Data 7, 300 (2020).
Song, W. et al. PyLipID: A python package for analysis of protein-lipid interactions from molecular dynamics simulations. J. Chem. Theory Comput. 18, 1188–1201 (2022).
Wang, X. et al. Molecular insights into differentiated ligand recognition of the human parathyroid hormone receptor 2. Proc. Natl. Acad. Sci. USA 118, e2101279118 (2021).
Duan, J. et al. Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy. Nat. Commun 11, 4121 (2020).
Cao, J. et al. A structural basis for amylin receptor phenotype. Science (1979) 375, eabm9609 (2022).
Zhou, F. et al. Structural basis for activation of the growth hormone-releasing hormone receptor. Nat Commun 11, 5205 (2020).
Zhao, F. et al. Structural insights into hormone recognition by the human glucosedependent insulinotropic polypeptide receptor. Elife 10, e68719 (2021).
Zhao, L. H. et al. Structure and dynamics of the active human parathyroid hormone receptor-1. Science (1979) 364, 148–153 (2019).
Ma, S. et al. Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors. Mol Cell 77, 669–680 (2020).
Wootten, D., Simms, J., Miller, L. J., Christopoulos, A. & Sexton, P. M. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc. Natl. Acad. Sci. USA 110, 5211–6 (2013).
Genheden, S., Essex, J. W. & Lee, A. G. G protein coupled receptor interactions with cholesterol deep in the membrane. Biochim. Biophys. Acta Biomembr. 1859, 268–281 (2017).
Guixà-González, R. et al. Membrane cholesterol access into a G-protein-coupled receptor. Nat. Commun 8, 14505 (2017).
Morra, G., Razavi, A. M., Menon, A. K. & Khelashvili, G. Cholesterol occupies the lipid translocation pathway to block phospholipid scrambling by a G protein-coupled receptor. Structure 30, 1208–1217 (2022).
Sejdiu, B. I. & Tieleman, D. P. Lipid-protein interactions are a unique property and defining feature of G protein-coupled receptors. Biophys. J. 118, 1887–1900 (2020).
Gater, D. L. et al. Two classes of cholesterol binding sites for the β2AR revealed by thermostability and NMR. Biophys. J. 107, 2305–12 (2014).
Aranda-García, D. et al. Large scale investigation of GPCR molecular dynamics data uncovers allosteric sites and lateral gateways. Nat. Commun 16, 2020 (2025).
Hedger, G. & Sansom, M. S. P. Lipid interaction sites on channels, transporters and receptors: Recent insights from molecular dynamics simulations. Biochim Biophys Acta Biomembr 1858, 2390–2400 (2016).
Hu, J. et al. Physiological temperature drives TRPM4 ligand recognition and gating. Nature 630, 509–515 (2024).
Vickery, O. N. & Stansfeld, P. J. CG2AT2: an Enhanced Fragment-Based Approach for Serial Multi-scale Molecular Dynamics Simulations. J. Chem. Theory Comput 17, 6472–6482 (2021).
Jo, S., Rui, H., Lim, J. B., Klauda, J. B. & Im, W. Cholesterol Flip-Flop: Insights from Free Energy Simulation Studies. J. Phys. Chem. B 114, 13342–13348 (2010).
Pedersen, K. B. et al. The Martini 3 lipidome: Expanded and refined parameters improve lipid phase behavior. ACS Cent. Sci. 11 1598–1610 (2025).
Cox, T. M. Eliglustat tartrate, an orally active glucocerebroside synthase inhibitor for the potential treatment of Gaucher disease and other lysosomal storage diseases. Curr. Opin. Investig. Drugs 11, 1169–81 (2010).
Chiu, P.-L., Orjuela, J. D., de Groot, B. L., Aponte Santamaría, C. & Walz, T. Structure and dynamics of cholesterol-mediated aquaporin-0 arrays and implications for lipid rafts. Elife 12, RP90851 (2024).
Chen, S. et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature 625, 92–100 (2024).
Damian, M. et al. Allosteric modulation of ghrelin receptor signaling by lipids. Nat. Commun 12, 3938 (2021).
Kjølbye, L. R. et al. Lipid Modulation of a Class B GPCR: Elucidating the Modulatory Role of PI(4,5)P2 Lipids. J. Chem. Inf. Model 62, 6788–6802 (2022).
Thakur, N. et al. Anionic phospholipids control mechanisms of GPCR-G protein recognition. Nat. Commun 14, 794 (2023).
Huang, W. et al. Structure of the neurotensin receptor 1 in complex with β-arrestin 1. Nature 579, 303–308 (2020).
Gomes, A. A. S. et al. Lipids modulate the dynamics of GPCR:β-arrestin interaction. Nature. Communications 16, 4982 (2025).
Zhang, H. et al. Structure of the full-length glucagon class B G-protein-coupled receptor. Nature 546, 259–264 (2017).
Jazayeri, A. et al. Extra-helical binding site of a glucagon receptor antagonist. Nature 533, 274–7 (2016).
Sun, B. et al. Structural determinants of dual incretin receptor agonism by tirzepatide. Proc. Natl. Acad. Sci. USA 119, e2116506119 (2022).
Tiemann, J. K. et al. MDverse, shedding light on the dark matter of molecular dynamics simulations. Elife 12, RP90061 (2024).
Cong, Z. et al. Molecular features of the ligand-free GLP-1R, GCGR and GIPR in complex with Gs proteins. Cell Discov. 10, 18 (2024).
Knapp, B., Ospina, L. & Deane, C. M. Avoiding false positive conclusions in molecular simulation: The importance of replicas. J. Chem. Theory Comput. 14, 6127–6138 (2018).
Sexton, R., Fazel, M., Schweiger, M., Pressé, S. & Beckstein, O. Bayesian nonparametric analysis of residence times for protein-lipid interactions in molecular dynamics simulations. J. Chem. Theory Comput. 21, 4203–4220 (2025).
Hay, D. L. & Pioszak, A. A. Receptor activity-modifying proteins (RAMPs): New insights and roles. Annu. Rev. Pharmacol. Toxicol. 56, 469–87 (2016).
Chetverikov, N., Janoušková-Randáková, A., Nelic, D. & Jakubík, J. Cholesterol differentially modulates the activity of opioid and muscarinic receptors via a common binding site. Biochem Pharm. 242, 117296 (2025).
Naglekar, A., Chattopadhyay, A. & Sengupta, D. Palmitoylation of the glucagon-like peptide-1 receptor modulates cholesterol interactions at the receptor–lipid microenvironment. J. Phys. Chem. B 127, 11000–11010 (2023).
Pándy-Szekeres, G. et al. GPCRdb in 2023: State-specific structure models using AlphaFold2 and new ligand resources. Nucleic Acids Res. 51, D395–D402 (2023).
Sali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Park, S. J., Kern, N., Brown, T., Lee, J. & Im, W. CHARMM-GUI PDB Manipulator: Various PDB structural modifications for biomolecular modeling and simulation. J. Mol. Biol. 435, 167995 (2023).
Berman, M.H. et al. The protein data bank. Nucleic Acids Res. 28, 235–242 (2000).
Kroon, P. C. et al. Martinize2 and Vermouth: Unified framework for topology generation. Elife 12, RP90627 (2023).
Periole, X., Cavalli, M., Marrink, S. J. & Ceruso, M. A. Combining an elastic network with a coarse-grained molecular force field: Structure, dynamics, and intermolecular recognition. J. Chem. Theory Comput. 5, 2531–43 (2009).
Lomize, A. L., Todd, S. C. & Pogozheva, I. D. Spatial arrangement of proteins in planar and curved membranes by PPM 3.0. Protein Sci. 31, 209–220 (2022).
Wassenaar, T. A., Ingólfsson, H. I., Böckmann, R. A., Tieleman, D. P. & Marrink, S. J. Computational lipidomics with insane: A versatile tool for generating custom membranes for molecular simulations. J. Chem. Theory Comput. 11, 2144–55 (2015).
Song, W., Yen, H. Y., Robinson, C. V. & Sansom, M. S. P. State-dependent lipid interactions with the A2a receptor revealed by MD simulations using in vivo-mimetic membranes. Structure 27, 392–403 (2019).
Borges-Araújo, L., Souza, P. C. T., Fernandes, F. & Melo, M. N. Improved parameterization of phosphatidylinositide lipid headgroups for the martini 3 coarse-grain force field. J. Chem. Theory Comput. 18, 357–373 (2022).
Borges-Araújo, L. et al. Martini 3 coarse-grained force field for cholesterol. J Chem Theory Comput 19, 7387–7404 (2023).
Qi, Y. et al. CHARMM-GUI Martini maker for coarse-grained simulations with the Martini force field. J. Chem. Theory Comput. 11, 4486–94 (2015).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007).
Parrinello, M. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl Phys. 52, 7182 (1981).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Humphrey, W., Dalke, A. & Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph 14, 27–28 (1996).
Schrödinger LLC. The PyMOL Molecular Graphics System, Version~1.8. (2015).
Cohen, J., Arkhipov, A., Braun, R. & Schulten, K. Imaging the migration pathways for O2, CO, NO, and Xe inside myoglobin. Biophys. J. 91, 1844–57 (2006).
Huang, J. & Mackerell, A. D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–45 (2013).
Nosé, S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 52, 255–268 (1984).
Hoover, W. G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A (Coll Park) 31, 1695–1697 (1985).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089 (1993).
Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: A linear constraint solver for molecular simulations. J. Comput Chem. 18, 1463–1472 (1997).
Kalayan, J., Chao, K., Rouse, S. & Gebbie-Rayet, J. Coarse-grained MD simulation provenance of membrane embedded GPCR using GROMACS and aiida-gromacs. https://doi.org/10.5281/ZENODO.14359056.
Chao, K., Kalayan, J., Gebbie-Rayet, J. & Rouse, S. Dataset supporting the publication of Human class B1 GPCR modulation by plasma membrane lipids. https://doi.org/10.5281/ZENODO.15619266.
Kim, S. et al. PubChem 2023 update. Nucleic Acids Res. 51, D1373–D1380 (2023).
Barneda, D., Cosulich, S., Stephens, L. & Hawkins, P. How is the acyl chain composition of phosphoinositides created and does it matter? Biochem. Soc. Trans. 47, 1291–1305 (2019).
Shayman, J. A., Hinkovska-Galcheva, V. & Shu, L. Inhibitors of glucosylceramide synthase. in Methods Mol. Biol. 2613, 271–288 (2023).
Klymchenko, A. S. Fluorescent probes for lipid membranes: From the cell surface to organelles. Acc Chem. Res. 56, 1–12 (2023).
Lucey, M. et al. Acylation of the incretin peptide exendin-4 directly impacts glucagon-like peptide-1 receptor signaling and traffickings. Mol. Pharmacol 100, 319–334 (2021).
Acknowledgements
We thank Joseph Barritt for helpful discussion. This work was supported by the following grants: UKRI Future Leaders Fellowship (MR/Y01975X/1): S.L.R.; MRC Project Grant (MR/X021467/1): A.T. (Y.M.); Wellcome Trust Discovery Award (301619/Z/23/Z): A.T. (A.I.O.), S.L.R. (G.H.)
Author information
Authors and Affiliations
Contributions
K.W.C., G.H., S.L.R., A.T., J.G.R., J.K.. Methodology: K.W.C., L.W., J.K., A.T.. Investigation: K.W.C., L.W., G.H., S.L.R., Y.M., A.I.O.. Visualization: K.W.C., L.W., G.H., S.L.R., A.T., A.I.O., Y.M.. Supervision: G.H., A.T., J.G.R., S.L.R.. Writing—original draft: K.W.C., G.H., J.K., J.G.R., A.T., S.L.R.. Writing—review & editing: L.W., A.I.O., J.K., J.G.R., G.H., A.T., S.L.R.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. EBM: Professor Michal Kolář. Primary Handling Editors: Dr. Nilanjan Banerjee and Dr. Laura Rodriguez Perez. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chao, K.W., Wong, L., Oqua, A.I. et al. Human class B1 GPCR modulation by plasma membrane lipids. Commun Biol (2026). https://doi.org/10.1038/s42003-025-09445-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-09445-2