Abstract
Biocatalysis represents a particularly promising approach for the degradation of recalcitrant fluorochemicals. However, the enzymatic activation of perfluorinated methylene groups remains a challenge, primarily due to the increase in C–F bond strength that correlates with the number of fluorine atoms attached to a single carbon atom. Here, we describe a new-to-nature hydrodefluorination reaction catalyzed by engineered transaminases for the biodegradation of difluorinated fused-ring ketones. Remarkable defluorination efficiency was achieved in most cases by employing a slight excess of 2-PrNH2 as amine donor. Experimental results provide conclusive evidence that the catalytic process commences with a reversible transamination step. The comprehensive examination of this promiscuous reaction provides constructive guidance for the development of emerging biocatalytic strategies, thereby facilitating the biodegradation of polyfluorinated compounds and contributing to environmental sustainability.
Similar content being viewed by others
Introduction
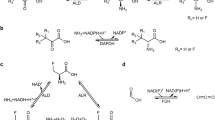
Despite the extensive applications of fluorochemicals1, they are often considered the quintessential example of industrial chemical pollution due to their toxicity, persistence, and bioaccumulation2,3. These issues are primarily attributable to the highly inert C–F bond4,5, which has a bond dissociation energy (BDE) up to 130 kcal·mol−1. Various methods have been developed to achieve the degradation of fluorochemicals, among which biodegradation is highly desirable as a cost-effective and environmentally friendly alternative6,7. In recent years, a number of responsible enzymes have been identified that are able to perform the defluorinations of monofluoro-substituted compounds, including uronate isomerase8, pyruvate dehydrogenase9, cytochrome P45010,11, and others12,13,14,15,16. Typically, the C–F bond in aliphatic fluorides becomes shorter and stronger as the number of fluorine atoms attached to the same carbon atom increases17,18. Consequently, the BDE of the C–F bond in a difluoromethylene group is higher than that of the C–F bond in a monofluoromethyl group. It is noteworthy that substances containing at least one perfluorinated methylene group (–CF2−) are classified as per- and polyfluorinated alkyl substances (PFASs) according to the 2021 OECD definition19,20. Enzymes can degrade most anthropogenic compounds composed of functional groups found in natural products, but the –CF2− group is inherently foreign to nature21. Despite a few advancements like fluoroacetate dehalogenase (FAcD) for difluoroacetate with low reactivity22,23,24 and nitrogenase for ‘strained’ difluorinated cyclopropene25, enzymatic activation of the difluoromethylene group remains a coveted but challenging goal (Fig. 1a).
β-Fluoroamines and α-fluoroketones are irreversible inhibitors of pyridoxal 5′-phosphate (PLP)-dependent transaminases (TAs), acting via elimination-conjugate addition pathways26,27. Inspired by this mechanism, Lavandera and co-workers developed catalytic hydrodefluorinations (HDFs) of monofluoro-substrates by removing the active-site nucleophile (Nu)28,29. However, this system underwent transaminations rather than defluorinations when difluorinated analogs were used as substrates, aligning with the inertness of the –CF2− group30. The distinct chemical pathways are determined by the action sites of the specific lysine: Cα-deprotonation initiates an elimination mechanism, while C4′-addition facilitates a transamination reaction (Fig. 1b). The reaction specificity of PLP-dependent enzymes has been demonstrated to be regulated by multiple factors, encompassing stereoelectronic effects, PLP protonation state, and active-site interactions31,32. As such, our theoretical framework suggests that the successful implementation of the sequential HDFs of perfluorinated methylene groups is contingent upon the identification of an appropriate PLP-dependent enzyme.
In this study, we present the development of an engineered TA-catalyzed sequential HDF system, utilizing difluorinated fused-ring ketones as substrates (Fig. 1c). Experimental evidence confirms that the process is initiated by transamination, and owing to the inherent reversibility of transamination, the amine intermediates are completely transformed into defluorinated products. This strategy exhibits remarkable sustainability and practicality, enabling efficient degradation of specialized fluorochemicals under mild conditions.
Results and discussion
To experimentally validate the feasibility of our proposed methodology, we systematically conducted optimization studies employing 2,2-difluoro-3,4-dihydronaphthalen-1(2H)-one (1a) as the model substrate in the presence of isopropylamine (2-PrNH2) as the amine donor. Among the TA library heterologously expressed in our lab, ATA117 Rd11, an engineered variant of ATA117 from Arthrobacter sp., which has been successfully implemented in the industrial-scale synthesis of Sitagliptin33, exhibited promising HDF activity of 1a (Fig. 2a, Table S1). The reaction achieved a total conversion rate of 57%, yielding a mixture of completely defluorinated product 2a and monofluorinated 3a.
a Model reaction. b View of the engineered active site (ATA117 Rd11, PDB: 5FR928). c The results of selected variants (the average of duplicates). Experimental conditions: 1a (6 mM), 2-PrNH2 (a13.2 mM, b15 mM), PLP (0.8 mM), solvent (1 mL 100 mM Kpi-buffer, pH 7.0 containing 2.5% v/v DMSO), crude cell extract (containing about 0.03 mM enzyme), 30 °C, 6 h. M0 = ATA117 Rd11, M1 = M122F, M2 = M122W, M3 = G224A, M4 = M122W-G224A, M5 = M122F-G224A.
To engineer the well-characterized TA into a highly efficient “defluorinase”, we implemented a semi-rational protein engineering strategy, namely “Focused Rational Iterative Site-specific Mutagenesis” (FRISM, Fig. S1)34,35. Based on the crystal structure analysis of ATA117 Rd11 (PDB: 5FR928), we identified key mutation hotspots within the active-site region, which were predicted to be the primary determinants of catalytic activity due to their spatial orientation and potential interactions with the substrate (Fig. 2b). Initially, amino acids with distinct steric profiles, including alanine (A), leucine (L) and phenylalanine (F) were introduced to systematically evaluate the impact of active-site volume on catalytic efficiency. Subsequently, based on the preliminary screening results, we constructed an extended mutant library by incorporating amino acids with similar steric properties but different functional groups: glycine (G) for alanine derivatives (A+), valine/cysteine/isoleucine/methionine (V/C/I/M) for leucine variants (L+), and tyrosine/tryptophan (Y/W) for phenylalanine analogs (F+). This progressive optimization approach enabled fine-tuning of both steric and electronic properties at the target positions (Table S2). To our delight, several mutations exhibited a significant increase in activity, with the G224A variant (M3) displaying the highest defluorination efficiency, reaching 76% conversion and yielding 57% of product 2a. Furthermore, the synergistic combination of the previously identified key mutations markedly enhanced the enzymatic activity. Complete defluorination of the difluorinated substrate was accomplished using the M122W-G224A biocatalyst (M5) and 2.5 equivalents of 2-PrNH2 as amine donor (Fig. 2c, Tables S3, S4). The variant displayed a significantly higher turnover number (TON) compared to ATA117 Rd11 (504 vs 72). Despite the mutation leading to a reduction in the KM value, the overall catalytic efficiency (kcat/KM) still exhibited a 4-fold enhancement (0.069 vs 0.017 mM−1·s−1, Table S5).
With the optimized reaction conditions in hand (Fig. 2c), we assessed the scope of the TA-catalyzed HDF reactions (Fig. 3). Our findings suggested that the TA/2-PrNH2 approach served as a versatile and applicable platform for the biodegradation of difluorinated fused-ring ketones, with a >90% defluorination rate in most cases. The chemoselective HDF of difluorinated tetralone derivatives (1a–1k), whether bearing electron-donating or electron-withdrawing substituents at different positions on the aryl moiety, predominantly yielded the corresponding completely defluorinated products. We were delighted to observe that the reaction also proceeded smoothly with heteroaryl substrate 1l, as well as with both the 4-substituted (1m) and 4-oxa (1n) substrates. Furthermore, the biocatalytic system showed compatibility with difluorinated 1-indanone (1o), though the transformation afforded the monofluorinated ketone as the major product. Based on the structural analysis of the substrates (1a–1o), steric hindrance is proposed as the primary factor governing their biodegradation activity. Surprisingly, the acyclic ketone 1p and monocyclic ketone 1q exhibited incompatibility with this system, undergoing transamination instead of participating in the desired HDF process. This result demonstrated that the fused-ring structure plays a pivotal role in facilitating the cleavage of the C–F bond, likely attributed to its rigid geometric configuration.
The catalytic potential of natural TAs was frequently harnessed in amino transfer reactions, driving the efficient synthesis of chiral amino compounds. Surprisingly, the model reaction did not afford the desired aminated compound but instead yielded 1-tetralone (2a) as the only detectable product. To explore the mechanism of this enzymatic new-to-nature reaction, we leveraged 19F NMR as a powerful tool to track the reaction progress by monitoring the chemical shifts for 1a (δ −110.4 ppm), 3a (δ −189.5 ppm), 4a (δ −101.5/102.2/109.3/110.0 ppm) and F− (δ −118.6 ppm) within the reaction mixture (Fig. 4a, S2). The presence of the F-signal corresponding to 4a was detected at the initial stage of the reaction, providing clear evidence that the process was initiated through a transamination mechanism. The signal corresponding to 4a completely vanished after a reaction time of 3 h, concurrently giving way to the emergence of a distinct signal attributed to 3a. Compound 3a represented a highly reactive intermediate that rapidly underwent HDF under the reaction conditions. After 6 h, only F− could be detected in the 19F NMR spectrum. The efficient progression of this sequential HDF process can be attributed to the distinct thermodynamic characteristics of its elementary steps: the reversible nature of the transamination facilitates the reaction equilibrium, whereas the subsequent defluorination, being thermodynamically favorable and irreversible, drives the overall transformation to completion. Then we used rac-4a as substrate under standard conditions, and found the formation of 2a (Fig. 4b). Remarkably, this transformation proceeded as a highly enantioselective kinetic resolution process, remaining 43% (+)-4a with exceptional enantiopurity (>99% ee). This result agreed well with the 19F NMR experiments.
We proposed the mechanism illustrated in Fig. 4c. To begin with, the coenzyme PLP undergoes conversion into pyridoxamine 5′-phosphate (PMP) in the presence of 2-PrNH2. The condensation reaction between PMP and ketone 1a forms the External Aldimine I, which is followed by tautomeric resonance to yield the key External Aldimine II via quinonoid intermediate. Nucleophilic addition of lysine to the imine bond occurs preferentially, thereby releasing PLP and generating 4a, the product of the transamination reaction. In the alternative reaction pathway, the irreversible elimination process occurs prior to the addition step. The progressive depletion of External Aldimine II thermodynamically drives the reversible transformation of its precursor 4a. The resulting enamine rapidly tautomerizes in water, yielding the corresponding ketone 3a. The monofluorinated intermediate 3a exhibits significantly enhanced reactivity within the biocatalytic system, facilitating its efficient participation in the subsequent catalytic cycle, which ultimately leads to the formation of the fully defluorinated product 2a.
During the study on protein engineering and reaction scope, we identified the formation of monofluoriated ketones 3. These compounds are structurally significant due to the presence of chiral centers at the α-position36. Regrettably, the stereocontrol efficiency of them proved to be suboptimal, with limited stereoselectivity observed. Based on the proposed reaction mechanism, we postulated that these compounds were derived from achiral enamines through isomerization, where the enamine intermediates had already lost their covalent attachment to PLP during their formation process.
Conclusion
In summary, we developed a versatile biodegradation platform for the consecutive HDFs of difluorinated ketones mediated by PLP-dependent enzyme. Through a semi-rational protein engineering strategy, we successfully obtained a mutant enzyme that demonstrates improved defluorination efficiency. 19F NMR was used to monitor the reaction progress, revealing that the new-to-nature HDF reaction was initiated by a reversible transformation process. A thorough mechanistic understanding of this reaction provided valuable insights for advancing enzyme engineering strategies, enabling efficient degradation of fluorochemicals while contributing significantly to environmental sustainability.
Methods
General procedure for the transaminase-catalyzed sequential hydrodefluorinations of 1a–1p 25 μL stock solutions (DMSO) with 1a–1p (6 μmol) were added into 965 μL crude cell extract (KPi-buffer 100 mM, pH 7.0, containing 0.8 mM PLP and about 0.03 mM enzyme). Then, 2.5–4.0 eq. 2-PrNH2 was added to the system. The mixture (about 1 mL total volume) was shaken at 800 rpm for 6–24 h at 30–35 °C (2.5 eq. 2-PrNH2, 30 °C and 6 h for 1a, 1b, 1d, 1n; 3.5 eq. 2-PrNH2, 35 °C and 12 h for 1c, 1e–1g, 1i–1k, 1m; 4.0 eq. 2-PrNH2, 35 °C and 24 h for 1h, 1l, 1o, 1p, 1q), then extracted with ethyl acetate for three times (3 × 1 mL). The conversion rates and ratios of 2/3 were determined by HPLC with Daicel Chiralcel OJ-H column (250 × 4.6 mm) using n-hexanes/isopropanol as the mobile phase.
Data availability
Data relating to supplementary methods, supplementary tables and figures, characterization data, NMR spectra, chromatographic data, and supplementary references are available in the Supplementary Information and from the corresponding author(s) upon request.
References
Müller, K., Faeh, C. & Diederich, F. Fluorine in pharmaceuticals: looking beyond intuition. Science 317, 1881–1886 (2007).
Han, J. et al. Chemical aspects of human and environmental overload with fluorine. Chem. Rev. 121, 4678–4742 (2021).
Evich, M. G. et al. Per- and polyfluoroalkyl substances in the environment. Science 375, eabg9065 (2022).
Amii, H. & Uneyama, K. C–F bond activation in organic synthesis. Chem. Rev. 109, 2119–2183 (2009).
Shen, Q. et al. Review of recent advances in C–F bond activation of aliphatic fluorides. J. Fluor. Chem. 179, 14–22 (2015).
Wackett, L. P. & Robinson, S. L. A prescription for engineering PFAS biodegradation. Biochem. J. 481, 1757–1770 (2024).
Wang, Y. & Liu, A. Carbon-fluorine bond cleavage mediated by metalloenzymes. Chem. Soc. Rev. 49, 4906–4925 (2020).
Williams, L., Nguyen, T., Li, Y., Porter, T. N. & Raushel, F. M. Uronate isomerase: a nonhydrolytic member of the amidohydrolase superfamily with an ambivalent requirement for a divalent metal ion. Biochemistry 45, 7453–7462 (2006).
Flournoy, D. S. & Frey, P. A. Pyruvate dehydrogenase and 3-fluoropyruvate: chemical competence of 2-acetylthiamin pyrophosphate as an acetyl group donor to dihydrolipoamide. Biochemistry 25, 6036–6043 (1986).
Li, S. & Wackett, L. P. Reductive dehalogenation by cytochrome P450CAM: substrate binding and catalysis. Biochemistry 32, 9355–9361 (1993).
Huang, Q. et al. Discovery of a P450-catalyzed oxidative defluorination mechanism toward chiral organofluorines: uncovering a hidden pathway. ACS Catal. 12, 265–272 (2022).
Porter, D. J. et al. Enzymatic elimination of fluoride from α-fluoro-β-alanine. Biochem. Pharmacol. 50, 1475–1484 (1995).
Solyanikova, I. P. et al. Conversion of 2-fluoromuconate to cis-dienelactone by purified enzymes of Rhodococcus opacus 1cp. Appl. Environ. Microbiol. 69, 5636–5642 (2003).
Walsh, C. Fluorinated substrate analogs: routes of metabolism and selective toxicity. Adv. Enzymol. Relat. Areas Mol. Biol. 55, 197–289 (1983).
Chan, P. W. Y. et al. Defluorination capability of l-2-haloacid dehalogenases in the HAD-like hydrolase superfamily correlates with active site compactness. ChemBioChem 23, e202100414 (2021).
Wu, L. & Deng, H. Defluorination of 4-fluorothreonine by threonine deaminase. Org. Biomol. Chem. 18, 6236–6240 (2020).
O’Hagan, D. Understanding organofluorine chemistry. An introduction to the C–F bond. Chem. Soc. Rev. 37, 308–319 (2008).
Blanksby, S. J. & Ellison, G. B. Bond dissociation energies of organic molecules. Acc. Chem. Res. 36, 255–263 (2003).
OECD. Reconciling Terminology of the Universe of Per- And Polyfluoroalkyl Substances: Recommendations and Practical Guidance. OECD Series on Risk Management No. 61 (OECD, 2021).
Schymanski, E. L. et al. Per- and polyfluoroalkyl substances (PFAS) in PubChem: 7 million and growing. Environ. Sci. Technol. 57, 16918–16928 (2023).
Walker, M. C. & Chang, M. C. Y. Natural and engineered biosynthesis of fluorinated natural products. Chem. Soc. Rev. 43, 6527–6536 (2014).
Yue, Y. et al. Comprehensive understanding of fluoroacetate dehalogenase-catalyzed degradation of fluorocarboxylic acids: a QM/MM approach. Environ. Sci. Technol. 55, 9817–9825 (2021).
Khusnutdinova, A. N. et al. Structural insights into hydrolytic defluorination of difluoroacetate by microbial fluoroacetate dehalogenases. FEBS J. 290, 4966–4983 (2023).
Farajollahi, S. et al. Defluorination of organofluorine compounds using dehalogenase enzymes from Delftia acidovorans (D4B). ACS Omega 9, 28546–28555 (2024).
Ni, F. et al. Reduction of fluorinated cyclopropene by nitrogenase. J. Am. Chem. Soc. 135, 10346–10352 (2013).
Nanavati, S. M. & Silverman, R. B. Design of potential anticonvulsant agents: mechanistic classification of GABA aminotransferase inactivators. J. Med. Chem. 32, 2413–2421 (1989).
Lippert, B., Metcalf, B. W. & Resvick, R. J. Enzyme-activated irreversible inhibition of rat and mouse brain 4-aminobutyric acid-α-ketoglutarate transaminase by 5-fluoro-4-oxo-pentanoic acid. Biochem. Biophys. Res. Commun. 108, 146–152 (1982).
Cuetos, A. et al. Catalytic promiscuity of transaminases: preparation of enantioenriched β-fluoroamines by formal tandem hydrodefluorination/deamination. Angew. Chem. Int. Ed. 55, 2947–3232 (2016).
García-Ramos, M., Cuetos, A., Kroutil, W., Grogan, G. & Lavandera, I. The reactivity of α-fluoroketones with PLP dependent enzymes: transaminases as hydrodefluorinases. ChemCatChem 13, 3967–3972 (2021).
García-Ramosa, M. & Lavandera, I. Transaminases as suitable catalysts for the synthesis of enantiopure β,β-difluoroamines. Org. Biomol. Chem. 20, 984–988 (2022).
Dunathan, H. C. Conformation and reaction specificity in pyridoxal phosphate enzymes. Proc. Natl. Acad. Sci. USA 55, 712–716 (1966).
Toney, M. D. Controlling reaction specificity in pyridoxal phosphate enzymes. Biochim. Biophys. Acta. 1814, 1407–1418 (2011).
Savile, C. K. et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science 329, 305–309 (2010).
Xu, J. et al. Stereodivergent protein engineering of a lipase to access all possible stereoisomers of chiral esters with two stereo-centers. J. Am. Chem. Soc. 141, 7934–7945 (2019).
Bao, Y., Xu, Y. & Huang, X. Focused rational iterative site-specific mutagenesis (FRISM): a powerful method for enzyme engineering. Mol. Catal. 553, 113755 (2024).
Lectard, S., Hamashima, Y. & Sodeoka, M. Recent advances in catalytic enantioselective fluorination reactions. Adv. Synth. Catal. 325, 2708–2732 (2010).
Acknowledgements
The financial support from the National Natural Science Foundation of China (No. 22201106, 22477112) and National Key Research and Development Program of China (2023YFA0914200) is gratefully acknowledged. The authors thank the Analysis and Testing Center at Zhejiang University of Technology for the assistance in the structural analysis of the compounds.
Author information
Authors and Affiliations
Contributions
X. Chen and X. Chu directed the project. X. Chen and Y.Z. performed the majority of the synthetic experiments. J.F. and Z.C. assisted with protein engineering experiments under the guidance of X.Z. X. Chen and X.Z. wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Chemistry thanks Peter Stockinger and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, X., Zhang, Y., Fu, J. et al. Transaminase-catalyzed sequential hydrodefluorinations of perfluorinated methylene groups. Commun Chem 8, 396 (2025). https://doi.org/10.1038/s42004-025-01781-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42004-025-01781-y