Abstract
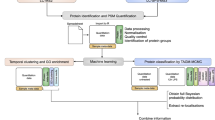
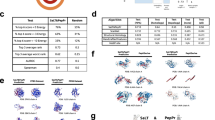
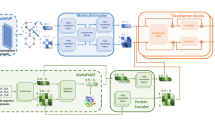
Despite the success of pretrained natural language processing (NLP) models in various fields, their application in computational biology has been hindered by their reliance on biological sequences, which ignores vital three-dimensional (3D) structural information incompatible with the sequential architecture of NLP models. Here we present a topological transformer (TopoFormer), which is built by integrating NLP models and a multiscale topology technique, the persistent topological hyperdigraph Laplacian (PTHL), which systematically converts intricate 3D protein–ligand complexes at various spatial scales into an NLP-admissible sequence of topological invariants and homotopic shapes. PTHL systematically transforms intricate 3D protein–ligand complexes into NLP-compatible sequences of topological invariants and shapes, capturing essential interactions across spatial scales. TopoFormer gives rise to exemplary scoring accuracy and excellent performance in ranking, docking and screening tasks in several benchmark datasets. This approach can be utilized to convert general high-dimensional structured data into NLP-compatible sequences, paving the way for broader NLP based research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The training dataset employed in this study comprises a comprehensive collection of protein–ligand complexes sourced from various PDBbind databases, specifically CASF-2007, CASF-2013, CASF-2016 and PDBbind v.2020. To ensure the dataset’s reliability and eliminate redundancies, a meticulous curation process was undertaken, resulting in a total of 19,513 non-overlapping complexes. All data used in this study can be downloaded from the official PDBbind website: http://www.pdbbind.org.cn/index.php. We also provide a comprehensive set of resources at https://github.com/WeilabMSU/TopoFormer. This includes topological embedded features used in both TopoFormer and TopoFormers, sequence-based features derived from the Transformer-CPZ28 and ESM33 models and all additional generated poses with their associated scores, which were crucial for the docking and screening tasks. Instructions for accessing the poses are also available via Zenodo at https://doi.org/10.5281/zenodo.10892799 (ref. 66).
Code availability
All source code and models are publicly available via Zenodo at https://doi.org/10.5281/zenodo.10892799 (ref. 66).
References
Fleming, N. How artificial intelligence is changing drug discovery. Nature 557, S55–S57 (2018).
Lyu, J. et al. Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 (2019).
Kitchen, D. B., Decornez, H., Furr, J. R. & Bajorath, J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discov. 3, 935–949 (2004).
Pinzi, L. & Rastelli, G. Molecular docking: shifting paradigms in drug discovery. Int. J. Mol. Sci. 20, 4331 (2019).
Pagadala, N. S., Syed, K. & Tuszynski, J. Software for molecular docking: a review. Biophys. Rev. 9, 91–102 (2017).
Wang, L. et al. Accurate and reliable prediction of relative ligand binding potency in prospective drug discovery by way of a modern free-energy calculation protocol and force field. J. Am. Chem. Soc. 137, 2695–2703 (2015).
Sliwoski, G., Kothiwale, S., Meiler, J. & Lowe, E. W. Computational methods in drug discovery. Pharmacol. Rev. 66, 334–395 (2014).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Baek, M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Song, Y. & Wang, L. Multiobjective tree-based reinforcement learning for estimating tolerant dynamic treatment regimes. Biometrics 80, ujad017 (2024).
Luo, J., Wei, W., Waldispühl, J. & Moitessier, N. Challenges and current status of computational methods for docking small molecules to nucleic acids. Eur. J. Med. Chem. 168, 414–425 (2019).
Lo, Yu-Chen, Rensi, S. E., Torng, W. & Altman, R. B. Machine learning in chemoinformatics and drug discovery. Drug Discov. Today 23, 1538–1546 (2018).
The Atomwise AIMS Program. AI is a viable alternative to high throughput screening: a 318-target study. Sci. Rep. 14, 7526 (2024).
Gómez-Sacristán, P., Simeon, S., Tran-Nguyen, V.-K., Patil, S. & Ballester, P. J. Inactive-enriched machine-learning models exploiting patent data improve structure-based virtual screening for PDL1 dimerizers. J. Adv. Res. (in the press); https://doi.org/10.1016/j.jare.2024.01.024
Hu, X. et al. Discovery of novel non-steroidal selective glucocorticoid receptor modulators by structure-and IGN-based virtual screening, structural optimization, and biological evaluation. Eur. J. Med. Chem. 237, 114382 (2022).
Vaswani, A. et al. Attention is all you need. In NIPS'17: Proc. 31st International Conference on Neural Information Processing Systems (eds von Luxburg, U. et al.) 6000–6010 (Curran Associates, 2017).
Devlin, J., Chang, M. W., Lee, K. & Toutanova, K. B. BERT: pre-training of deep bidirectional transformers for language understanding. In Proc. 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies 1, 4171–4186 (Association for Computational Linguistics, 2019).
Ouyang, L. et al. Training language models to follow instructions with human feedback. Adv. Neural Inf. Process. Syst. 35, 27730–27744 (2022).
Singh, R., Sledzieski, S., Bryson, B., Cowen, L. & Berger, B. Contrastive learning in protein language space predicts interactions between drugs and protein targets. Proc. Natl Acad. Sci. USA 120, e2220778120 (2023).
Saar, K. L. et al. Turning high-throughput structural biology into predictive inhibitor design. Proc. Natl Acad. Sci. USA 120, e2214168120 (2023).
Cang, Z., Mu, L. & Wei, G.-W. Representability of algebraic topology for biomolecules in machine learning based scoring and virtual screening. PLoS Comput. Biol. 14, e1005929 (2018).
Nguyen, D. D., Cang, Z. & Wei, G.-W. A review of mathematical representations of biomolecular data. Phys. Chem. Chem. Phys. 22, 4343–4367 (2020).
Wang, R., Nguyen, D. D. & Wei, G.-W. Persistent spectral graph. Int. J. Numer. Methods Biomed. Eng. 36, e3376 (2020).
Meng, Z. & Xia, K. Persistent spectral–based machine learning (PerSpect ML) for protein-ligand binding affinity prediction. Sci. Adv. 7, eabc5329 (2021).
Chen, D., Liu, J., Wu, J. & Wei, G.-W. Persistent hyperdigraph homology and persistent hyperdigraph Laplacians. Found. Data Sci. 5, 558–588 (2023).
Zomorodian, A. & Carlsson, G. Computing persistent homology. Discrete Comput. Geom. 33, 249–274 (2005).
Chen, D., Zheng, J., Wei, G.-W. & Pan, F. Extracting predictive representations from hundreds of millions of molecules. J. Phys. Chem. Lett. 12, 10793–10801 (2021).
Ruff, K. M. & Pappu, R. V. AlphaFold and implications for intrinsically disordered proteins. J. Mol. Biol. 433, 167208 (2021).
Li, Y., Han, L., Liu, Z. & Wang, R. Comparative assessment of scoring functions on an updated benchmark: 2. Evaluation methods and general results. J. Chem. Inf. Model. 54, 1717–1736 (2014).
Cheng, T., Li, X., Li, Y., Liu, Z. & Wang, R. Comparative assessment of scoring functions on a diverse test set. J. Chem. Inf. Model. 49, 1079–1093 (2009).
Su, M. et al. Comparative assessment of scoring functions: the CASF-2016 update. J. Chem. Inf. Model. 59, 895–913 (2018).
Trull, T. J. & Ebner-Priemer, U. W. Using experience sampling methods/ecological momentary assessment (ESM/EMA) in clinical assessment and clinical research: introduction to the special section. Psychol. Assess. 21, 457–462 (2009).
Karlov, D. S., Sosnin, S., Fedorov, M. V. & Popov, P. graphDelta: MPNN scoring function for the affinity prediction of protein–ligand complexes. ACS Omega 5, 5150–5159 (2020).
Sánchez-Cruz, N., Medina-Franco, J., Mestres, J. & Barril, X. Extended connectivity interaction features: improving binding affinity prediction through chemical description. Bioinformatics 37, 1376–1382 (2021).
Wang, Z. et al. Onionnet-2: a convolutional neural network model for predicting protein-ligand binding affinity based on residue-atom contacting shells. Front. Chem. 9, 753002 (2021).
Rezaei, M. A., Li, Y., Wu, D., Li, X. & Li, C. Deep learning in drug design: protein-ligand binding affinity prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 19, 407–417 (2020).
Wang, S. et al. Se-onionnet: a convolution neural network for protein–ligand binding affinity prediction. Front. Genet. 11, 607824 (2021).
Jones, D. et al. Improved protein–ligand binding affinity prediction with structure-based deep fusion inference. J. Chem. Inf. Model. 61, 1583–1592 (2021).
Boyles, F., Deane, C. M. & Morris, G. M. Learning from the ligand: using ligand-based features to improve binding affinity prediction. Bioinformatics 36, 758–764 (2020).
Liu, Z. et al. PDB-wide collection of binding data: current status of the PDBbind database. Bioinformatics 31, 405–412 (2015).
Wang, C. & Zhang, Y. Improving scoring-docking-screening powers of protein–ligand scoring functions using random forest. J. Comput. Chem. 38, 169–177 (2017).
Gentile, F. et al. Automated discovery of noncovalent inhibitors of SARS-Cov-2 main protease by consensus deep docking of 40 billion small molecules. Chem. Sci. 12, 15960–15974 (2021).
Méndez-Lucio, O., Ahmad, M., del Rio-Chanona, E. A. & Wegner, J. K. A geometric deep learning approach to predict binding conformations of bioactive molecules. Nat. Mach. Intell. 3, 1033–1039 (2021).
Zheng, L. et al. Improving protein–ligand docking and screening accuracies by incorporating a scoring function correction term. Brief. Bioinform. 23, bbac051 (2022).
Bao, J., He, X. & Zhang, J. Z. H. DeepBSP—a machine learning method for accurate prediction of protein–ligand docking structures. J. Chem. Inf. Model. 61, 2231–2240 (2021).
Shen, C. et al. Boosting protein–ligand binding pose prediction and virtual screening based on residue–atom distance likelihood potential and graph transformer. J. Med. Chem. 65, 10691–10706 (2022).
Nguyen, D. D. & Wei, G.-W. AGL-Score: algebraic graph learning score for protein–ligand binding scoring, ranking, docking, and screening. J. Chem. Inf. Model. 59, 3291–3304 (2019).
Liu, X., Feng, H., Wu, J. & Xia, K. Dowker complex based machine learning (DCML) models for protein-ligand binding affinity prediction. PLoS Comput. Biol. 18, e1009943 (2022).
Tran-Nguyen, V.-K., Junaid, M., Simeon, S. & Ballester, P. J. A practical guide to machine-learning scoring for structure-based virtual screening. Nat. Protoc. 18, 3460–3511 (2023).
Moon, S., Zhung, W., Yang, S., Lim, J. & Kim, W. Y. PIGNet: a physics-informed deep learning model toward generalized drug–target interaction predictions. Chem. Sci. 13, 3661–3673 (2022).
Tran-Nguyen, V.-K., Bret, G. & Rognan, D. True accuracy of fast scoring functions to predict high-throughput screening data from docking poses: the simpler the better. J. Chem. Inf. Model. 61, 2788–2797 (2021).
Tran-Nguyen, V.-K. & Ballester, P. J. Beware of simple methods for structure-based virtual screening: the critical importance of broader comparisons. J. Chem. Inf. Model. 63, 1401–1405 (2023).
Tran-Nguyen, V.-K., Simeon, S., Junaid, M. & Ballester, P. J. Structure-based virtual screening for PDL1 dimerizers: evaluating generic scoring functions. Curr. Res. Struct. Biol. 4, 206–210 (2022).
Shen, C. et al. A generalized protein–ligand scoring framework with balanced scoring, docking, ranking and screening powers. Chem. Sci. 14, 8129–8146 (2023).
Jones, G., Willett, P., Glen, R. C., Leach, A. R. & Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267, 727–748 (1997).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Tran-Nguyen, V.-K., Jacquemard, C. & Rognan, D. LIT-PCBA: an unbiased data set for machine learning and virtual screening. J. Chem. Inf. Model. 60, 4263–4273 (2020).
Horak, D. & Jost, J. Spectra of combinatorial Laplace operators on simplicial complexes. Adv. Math. 244, 303–336 (2013).
Eckmann, B. Harmonische funktionen und randwertaufgaben in einem komplex. Comment. Math. Helv. 17, 240–255 (1944).
Chen, J., Zhao, R., Tong, Y. & Wei, G.-W. Evolutionary de Rham-Hodge method. Discrete Continuous Dyn. Syst. Ser. B. 26, 3785–3821 (2021).
Mémoli, F., Wan, Z. & Wang, Y. Persistent Laplacians: properties, algorithms and implications. SIAM J. Math. Data Sci. 4, 858–884 (2022).
Edelsbrunner, H., Letscher, D. & Zomorodian, A. Topological persistence and simplification. Discrete Comput. Geom. 28, 511–533 (2002).
Liu, J., Li, J. & Wu, J. The algebraic stability for persistent Laplacians. Preprint at arXiv https://doi.org/10.48550/arXiv.2302.03902 (2023).
He, K. et al. Masked autoencoders are scalable vision learners. In Proc. 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 15979–15988 (IEEE, 2022).
Chen, D. WeilabMSU/TopoFormer: TopoFormer. Zenodo https://doi.org/10.5281/zenodo.10892799 (2024).
Sunseri, J. & Koes, D. R. Virtual screening with Gnina 1.0. Molecules 26, 7369 (2021).
Yang, C. & Zhang, Y. Delta machine learning to improve scoring-ranking-screening performances of protein–ligand scoring functions. J. Chem. Inf. Model. 62, 2696–2712 (2022).
Wójcikowski, M., Kukiełka, M., Stepniewska-Dziubinska, M. M. & Siedlecki, P. Development of a protein–ligand extended connectivity (PLEC) fingerprint and its application for binding affinity predictions. Bioinformatics 35, 1334–1341 (2019).
Stepniewska-Dziubinska, M. M., Zielenkiewicz, P. & Siedlecki, P. Development and evaluation of a deep learning model for protein–ligand binding affinity prediction. Bioinformatics 34, 3666–3674 (2018).
Acknowledgements
This work was supported in part by NIH grant nos. R01GM126189, R01AI164266 and R35GM148196, National Science Foundation grant nos. DMS2052983 and IIS-1900473, Michigan State University Research Foundation, and Bristol-Myers Squibb grant no. 65109. The work of J.L. was performed while visiting Michigan State University.
Author information
Authors and Affiliations
Contributions
D.C. designed the project, modified the method, wrote the code, performed computational studies, wrote the first draft and revised the manuscript. J.L. wrote the methods section and revised the manuscript. G.-W.W. conceptualized and supervised the project, acquired funding and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Machine Intelligence thanks Emil Alexov, Pedro Ballester and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10, Tables 1–7, Evaluation metrics, Hyperparameter selection and optimization, Topological objects, Vietoris–Rips hyperdigraph and alpha hyperdigraph.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, D., Liu, J. & Wei, GW. Multiscale topology-enabled structure-to-sequence transformer for protein–ligand interaction predictions. Nat Mach Intell 6, 799–810 (2024). https://doi.org/10.1038/s42256-024-00855-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42256-024-00855-1
This article is cited by
-
A unified time-frequency foundation model for sleep decoding
Nature Communications (2026)
-
Nanoparticle-mediated targeting chimeras transform targeted protein degradation
Nature Nanotechnology (2026)
-
Harnessing pre-trained models for accurate prediction of protein-ligand binding affinity
BMC Bioinformatics (2025)
-
Active phase discovery in heterogeneous catalysis via topology-guided sampling and machine learning
Nature Communications (2025)
-
Geometry based prediction of tau protein sites and motifs associated with misfolding and aggregation
Scientific Reports (2025)