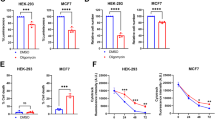
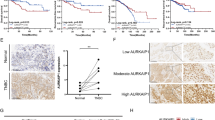
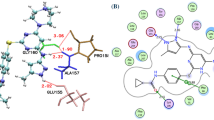
Abstract
Aurora kinase A/AURKA is a serine/threonine kinase frequently overexpressed in cancer. Recent discoveries pointed to subcellular pools of AURKA, including at mitochondria. There, AURKA induces organelle clearance by mitophagy together with the autophagy mediator LC3, and its receptor PHB2.
Here, we show that the natural product capsaicin modifies the AURKA/PHB2 interaction. We synthesize 16 capsaicin analogs, and Förster’s Resonance Energy Transfer/Fluorescence Lifetime Imaging Microscopy (FRET/FLIM) in breast cancer cells reveals that compounds 12 and 13 increase the AURKA/PHB2 interaction. Molecular docking shows that they bind to the inhibitory pocket of PHB2 and to the AURKA active site. We demonstrate that compound 13 specifically inhibits mitophagy while leaving AURKA activation unaltered at centrosomes. Our results demonstrate that compound 13 is a PHB ligand acting on the AURKA/PHB2 interaction. Thanks to its specificity, it may lead to the development of anticancer drugs targeting the mitochondrial functions of AURKA.
Similar content being viewed by others
Data availability
Source microscopy data are available on Zenodo52 (https://doi.org/10.5281/zenodo.17610406). All other data are available from the corresponding authors (L.D. and G.B.) upon request.
References
Wang, T., Ma, F. & Qian, H. Defueling the cancer: ATP synthase as an emerging target in cancer therapy. Mol. Ther. Oncol. 23, 82–95 (2021).
Pickrell, A. M. & Youle, R. J. The roles of PINK1, parkin and mitochondrial fidelity in Parkinson’s disease. Neuron 85, 257–273 (2015).
Wang, S. et al. The mitophagy pathway and its implications in human diseases. Signal Transduct. Target Ther. 8, 1–28 (2023).
Wei, Y., Chiang, W.-C., Sumpter, R., Mishra, P. & Levine, B. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell 168, 224–238.e10 (2017).
Wang, D. et al. Prohibitin ligands: a growing armamentarium to tackle cancers, osteoporosis, inflammatory, cardiac and neurological diseases. Cell. Mol. Life Sci. 77, 3525–3546 (2020).
Bertolin, G. et al. Mitochondrial Aurora kinase A induces mitophagy by interacting with MAP1LC3 and prohibitin 2. Life Sci. Alliance https://doi.org/10.26508/lsa.202000806 (2021).
Kashatus, D. F. et al. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat. Cell Biol. 13, 1108–1115 (2011).
Nikonova, A. S., Astsaturov, I., Serebriiskii, I. G., Dunbrack, R. L. & Golemis, E. A. Aurora A kinase (AURKA) in normal and pathological cell division. Cell. Mol. Life Sci. 70, 661–687 (2013).
Bertolin, G., et al. Aurora kinase A localises to mitochondria to control organelle dynamics and energy production. eLife 7, e38111 (2018).
Grant, R. et al. Constitutive regulation of mitochondrial morphology by Aurora A kinase depends on a predicted cryptic targeting sequence at the N-terminus. Open Biol. 8, 170272 (2018).
Bertolin, G. & Tramier, M. Insights into the non-mitotic functions of Aurora kinase A: more than just cell division. Cell. Mol. Life Sci. 77, 1031–1047 (2020).
Chouha, N. et al. Development of fluorizoline analogues as prohibitin ligands that modulate C-RAF signaling, p21 expression and melanogenesis. Eur. J. Med. Chem. 242, 114635 (2022).
Tabti, R. et al. Development of prohibitin ligands against osteoporosis. Eur. J. Med. Chem. 210, 112961 (2021).
Yurugi, H. et al. A subset of flavaglines inhibits KRAS nanoclustering and activation. J. Cell Sci. 133, jcs244111 (2020).
Elderwish, S., Audebrand, A., Nebigil, C. G. & Désaubry, L. Discovery of 3,3′-pyrrolidinyl-spirooxindoles as cardioprotectant prohibitin ligands. Eur. J. Med. Chem. 186, 111859 (2020).
Djehal, A. et al. Targeting prohibitin with small molecules to promote melanogenesis and apoptosis in melanoma cells. Eur. J. Med. Chem. 155, 880–888 (2018).
Padilla-Parra, S., Audugé, N., Coppey-Moisan, M. & Tramier, M. Quantitative FRET analysis by fast acquisition time domain FLIM at high spatial resolution in living cells. Biophys. J. 95, 2976–2988 (2008).
Leray, A., Padilla-Parra, S., Roul, J., Héliot, L. & Tramier, M. Spatio-temporal quantification of FRET in living cells by fast time-domain FLIM: a comparative study of non-fitting methods [corrected]. PLoS ONE 8, e69335 (2013).
Kuramori, C. et al. Capsaicin binds to prohibitin 2 and displaces it from the mitochondria to the nucleus. Biochem. Biophys. Res. Commun. 379, 519–525 (2009).
Janusz, J. M. et al. Vanilloids. 1. Analogs of capsaicin with antinociceptive and antiinflammatory activity. J. Med. Chem. 36, 2595–2604 (1993).
Padilla-Parra, S. et al. Quantitative comparison of different fluorescent protein couples for fast FRET-FLIM acquisition. Biophys. J. 97, 2368–2376 (2009).
Nguyen, T. A., Puhl, H. L., Hines, K., Liput, D. J. & Vogel, S. S. Binary-FRET reveals transient excited-state structure associated with activity-dependent CaMKII - NR2B binding and adaptation. Nat. Commun. 13, 6335 (2022).
Amantini, C. et al. Capsaicin-induced apoptosis of glioma cells is mediated by TRPV1 vanilloid receptor and requires p38 MAPK activation. J. Neurochem. 102, 977–990 (2007).
Xu, S. et al. Capsaicin induces mitochondrial dysfunction and apoptosis in anaplastic thyroid carcinoma cells via TRPV1-mediated mitochondrial calcium overload. Cell. Signal. 75, 109733 (2020).
Yang, W. et al. Non-classical ferroptosis inhibition by a small molecule targeting PHB2. Nat. Commun. 13, 7473 (2022).
Schreiber, S. L. The rise of molecular glues. Cell 184, 3–9 (2021).
Bertolin, G. et al. A FRET biosensor reveals spatiotemporal activation and functions of aurora kinase A in living cells. Nat. Commun. 7, 12674 (2016).
Cheetham, G. M. T. Crystal structure of aurora-2, an oncogenic serine/threonine kinase. J. Biol. Chem. 277, 42419–42422 (2002).
Bayliss, R., Sardon, T., Vernos, I. & Conti, E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. cell 12, 851–862 (2003).
Zhang, Y. et al. Identification of the auto-inhibitory domains of Aurora-A kinase. Biochem. Biophys. Res. Commun. 357, 347–352 (2007).
Damodaran, A. P., Vaufrey, L., Gavard, O. & Prigent, C. Aurora A kinase is a priority pharmaceutical target for the treatment of cancers. Trends Pharmacol. Sci. 38, 687–700 (2017).
Sharma, R. K., Chafik, A. & Bertolin, G. Aurora kinase A/AURKA functionally interacts with the mitochondrial ATP synthase to regulate energy metabolism and cell death. Cell Death Discov. 9, 1–12 (2023).
Islam, A., Su, A. J., Zeng, Z.-M., Chueh, P. J. & Lin, M.-H. Capsaicin targets tNOX (ENOX2) to inhibit G1 cyclin/CDK complex, as assessed by the Cellular Thermal Shift Assay (CETSA). Cells 8, 1275 (2019).
Jia, G., Cang, S., Ma, P. & Song, Z. Capsaicin: a “hot” KDM1A/LSD1 inhibitor from peppers. Bioorg. Chem. 103, 104161 (2020).
Chen, H. Y. et al. Capsaicin inhibited aggressive phenotypes through downregulation of tumor-associated NADH oxidase (tNOX) by POU domain transcription factor POU3F2. Molecules 21, 733 (2016).
Yang, F. et al. Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat. Chem. Biol. 11, 518–524 (2015).
Durel, B., Kervrann, C. & Bertolin, G. Quantitative dSTORM super-resolution microscopy localizes Aurora kinase A/AURKA in the mitochondrial matrix. Biol. Cell 113, 458–473 (2021).
Bertolin, G., Marchand, G. L., & Tramier, M. Real-time monitoring of aurora kinase a activation using conformational fret biosensors in live cells. J. Vis. Exp. 161, e61611 (2020).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Kim, S. et al. PubChem 2023 update. Nucleic Acids Res. 51, D1373–D1380 (2023).
Kwon, S., Jung, N., Yang, J., and Seok, C. GalaxySagittarius-AF: predicting targets for drug-like compounds in the extended human 3D proteome. J. Mol. Biol. 436, 168617 (2024).
Berman, H. M. et al. The protein data bank. Nucleic Acids Res. 28, 235–242 (2000).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Yan, Y., Tao, H., He, J. & Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 15, 1829–1852 (2020).
Eswar, N. et al. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics https://doi.org/10.1002/0471250953.bi0506s15 (2006).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Wiederstein, M. & Sippl, M. J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35, W407–W410 (2007).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).
Wallace, A. C., Laskowski, R. A. & Thornton, J. M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8, 127–134 (1995).
Goedhart, J. SuperPlotsOfData – a web app for the transparent display and quantitative comparison of continuous data from different conditions. https://doi.org/10.1091/mbc.E20-09-0583 (2021).
Lord, S. J., Velle, K. B., Mullins, R. D. & Fritz-Laylin, L. K. SuperPlots: Communicating reproducibility and variability in cell biology. J. Cell Biol. 219, e202001064 (2020).
Caron, C., Bertolin, G. & Djehal, C. et al. Raw images (Zenodo) https://doi.org/10.5281/ZENODO.17610405 (2025).
Acknowledgements
We thank S. Dutertre, X. Pinson, and G. Le Marchand at the Microscopy Rennes Imaging Center (MRic, BIOSIT, Biogenouest) for assistance with FLIM experiments. MRic is a member of the national infrastructure France-BioImaging, supported by the French National Research Agency (ANR-24-INBS-0005 FBI BIOGEN). We thank the Plateforme de chimie biologique integrative (PCBIS, University of Strasbourg) for assistance with cell proliferation and viability experiments. We are grateful to N. Jolivet (IGDR, Rennes) for critical reading of the manuscript, to C. Bertin (IGDR, Rennes) for technical assistance, and to all lab members for helpful comments. This work was supported by the Centre National de la Recherche Scientifique (CNRS), the University of Rennes, the Ligue Contre le Cancer, Comités d’Ille et Vilaine et du Finistère to G.B. C.C was supported by a PhD fellowship from the Ligue Nationale Contre le Cancer (grant n. IP/SC – 17653) and by the Fondation ARC pour la Recherche sur le Cancer.
Author information
Authors and Affiliations
Contributions
CRediT taxonomy: conceptualization (L.D. and G.B.) data curation (A.D., C.C., D.G., V.P., K.F., A.F., L.D., and G.B.), Formal analysis (A.D., C.C., D.G, V.P., K.F., A.F., L.D., and G.B.), Funding acquisition (L.D. and G.B.), Investigation (A.D., C.C., D.G., V.P., K.F., A.F., L.D., and G.B.), Methodology (A.F., L.D., and G.B.), Project administration (L.D. and G.B.), Resources (L.D. and G.B.), Software (C.C., D.G., V.P., A.F., and G.B.), Supervision (A.F, L.D., and G.B.), Validation (L.D. and G.B.), Visualization (C.C., D.G., V.P., A.F., L.D., and G.B.), Writing–original, review and editing (C.C., D.G, A.F., L.D., and G.B.).
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing or financial interests. G. Bertolin is an Editorial Board Member for Communications Biology, but was not involved in the editorial review of, nor the decision to publish this article.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Laura Rodríguez Pérez.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Djehal, A., Caron, C., Giordano, D. et al. Development of capsaicin-derived prohibitin ligands to modulate the Aurora kinase A/PHB2 interaction and mitophagy in cancer cells. Commun Biol (2026). https://doi.org/10.1038/s42003-026-09573-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-026-09573-3