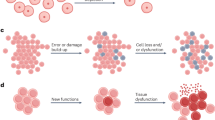
Abstract
The mechanisms of aging are becoming increasingly well mapped; however, there remains ongoing debate about the ultimate and proximate causes of aging. The recent development of highly precise aging clocks led to a resurgence of arguments in support of a biological program of aging. However, the declining force of natural selection after the onset of reproduction means that cellular function could deteriorate without requiring a specific program. Here, we argue that aging clocks do not imply an intrinsic program but rather reflect the stochastic accumulation of molecular errors and damage. Damage accumulates due to insufficient maintenance and repair and contributes to system-wide entropy. In support of this, cross-species comparisons indicate that enhanced DNA repair capacity is a key determinant of exceptional longevity in mammals. By better understanding the nature of the stochasticity that governs the aging process, we will have a stronger mechanistic basis for developing geroprotective interventions to promote healthy aging in humans.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
15 December 2025
A Correction to this paper has been published: https://doi.org/10.1038/s43587-025-01050-6
References
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: an expanding universe. Cell 186, 243–278 (2023).
Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin.74, 12–49 (2024).
Ahadi, S. et al. Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat. Med. 26, 83–90 (2020).
Zhang, W. B. et al. Extended twilight among isogenic C. elegans causes a disproportionate scaling between lifespan and health. Cell Syst. 3, 333–345 (2016).
Li, X. et al. Systems properties and spatiotemporal regulation of cell position variability during embryogenesis. Cell Rep. 26, 313–321 (2019).
Ruby, J. G. et al. Estimates of the heritability of human longevity are substantially inflated due to assortative mating. Genetics 210, 1109–1124 (2018).
Shenhar, B. et al. Heritability of human lifespan is about 50% when confounding factors are addressed. Preprint at bioRxiv https://doi.org/10.1101/2025.04.20.649385 (2025).
Issa, J. Aging and epigenetic drift: a vicious cycle. J. Clin. Invest. 124, 24–29 (2014).
Tarkhov, A. E. et al. Nature of epigenetic aging from a single-cell perspective. Nat. Aging 4, 854–870 (2024).
Schumacher, B., Pothof, J., Vijg, J. & Hoeijmakers, J. H. J. The central role of DNA damage in the ageing process. Nature 592, 695–703 (2021).
Ren, P., Zhang, J. & Vijg, J. Somatic mutations in aging and disease. GeroScience 46, 5171–5189 (2024).
Cagan, A. et al. Somatic mutation rates scale with lifespan across mammals. Nature 604, 517–524 (2022).
Hamilton, W. D. The moulding of senescence by natural selection. J. Theor. Biol. 12, 12–45 (1966).
Moorad, J., Promislow, D. & Silvertown, J. Evolutionary ecology of senescence and a reassessment of Williams’ ‘extrinsic mortality’ hypothesis. Trends Ecol. Evol. 34, 519–530 (2019).
Rose, M. R. Evolutionary Biology of Aging (Oxford Univ. Press, 1994).
Medawar, P. B. An unsolved problem of biology. Med. J. Aust. https://doi.org/10.5694/j.1326-5377.1953.tb84985.x (1952).
Williams, G. C. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11, 398–411 (1957).
Lemaître, J.-F., Moorad, J., Gaillard, J., Maklakov, A. A. & Nussey, D. H. A unified framework for evolutionary genetic and physiological theories of aging. PLoS Biol. 22, e3002513 (2024).
Bulut-Karslioglu, A. et al. Inhibition of mTOR induces a paused pluripotent state. Nature 540, 119–123 (2016).
Iyer, D. P. et al. mTOR activity paces human blastocyst stage developmental progression. Cell 187, 6566–6583 (2024).
Szathmáry, E. The origin of replicators and reproducers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1761–1776 (2006).
Piette, B. M. A. G. & Heddle, J. G. A peptide–nucleic acid replicator origin for life. Trends Ecol. Evol. 35, 397–406 (2020).
Miller, S. L. A production of amino acids under possible primitive earth conditions. Science 117, 528–529 (1953).
Miller, S. L. & Urey, H. C. Organic compound synthesis on the primitive earth. Science 130, 245–251 (1959).
Dattani, J. & Barahona, M. Stochastic models of gene transcription with upstream drives: exact solution and sample path characterization. J. R. Soc. Interface 14, 20160833 (2017).
Seeger, M., Flöttmann, M. & Klipp, E. A dynamical stochastic model of yeast translation across the cell cycle. Heliyon 9, e13101 (2023).
Leote, A. C., Lopes, F. & Beyer, A. Loss of coordination between basic cellular processes in human aging. Nat. Aging 4, 1432–1445 (2024).
Cohen, A. A., Coste, C. F. D., Li, X., Bourg, S. & Pavard, S. Are trade-offs really the key drivers of ageing and life span? Funct. Ecol. 34, 153–166 (2020).
Wensink, M. J. & Cohen, A. A. The Danaid theory of aging. Front. Cell Dev. Biol. 9, 671208 (2022).
Gems, D. & Kern, C. C. Biological constraint, evolutionary spandrels and antagonistic pleiotropy. Ageing Res. Rev. 101, 102527 (2024).
Lynch, M. Evolutionary layering and the limits to cellular perfection. Proc. Natl Acad. Sci. USA 109, 18851–18856 (2012).
Mitteldorf, J. An epigenetic clock controls aging. Biogerontology 17, 257–265 (2016).
Pamplona, R., Jové, M., Gómez, J. & Barja, G. Programmed versus non-programmed evolution of aging. What is the evidence? Exp. Gerontol. 175, 112162 (2023).
Goldsmith, T. C. Evolution of aging theories: why modern programmed aging concepts are transforming medical research. Biochemistry 81, 1406–1412 (2016).
Weismann, A. Ueber Die Dauer Des Lebens; Ein Vortrag (G. Fischer, 1882).
Libertini, G. The programmed aging paradigm: how we get old. Biochemistry 79, 1004–1016 (2014).
Lidsky, P. V. & Andino, R. Could aging evolve as a pathogen control strategy? Trends Ecol. Evol. 37, 1046–1057 (2022).
Kirkwood, T. B. L. & Melov, S. On the programmed/non-programmed nature of ageing within the life history. Curr. Biol. 21, R701–R707 (2011).
Cohen, A. A. Physiological and comparative evidence fails to confirm an adaptive role for aging in evolution. Curr. Aging Sci. 8, 14–23 (2015).
de Magalhães, J. P. Ageing as a software design flaw. Genome Biol. 24, 51 (2023).
Gems, D., Virk, R. S. & de Magalhães, J. P. Epigenetic clocks and programmatic aging. Ageing Res. Rev. 101, 102546 (2024).
de Magalhães, J. P. Programmatic features of aging originating in development: aging mechanisms beyond molecular damage? FASEB J. 26, 4821–4826 (2012).
Maklakov, A. A. & Chapman, T. Evolution of ageing as a tangle of trade-offs: energy versus function. Proc. Biol. Sci. 286, 20191604 (2019).
Horvath, S. & Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19, 371–384 (2018).
Jaenisch, R. & Bird, A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254 (2003).
Smith, Z. D., Hetzel, S. & Meissner, A. DNA methylation in mammalian development and disease. Nat. Rev. Genet. 26, 7–30 (2024).
Seisenberger, S., et al. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20110330 (2013).
Vanyushin, B. F., Nemirovsky, L. E., Klimenko, V. V., Vasiliev, V. K. & Belozersky, A. N. The 5-methylcytosine in DNA of rats. Tissue and age specificity and the changes induced by hydrocortisone and other agents. Gerontologia 19, 138–152 (1973).
Wilson, V. L. & Jones, P. A. DNA methylation decreases in aging but not in immortal cells. Science 220, 1055–1057 (1983).
Issa, J. P. et al. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat. Genet. 7, 536–540 (1994).
Rakyan, V. K. et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 20, 434–439 (2010).
Christensen, B. C., et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 5, e1000602 (2009).
Bocklandt, S., et al. Epigenetic predictor of age. PLoS ONE 6, e14821 (2011).
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14, R115 (2013).
Koch, C. M. & Wagner, W. Epigenetic-aging-signature to determine age in different tissues. Aging 3, 1018–1027 (2011).
Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367 (2013).
Levine, M. E. et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 10, 573–591 (2018).
Lu, A. T. et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11, 303–327 (2019).
Föhr, T. et al. Does the epigenetic clock GrimAge predict mortality independent of genetic influences: an 18 year follow-up study in older female twin pairs. Clin. Epigenetics 13, 128 (2021).
Moqri, M. et al. Biomarkers of aging for the identification and evaluation of longevity interventions. Cell 186, 3758–3775 (2023).
Meyer, D. H. & Schumacher, B. BiT age: a transcriptome-based aging clock near the theoretical limit of accuracy. Aging Cell 20, e13320 (2021).
Schumacher, B., Gallrein, C. & Meyer, D. Neuron-type specific aging-rate reveals age decelerating interventions preventing neurodegeneration. Preprint at Research Square https://doi.org/10.21203/rs.3.rs-4360587/v1 (2024).
Kriukov, D. et al. ComputAgeBench: epigenetic aging clocks benchmark. In Proc. 31st ACM SIGKDD Conf. on Knowledge Discovery and Data Mining V.2 https://doi.org/10.1145/3711896.3737382 (ACM, 2025).
Lu, A. T. et al. Universal DNA methylation age across mammalian tissues. Nat. Aging 3, 1144–1166 (2023).
Seale, K., Horvath, S., Teschendorff, A., Eynon, N. & Voisin, S. Making sense of the ageing methylome. Nat. Rev. Genet. 23, 585–605 (2022).
Raj, K. & Horvath, S. Current perspectives on the cellular and molecular features of epigenetic ageing. Exp. Biol. Med. 245, 1532–1542 (2020).
Blagosklonny, M. V. Aging is not programmed: genetic pseudo-program is a shadow of developmental growth. Cell Cycle 12, 3736–3742 (2013).
Meyer, D. H. & Schumacher, B. Aging clocks based on accumulating stochastic variation. Nat. Aging 4, 871–885 (2024).
Chan, J., Rubbi, L. & Pellegrini, M. DNA methylation entropy is a biomarker for aging. Aging 17, 685–698 (2025).
Tong, H. et al. Quantifying the stochastic component of epigenetic aging. Nat. Aging 4, 886–901 (2024).
Kabacik, S. et al. The relationship between epigenetic age and the hallmarks of aging in human cells. Nat. Aging 2, 484–493 (2022).
Perez, K., et al. DNA repair-deficient premature aging models display accelerated epigenetic age. Aging Cell 23, e14058 (2024).
Yang, J. H. et al. Loss of epigenetic information as a cause of mammalian aging. Cell 186, 305–326 (2023).
Moqri, M. et al. PRC2-AgeIndex as a universal biomarker of aging and rejuvenation. Nat. Commun. 15, 5956 (2024).
Laugesen, A., Højfeldt, J. W. & Helin, K. Role of the Polycomb repressive complex 2 (PRC2) in transcriptional regulation and cancer. Cold Spring Harb. Perspect. Med. 6, a026575 (2016).
Corley, M. & Kroll, K. L. The roles and regulation of Polycomb complexes in neural development. Cell Tissue Res. 359, 65–85 (2015).
Horvath, S., Zhang, J., Haghani, A., Lu, A. T. & Fei, Z. Fundamental equations linking methylation dynamics to maximum lifespan in mammals. Nat. Commun. 15, 8093 (2024).
McLain, A. T. & Faulk, C. The evolution of CpG density and lifespan in conserved primate and mammalian promoters. Aging 10, 561–572 (2018).
Mayne, B., Berry, O., Davies, C., Farley, J. & Jarman, S. A genomic predictor of lifespan in vertebrates. Sci. Rep. 9, 17866 (2019).
Jenkinson, G., Pujadas, E., Goutsias, J. & Feinberg, A. P. Potential energy landscapes identify the information-theoretic nature of the epigenome. Nat. Genet. 49, 719–729 (2017).
Koch, Z., Li, A., Evans, D. S., Cummings, S. & Ideker, T. Somatic mutation as an explanation for epigenetic aging. Nat. Aging 5, 709–719 (2025).
Sziráki, A., Tyshkovskiy, A. & Gladyshev, V. N. Global remodeling of the mouse DNA methylome during aging and in response to calorie restriction. Aging Cell 17, e12738 (2018).
Wang, T. et al. Epigenetic aging signatures in mice livers are slowed by dwarfism, calorie restriction and rapamycin treatment. Genome Biol. 18, 57 (2017).
Bertucci, E. M., Mason, M. W., Rhodes, O. E. & Parrott, B. B. The aging DNA methylome reveals environment-by-aging interactions in a model teleost. Preprint at bioRxiv https://doi.org/10.1101/2021.03.01.433371 (2021).
Koike, Y., et al. Age-related demethylation of the TDP-43 autoregulatory region in the human motor cortex. Commun. Biol. 4, 1107 (2021).
Meyer, D. H. & Schumacher, B. The stochasticity of biological aging. Nat. Med. 31, 1735–1736 (2025).
Rieckher, M., Garinis, G. A. & Schumacher, B. Molecular pathology of rare progeroid diseases. Trends Mol. Med. 27, 907–922 (2021).
Panier, S., Wang, S. & Schumacher, B. Genome instability and DNA repair in somatic and reproductive aging. Annu. Rev. Pathol. 19, 261–290 (2024).
Milholland, B. et al. Differences between germline and somatic mutation rates in humans and mice. Nat. Commun. 8, 15183 (2017).
Bujarrabal-Dueso, A. et al. The DREAM complex functions as conserved master regulator of somatic DNA-repair capacities. Nat. Struct. Mol. Biol. 30, 475–488 (2023).
Bujarrabal-Dueso, A. Targeting DNA damage in ageing: towards supercharging DNA repair. Nat. Rev. Drug Discov. https://doi.org/10.1038/s41573-025-01212-6 (2025).
de Lima Camillo, L. P., Lapierre, L. R. & Singh, R. A pan-tissue DNA-methylation epigenetic clock based on deep learning. NPJ Aging 8, 4 (2022).
Wang, S., Meyer, D. H. & Schumacher, B. Inheritance of paternal DNA damage by histone-mediated repair restriction. Nature 613, 365–374 (2023).
Gladyshev, V. N. The ground zero of organismal life and aging. Trends Mol. Med. 27, 11–19 (2021).
Totska, K., Barata, J. C. V. V., Sandt, W., Meyer, D. H. & Schumacher, B. Age deceleration and reversal gene patterns in dauer diapause. Preprint at bioRxiv https://doi.org/10.1101/2025.04.14.648662 (2025).
Kawamura, K. et al. Resilience and restoration from fasting–refeeding mediated by a nutrient-regulated linker histone. Preprint at bioRxiv https://doi.org/10.1101/2025.04.14.648802 (2025).
Benjamin, H. Biologic versus chronologic age. J. Gerontol. 2, 217–227 (1947).
Hollingsworth, J. W., Ishii, G. & Conard, R. A. Skin aging and hair graying in Hiroshima. Geriatrics 16, 27–36 (1961).
Hollingsworth, J. W., Hashizume, A. & Jablon, S. Correlations between tests of aging in Hiroshima subjects—an attempt to define ‘physiologic age’. Yale J. Biol. Med. 38, 11–26 (1965).
Hollingsworth, J. W., Hamilton, H. B. & Ishii, G. Age-related changes in erythrocyte agglutinability in Hiroshima subjects. J. Appl. Physiol. 16, 1093–1096 (1961).
Webster, I. W. & Logie, A. R. A relationship between functional age and health status in female subjects. J. Gerontol. 31, 546–550 (1976).
Furukawa, T., Inoue, M., Kajiya, F., Inada, H. & Takasugi, S. Assessment of biological age by multiple regression analysis. J. Gerontol. 30, 422–434 (1975).
Costa, P. T. & McCrae, R. R. Measures and markers of biological aging: ‘a great clamoring… of fleeting significance’. Arch. Gerontol. Geriatr. 7, 211–214 (1988).
Costa, P. T. & McCrae, R. R. in Principles of Geriatric Medicine 30–37 (McGraw-Hill, 1985).
Wilson, D. L. Aging hypotheses, aging markers and the concept of biological age. Exp. Gerontol. 23, 435–438 (1988).
Salthouse, T. A. in Age, Health, and Employment 78–92 (Prentice-Hall, 1986).
Hochschild, R. Improving the precision of biological age determinations. Part 1: a new approach to calculating biological age. Exp. Gerontol. 24, 289–300 (1989).
Zhang, Q. et al. Improved precision of epigenetic clock estimates across tissues and its implication for biological ageing. Genome Med. 11, 54 (2019).
Sluiskes, M. H., et al. Clarifying the biological and statistical assumptions of cross-sectional biological age predictors: an elaborate illustration using synthetic and real data. BMC Med. Res. Methodol. 24, 58 (2024).
Belsky, D. W., et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. eLife 11, e73420 (2022).
Kriukov, D., Kuzmina, E., Efimov, E., Dylov, D. V. & Khrameeva, E. E. Epistemic uncertainty challenges aging clock reliability in predicting rejuvenation effects. Aging Cell 23, e14283 (2024).
Higgins-Chen, A. T. et al. A computational solution for bolstering reliability of epigenetic clocks: implications for clinical trials and longitudinal tracking. Nat. Aging 2, 644–661 (2022).
Ying, K. et al. Causality-enriched epigenetic age uncouples damage and adaptation. Nat. Aging 4, 231–246 (2024).
Schrödinger, E. What Is Life? The Physical Aspect of the Living Cell & Mind and Matter (Cambridge Univ. Press, 1974).
Clausius, R. Ueber verschiedene für die Anwendung bequeme Formen der Hauptgleichungen der mechanischen Wärmetheorie. Ann. Phys. 201, 353–400 (1865).
Ben-Naim, A. Entropy and time. Entropy 22, 430 (2020).
Bortz, W. M. 2nd. Aging as entropy. Exp. Gerontol. 21, 321–328 (1986).
Sacher, G. The complementarity of entropy terms for the temperature dependence of development and aging. Ann. N. Y. Acad. Sci. 138, 680–712 (1967).
Nikopoulou, C., Parekh, S. & Tessarz, P. Ageing and sources of transcriptional heterogeneity. Biol. Chem. 400, 867–878 (2019).
Cooney, C. A. Are somatic cells inherently deficient in methylation metabolism? A proposed mechanism for DNA methylation loss, senescence and aging. Growth Dev. Aging 57, 261–273 (1993).
Bronner, C., Alhosin, M., Hamiche, A. & Mousli, M. Coordinated dialogue between UHRF1 and DNMT1 to ensure faithful inheritance of methylated DNA patterns. Genes 10, 65 (2019).
Zhou, W. et al. DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat. Genet. 50, 591–602 (2018).
Takahashi, S. et al. Embryonic genome instability upon DNA replication timing program emergence. Nature 633, 686–694 (2024).
Scherer, M., et al. Quantitative comparison of within-sample heterogeneity scores for DNA methylation data. Nucleic Acids Res. 48, e46 (2020).
Loyfer, N. et al. A DNA methylation atlas of normal human cell types. Nature 613, 355–364 (2023).
Meaburn, E. L., Schalkwyk, L. C. & Mill, J. Allele-specific methylation in the human genome: implications for genetic studies of complex disease. Epigenetics 5, 578–582 (2010).
Landau, D. A. et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell 26, 813–825 (2014).
Landan, G. et al. Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nat. Genet. 44, 1207–1214 (2012).
Xie, H. et al. Genome-wide quantitative assessment of variation in DNA methylation patterns. Nucleic Acids Res. 39, 4099–4108 (2011).
Bertucci-Richter, E. M., Shealy, E. P. & Parrott, B. B. Epigenetic drift underlies epigenetic clock signals, but displays distinct responses to lifespan interventions, development, and cellular dedifferentiation. Aging 16, 1002–1020 (2024).
Zhang, X. & Wang, X. MeConcord: a new metric to quantitatively characterize DNA methylation heterogeneity across reads and CpG sites. Bioinformatics 38, i307–i315 (2022).
Acknowledgements
B.S. acknowledges funding from the Deutsche Forschungsgemeinschaft (Reinhart Koselleck-Project 524088035, FOR 5504 project 496650118, FOR 5762 project 531902955, SFB 1678, SFB 1607, CECAD EXC 2030-390661388, ANR-DFG project 545378328, the DFG-ISF project 561031107 and DFG project grants 558166204, 540136447, 496914708, 437825591, 437407415 and 418036758), the Deutsche Krebshilfe (70114555), Deutsche José Carreras Leukämie-Stiftung (DJCLS 04 R/2023), a John Templeton Foundation Grant (61734), the European Research Council (ERC-2023-SyG 101118919) and the Hevolution Foundation (HF-GRO-23-1199212-35). D.H.M. acknowledges funding from the Deutsche Forschungsgemeinschaft (DFG project grant 570621149) and from the Koeln Fortune Program / Faculty of Medicine, University of Cologne. A.A.M. acknowledges funding from the Leverhulme Trust RPG-2023-068 and the Natural Environment Research Council (NE/W001020/1).
Author information
Authors and Affiliations
Contributions
Conceptualization and writing: D.H.M., A.A.M. and B.S.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Aging thanks Steven Cummings, Vera Gorbunova, and Alan Cohen for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Meyer, D.H., Maklakov, A.A. & Schumacher, B. Aging by the clock and yet without a program. Nat Aging 5, 1946–1956 (2025). https://doi.org/10.1038/s43587-025-00975-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43587-025-00975-2
This article is cited by
-
The regeneration model of aging and its practical implications
Discover Medicine (2026)
-
A sex-adjusted 7-biomarker clinical aging clock for translational preventative medicine
Scientific Reports (2025)