Abstract
The growing number of health-data breaches, the use of genomic databases for law enforcement purposes and the lack of transparency of personal genomics companies are raising unprecedented privacy concerns. To enable a secure exploration of genomic datasets with controlled and transparent data access, we propose a citizen-centric approach that combines cryptographic privacy-preserving technologies, such as homomorphic encryption and secure multi-party computation, with the auditability of blockchains. Our open-source implementation supports queries on the encrypted genomic data of hundreds of thousands of individuals, with minimal overhead. We show that real-world adoption of our system alleviates widespread privacy concerns and encourages data access sharing with researchers.
Similar content being viewed by others
Main
The decreasing costs of DNA analyses and the promise of the value of large genomic datasets have ushered us into the era of population-scale genomics. This development is driven by companies as well as governments. The direct-to-consumer personal genomics market has grown dramatically over the past few years1 and is expected to triple by the end of 2025. Similarly, many countries around the globe are pursuing large-scale population genomics initiatives that aim at collecting massive amounts of genomic data from their citizens.
However, unresolved privacy issues make data sharing on a large scale extremely difficult and time-consuming. Individuals are particularly sensitive and concerned about genomic-data privacy, mainly because of the real or perceived risks of genetic discrimination that deter them from using direct-to-consumer genetic testing services2,3.
We surveyed 442 individuals interested in genetic testing and found that data privacy is one of the top two disincentives, together with cost (Fig. 1a). In particular, individuals want to have control over the use of their genomic data and to be able to share access to their data with researchers, without risking misuse (Fig. 1b). These are guarantees that population-genomics initiatives and personal-genomics companies are not able to provide today.
a, The factors that deter individuals from genetic testing. b, The importance of different security features to survey participants.
In the last few years, several researchers from the information security community have proposed solutions that try to meet various privacy guarantees. Some focus on providing secure storage for genomic data4, whereas others propose methods for how to securely carry out specific computations5,6. Most of these solutions have not yet been adopted in practice, mainly because of their academic demonstrative nature, limited application scope and lack of validation on real use cases. Only a few solutions have reached the level of maturity that is necessary to be used in operational settings7,8,9. None of them consider data privacy from an individual’s perspective. However, the issue of trust goes beyond the provision of secure storage and processing, as it is also intimately related to the question of transparency and control of data sharing and use. These systems mostly address hospitals and research institutions’ privacy and security concerns and do not consider individual citizens as data providers. In particular, they do not address the access control, consent management or accountability requirements sought by individuals.
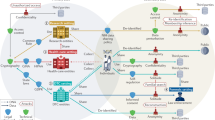
Guided by the results of our survey, our proposed solution describes a practical system for privacy-preserving personal genomics centered on individuals’ privacy requirements and their demand for more control and transparency over their data and its use. This work implements and extends the ideas, which we recently outlined10, by providing a concrete and tangible instantiation that efficiently integrates several privacy-preserving technologies, including homomorphic encryption, equality-preserving encryption, differential privacy and blockchain technology (Methods) into an end-to-end system that enables secure and auditable genomic data discovery and analysis (Fig. 2).
Individual data providers are shown on the left and data queriers (for example, researchers) on the right. In the middle are computing nodes that perform privacy-preserving computations by executing encrypted queries on encrypted data. The computing nodes also maintain a blockchain that stores data-access-sharing policies.
The proposed system is designed such that it empowers individual data providers to control the sharing of access to their personal data in a controlled, auditable and secure manner. The system operates in two phases. In the first, the data-preparation phase, a set of independent, yet collaborative, computing nodes generate one public–private cryptographic key pair each and combine their public keys into a single collective public encryption key for an additively homomorphic encryption scheme. The same computing nodes also set up a permissioned blockchain that can be updated based on a ‘majority-voting’ consensus protocol. When individuals (or data providers) want to upload their data to the system, they use the collective encryption key to individually encrypt their genetic variants and clinical attributes. They also generate, encrypt and upload dummy data together with binary flags that distinguish it from real data. The uploaded data is then re-encrypted by the computing nodes from the homomorphic to equality-preserving encryption scheme to enable the execution of equality-matching queries and then moved to a storage unit (for example, a cloud provider), while the flags remain homomorphically encrypted. At the same time, the data providers write their data-access policy, which specifies who can query and access their data, to the system’s blockchain, where it is time-stamped and stored immutably.
Once the data is on the platform, the second phase, the data-discovery/access phase, can take place. In this phase, the researcher can perform one or more of the following operations, depending on the data-access policies attached to the data. Researchers can discover cohorts by running combinatorial queries, composed of clinical and genetic inclusion/exclusion criteria, on the uploaded encrypted data to find individuals whose data match the search criteria. Furthermore, researchers can perform aggregate-data analyses on the encrypted data. For example, a researcher can ask the system to securely compute descriptive statistics for a cohort of individuals, such as the size of the cohort itself, the distribution of the individuals in the cohort for any combination of clinical and other attributes (for example, age, gender, treatment, geographical zone, phenotype and genotype), the frequency of a given genetic variant for different subgroups or the co-occurrence of multiple genetic variants. It is also possible to securely compute 2-by-2 contingency tables for the chi-squared association tests that are commonly used in genome-wide association studies (GWASs). Finally, researchers who need to conduct data analysis beyond what can be provided by the secure aggregate-data analysis functionality, can access cleartext individual-level data in a transparent and auditable fashion that ensures accountability. This is enabled by the use of a blockchain, which functions as an immutable ledger that logs data-access requests and permissions. This phase of the protocol is particularly important in relation to the citizen-centric aspect of the system because it enables the dynamic consent management and transparent data-access control that is crucial to alleviate the prevalent privacy concerns and build trust. The details of the protocols are described in the Methods and Supplementary Figs. 10–18.
The security of our platform is based on the cryptographic guarantees provided by the underlying decentralized protocols. Our threat model assumes honest-but-curious data storage units and computing nodes and potentially malicious data queriers. All input-sensitive variables are encrypted under a collectively maintained key, such that they cannot be decrypted without the cooperation of all computing nodes, thus guaranteeing confidentiality and avoiding single points of failure. Paired with the dummy data strategy, our platform protects the confidentiality of query results, as only the authorized data queriers can decrypt the query results thanks to a distributed key-switching protocol (Methods). Conversely, to avoid re-identification (or attribute disclosure) attacks, our platform also enables the application of differentially private noise to the results and, due to the proposed dummy strategy, it also guarantees the confidentiality of the data against potentially compromised storage units and participating computing nodes. Nevertheless, it is also possible to cope with malicious data storage units and computing nodes by using protocols that produce and publish zero-knowledge proofs for all the computations performed by the computing nodes11; the proofs can thus be verified by any entity to assess that no party has deviated from the correct behavior. This solution yields a hardened and resilient query protocol, but the cost of producing those proofs results in higher and still impractical computation costs.
To demonstrate the practicality of our privacy-preserving approach for population genomics, we implemented an operational prototype of the described system and benchmarked it on a simulated dataset of 150,000 individual records derived from The Cancer Genome Atlas (TCGA) dataset that has a total of 28 billion genetic variants (more details on the dataset are provided in the Methods). The performance results in Fig. 3 show that a researcher can discover individuals from such a dataset in a few minutes, with queries involving genetic and clinical attributes over the encrypted space; furthermore, the query response time grows linearly with the number of individuals and query items (clinical and genetic criteria used to build the query), whereas the overhead introduced by the encryption with respect to the cleartext is negligible (a few seconds), linear in the size of the matching set, and virtually constant for the database size or the number of query items. Even large queries that consist of hundreds of items are executed in just a few minutes with an average overhead of only 1% to 3% compared to querying cleartext data. For the computation of GWAS subqueries, the system enables an amortized response time of ~21 ms per variable and matching individual, where the overhead of the encrypted computations is kept below 6% (Supplementary Fig. 5). The response time can be further reduced by running the subqueries in parallel. Benchmark results for the query–response time breakdown (Supplementary Fig. 4) show that the database processing time (unaffected by the encryption) is the largest component. The efficiency of the raw-data-access functionality is affected by the blockchain performance, but is primarily determined by the response time of individuals to raw-data-access requests. The delay due to the writing of access requests and permissions is negligible, as the permissioned blockchain that is part of our system achieves a throughput of thousands of transactions per second (Supplementary Fig. 6). Finally, the preparation phase is relatively time-consuming (Supplementary Figs. 1–3), but it must be executed only once for every new data provider who uploads data.
a–d, Performance with a dataset of 150,000 individuals, each with 15,000 to 200,000 genetic variants, when varying the number of database and computing nodes (database size of 28 billion genetic variants and a query with 10 items) (a), varying the size of the database (three nodes and a query with 10 items) (b), varying the number of query items (three nodes and a database of 28 billion genetic variants) (c) and varying the result size (three nodes, a database of 28 billion genetic variants and a query with 10 items) (d). The difference between the protected encrypted data and cleartext data is shown in parentheses for each data point (in seconds).
The next step is a real-world deployment of our system. We are collaborating with a personal genome sequencing company that is making the technologies presented here available to their customers (Supplementary Fig. 8). We surveyed individuals who already used the blockchain component of the system and found that the offered data-privacy protections helped convince many of them to undergo genetic testing. Additionally, we found that the use of these technologies made many individuals more inclined to participate in research in the future (Supplementary Fig. 7). Separately, the privacy-preserving computing component of the platform is currently being evaluated by hospitals12 (Supplementary Fig. 9). We are now working towards an integrated deployment of both components. We expect that a fully integrated real-world deployment will reveal certain opportunities as well as challenges. At this point, we can only hypothesize about these. For example, a citizen-centric system might facilitate compliance with different privacy laws because it maximizes an individual’s control over personal genomic data. However, a possible drawback of this approach might be the limited reproducibility of studies when researchers do not have uniform access permissions. Finally, open questions remain about governance policies of the platform with respect to questions such as query budgets assigned to researchers and identity verification. We hope to gain insight into these practical challenges in future work.
Methods
Core cryptographic techniques
Our system relies on the combination of multiple cryptographic techniques. We briefly describe them here, while more detailed technical descriptions are provided in the Supplementary Information.
Collective data encryption
Data are secured by distributing trust between multiple independent parties (for example, different organizations) that operate computing nodes. Each node generates a public–private cryptographic key pair. The individual public keys are combined to generate a collective encryption key that is used to encrypt genetic variants and clinical attributes as well as investigator queries. During the execution of queries, the nodes collectively perform computations on the encrypted data and queries.
Homomorphic encryption
Homomorphic encryption enables computations on encrypted data, without having to decrypt it. For example, individuals who are affected by a rare disease are labeled by a 1 and those who are not affected are labeled by a 0. If these numbers are encrypted with homomorphic encryption, they will look like indistinguishable pseudorandom numbers, and it will be unfeasible to tell individuals apart without the decryption key. However, if the encrypted numbers are summed together, they will produce a result that, when decrypted, corresponds to the number of individuals affected by the rare disease.
Equality-preserving encryption
Equality-preserving encryption guarantees that numbers encrypted with the same key always generate the same ciphertext. This type of encryption enables equality-matching operations on encrypted attributes, without revealing them. However, if it is not used appropriately, equality-preserving encryption is vulnerable to frequency attacks due to its equality-preserving property. For example, if the encrypted attributes are ‘yes’ or ‘no’, indicating the presence or absence of a rare disease, then it can be inferred that the encrypted attribute that appears less often is ‘yes’.
Dummy-data generation
To avoid frequency attacks on data encrypted with an equality-preserving encryption scheme, the system introduces dummy individuals with data generated such that the frequency differences are evened out (uniform distribution). To prevent dummy individuals from affecting the query result yet still be indistinguishable from the real individuals, we label them with binary flags, which are probabilistically encrypted under the original additively homomorphic encryption scheme. We assign a 0 (‘false’) to dummy individuals and a 1 (‘true’) to the real ones.
Differential privacy
To prevent inferences stemming from released query results, the computing nodes can jointly obfuscate such results by adding noise sampled from a given distribution so as to satisfy the notion of differential privacy. Differential privacy guarantees that it is not possible to use the query results to infer whether the data of any given individual were used in the computation. Moreover, it introduces the notion of a privacy budget that prevents investigators from running repeated queries that aim at reconstructing the attributes of individuals. For example, the result of a query that returns the number of individuals with certain genetic variants is obfuscated with some noise, and it introduces a cost in terms of privacy budget. An investigator will be able to run queries until their budget is depleted.
Blockchain recording
Every activity in the platform, such as data upload, query execution, data-access request and consent to share data, generates a transaction on a blockchain that is maintained by the computing nodes. A blockchain is an immutable, collectively maintained database. New entries can be added to the blockchain only if they have been approved by a majority of the nodes that maintain the blockchain. Entries are bundled into time-stamped blocks and each block references its preceding block, which creates a sequential ordering that prevents malicious deletion or reordering of data stored in the chain. This enables tamper-proof, auditable record keeping.
System and threat models
We model the proposed system with the following four parties: (1) data providers, that is, individual citizens who use the platform to securely expose their clinical and genetic data for population genomic studies, (2) data queriers (for example, researchers), who use the platform to find individuals with interesting clinical and genetic characteristics so as to recruit them in clinical research studies or pharmaceutical trials, (3) storage units, which are responsible for securely storing the clinical and genomic data of data providers (data providers can choose any of the available storage units for storing their data) and (4) computing nodes, which are a set of independent governmental, academic or commercial institutions hosting one or several servers that are responsible for jointly and securely processing data discovery requests from data queriers.
According to the reported definitions of possible adversarial behaviors, we assume that storage units and computing nodes are honest-but-curious, non-colluding parties. This means that they honestly follow the protocol, but might try to infer sensitive information about data providers and data queriers from information processed or stored at their premises during the protocol. We assume that data providers are honest; that is, they only expose their real and accurate clinical and genomic data without trying to fool the system with fake information. Finally, we consider data queriers as potentially misbehaving parties, as their credentials can be stolen by malicious attackers who can use the system in an unethical way in order to obtain sensitive information about data providers.
Data encoding
The proposed system enables data providers to securely share their clinical and genomic data.
Genomic data encoding
We consider genomic variants regardless of the format. A variant is uniquely identified by its chromosomal position (CHR, POS), the reference allele (REF) and the alternate allele (ALT), which we refer to as variant metadata. In general, the variant metadata is public and considered to be non-sensitive. What is sensitive and must be protected is the association of variant metadata with an individual genotype, that is, the value that the variant takes for a given individual. In particular, we consider that an individual variant genotype is sensitive when it is ‘mutated’, that is, if at least one of the two alleles composing the genotype has changed from the reference to the alternate allele. Therefore, a variant genotype can be encoded as strings of the form ‘CHR:POS:REF>ALT’ which means that, at the given chromosomal position (CHR:POS), the individual carries at least one allele that has mutated from the reference to the alternate (REF>ALT). To encrypt such a string, we transform it into a 64-bit integer with the following convention: 1 bit indicating that the code encodes a genetic variant, 5 bits for the chromosome identifier, 28 bits for the position, 15 bits for the reference allele and 15 bits for the alternate allele.
Clinical data encoding
We consider self-reported clinical data. This can include structured information on diagnoses, medications, procedures, simple findings and demographics. We refer to each piece of clinical information as a ‘clinical attribute’. In general, clinical attributes can be encoded by using standard medical ontologies and terminologies such as ICD10 and SNOMED-CT. As for the genomic encoding, we convert alphanumeric concept codes into 64-bit integers.
Privacy-preserving and secure data-sharing protocol
The privacy-preserving and secure data-sharing protocol enabled by our platform consists of four phases: the platform initialization phase (performed only once), a data-preparation phase that is performed by each data provider each time clinical and genetic variables are uploaded to the platform, and a data-discovery phase and a data-access phase that are performed each time a researcher wants to run data-discovery queries or request access to individual-level records.
Platform initialization
During the initialization phase (Supplementary Fig. 12), each computing node generates a pair of public/private keys for the EC-ElGamal additively homomorphic encryption scheme along with a secret key. Then, all computing nodes combine their public keys to generate a single public key that is used by data providers to encrypt their data before uploading them to the platform. We denote this key the ‘collective encryption key’. This ‘joint’ key-generation technique ensures that data encrypted under the collective key are protected unless all computing nodes are compromised and their individual private keys are stolen. The more computing nodes participate in the generation of the collective key, the higher is the overall security of the system. Finally, along with the key generation, the computing nodes set up a private permissioned blockchain to be used as an auditable and immutable log for the operations executed on the platform by the different parties.
Data preparation
The data-preparation phase consists of four steps, also represented by the sequence diagram in Supplementary Fig. 13.
Generation of dummy data
The data preparation phase starts with a dummy data generation algorithm executed by data providers. A dummy data-generation algorithm takes as input the set of clinical attributes and genetic variants and outputs a set of dummy individuals with a plausible set of observations specifically selected to flatten the global joint distribution of observations. To prevent the fake observations from affecting the data-discovery process, a binary flag is assigned to each individual and appended to the corresponding set of observations. Such a flag is set to 1 for real individuals and 0 for dummy individuals. The purpose of dummy data generation is to protect against frequency attacks. An attacker with knowledge of the population statistics would be able to map the histogram of the encrypted codes to the known frequencies of occurrence of each code, hence breaking the encryption. The goal is therefore to maximize the confusion of an attacker with access to the encrypted real and dummy observations trying to map encrypted observation codes with cleartext observation codes. This can be achieved by flattening the empirical joint distribution of the observed codes, by producing dummies that ‘waterfill’ their distribution. To guarantee that the dummy individuals are indistinguishable from real ones, the former is generated with a set of codes that is a random permutation of the codes present in each real individual. This strategy minimizes the number of needed dummies while maximizing the attacker’s confusion. When the whole database is known in advance by the data provider, it is possible to exactly adopt the empirical distribution to make it perfectly flat per cluster. Contrarily, in our system model, each data provider is just one individual submitting one set of codes, without knowing the exact histogram of the other codes in the database. Hence, as the exact empirical distribution is not available, the system instead uses the publicly available frequency/prevalence for each of the available codes and applies the clustering based on this estimated distribution. This target distribution is published and communicated to each data provider. Its computation can also be published on the blockchain for auditability. When a data provider wants to submit an observed set of codes to the system, they produce a fixed number of dummies as random permutations of the codes. These permuted records approximately follow the ‘reversed’ population distribution that will guarantee that the union of the dummy and real individuals will converge to a flat distribution. The dummy ratio (number of dummies per real individual) has to be fixed for all data providers; otherwise, it would leak information on the likelihood of the real observations. This ratio can be adjusted as the weighting term between the real and the dummy distribution to make them sum up to a flat uniform distribution. As a completely flat target uniform distribution would result in an extremely high ratio, we group the observations in clusters of similar (published) observable frequency, and flatten the distribution per cluster, guaranteeing enough entropy within the cluster contents (the number of observation codes in each cluster is lower bounded by a system parameter). In our test databases (data from the TCGA database; see the final paragraph in Supplementary Section 3.1), optimal clustering strategies lead to a dummy ratio similar to the one obtained when the whole database is known in advance.
Data encryption and upload
After dummy data generation, the data provider generates a symmetric encryption key and uses it to encrypt the original files containing the genetic variants and self-reported clinical attributes. The data provider then uses the collective encryption key to encrypt the symmetric encryption key, the codes of their observations and those from their dummies, and the set of binary flags. The data upload generates a blockchain transaction that is verified by all computing nodes and immutably stored on the blockchain. The transaction contains the data-access policy and the mapping between the data provider’s pseudo-identity and the set of pointers to the uploaded encrypted files storing the genetic and clinical variables.
Data re-encryption
The computing node receiving the data initiates a distributed re-encryption protocol on the encrypted observation codes. This protocol switches the encryption of the codes from homomorphic to equality-preserving, thus enabling the execution of equality-matching queries on the re-encrypted codes. Homomorphically encrypted flags and symmetrically encrypted files are not involved in this protocol and are sent for storage to the storage unit. The use of equality-preserving encryption reveals the distribution of the encrypted observation codes. However, thanks to the presence of the previously generated dummy individuals and their encrypted observation codes, it is impossible for the adversary to perform frequency attacks—the overall distribution of encrypted codes is uniform, and codes are indistinguishable from each other. This protection is guaranteed as long as the binary flags remain homomorphically encrypted and dummy individuals cannot be told apart from real individuals. The distributed re-encryption protocol is explained in detail in Supplementary Fig. 17.
Data storage
Once the re-encryption of clinical and genetic variables is completed, the computing node that initiated the protocol sends them to the storage unit for storage under the star-schema data model. Supplementary Fig. 10 and Fig. 11 show a toy example with encrypted data and the addition of a dummy individual necessary to flatten the distribution of encrypted codes.
Data discovery
Depending on the data-access policy specified by the data providers and the rights of the researcher, this phase can comprise operations of data discovery, aggregate-data analysis and raw-data access. These three operations are described in the following sections.
The cohort discovery operation enables a researcher to run combinatorial Boolean queries, made of clinical and genetic inclusion/exclusion criteria, on the uploaded, encrypted data so as to find individuals in the system who match the search criteria. This operation consists of a secure protocol made of three steps, represented by the sequence diagram in Supplementary Fig. 14.
Query generation
First, the data querier generates their own pair of public/private keys and builds a query by logically combining (that is, through Boolean AND and OR operators) clinical and genomic codes from the list of all possible codes present in the system. The selected codes are then encrypted with the collective encryption key and sent along with the data querier’s public key to one of the computing nodes. The query upload generates a blockchain transaction that gets verified by all computing nodes of the platform and is immutably stored on the blockchain.
Query re-encryption
To match the codes of the observations uploaded by the data providers, the codes used in the query are also re-encrypted from the initial homomorphic scheme to the equality-preserving scheme. To this end, the computing node receiving the query initiates the same distributed re-encryption algorithm as during the data-preparation phase. Once the protocol is over, it forwards the query with the re-encrypted codes to the storage unit.
Query execution
The storage unit executes the query relying on the equality-preserving properties of the encrypted codes in the query and in the database. The identifiers and the encrypted binary flags for the individuals who have encrypted observation codes that match the ones in the query are retrieved and returned to the computing nodes that initiated the request.
Next, the researcher can either ask to obtain the result of an aggregate-data analysis run over the identified cohort or to directly access the individual-level raw information. The type of data that the researcher can obtain depends on the most conservative individual privacy policy stored on the blockchain for the individuals in the identified cohort. Individuals can grant access to their data either in an aggregate-level (and potentially obfuscated) form only or also in the raw individual-level form.
Aggregate-data analysis
The aggregate-data analysis operation enables the researcher to perform some privacy-preserving computations on an identified cohort without having to access the unencrypted raw individual-level data. In particular, owing to the additively homomorphic property of the encryption scheme used to encrypt the dummy/real binary flags, the system enables the computation of any count-based statistics. The basic operation for computing such statistics is represented by a secure count protocol that consists of four steps (described in the following subsections and represented by the sequence diagram in Supplementary Fig. 15).
Secure count computation
The size of a cohort can be computed by homomorphically summing the encrypted flags. Note that, in this case, the dummy individuals fetched by the query are automatically filtered out from the result as their encrypted binary flags sum to the encryption of zero.
Secure result obfuscation
Upon registration to the platform, a data querier is assigned a query budget (εq) that is stored on the blockchain and consumed each time he sends a new query to the system. The goal of this query budget is to limit the total number of queries that can be sent by the same data querier and, as a consequence, the amount of undisclosed sensitive information about data providers that could potentially be inferred from query results. Depending on the authorization level of the data querier, computing nodes can run a distributed obfuscation protocol so that the released counts cannot be used to infer further sensitive information about data providers. This protocol enables a set of computing nodes to collectively and homomorphically add to the query result a noise value, sampled from a probability distribution that satisfies the differential privacy requirements. The protocol operates in two phases: an initialization phase executed during the platform set-up and a runtime phase executed for each new query. In the initialization phase, a randomly selected computing node generates a probability distribution curve (for example, Laplacian distribution or Gaussian distribution) based on the publicly defined differential privacy parameter (ε). The same computing node samples a list of integer noise values that approximate such a distribution and stores the list along with the distribution parameters on the system blockchain. In this way, every party involved in the protocol can verify the correctness of the sampling procedure. In the runtime phase, the computing node that receives the encrypted query results from the storage unit fetches the list of sampled noise values from the blockchain, encrypts them with the collective public key, and starts a distributed secure shuffling sub-protocol on this list. Once the distributed secure shuffling is over, the same computing node that initiated the sub-protocol selects the first element in the list and homomorphically adds it to the encrypted query result, thus obtaining a differentially private obfuscated query result.
Key switching
For a data querier to decrypt the query result, its encryption needs to be switched from the collective public key to the data querier’s public key. Computing nodes jointly run a distributed key-switching protocol that enables them to perform this operation without ever decrypting the data. The key-switching distributed protocol is described in detail in Supplementary Fig. 18.
Result decryption
The data querier uses their private key to decrypt the query results.
Raw-data access
The raw-data access operation enables the data querier to access the raw individual-level data necessary to perform analyses that go beyond the count-based statistics provided by the aggregate-data analysis operation. This relies on a protocol that consists of eight steps, represented by the sequence diagram in Supplementary Fig. 16.
Masking of identifiers
The raw-data-access protocol starts after the cohort discovery operation, with the data querier who wants to obtain the identifiers of the discovered individuals. The identifiers are returned after having been masked by a homomorphic scalar multiplication with the corresponding encrypted flag. The masking is necessary for concealing the identifiers of the dummy individuals from the data querier, in such a way that each of them will decrypt to zero. The query execution generates a blockchain transaction that contains the query definition and the identifiers of the selected individuals, and it is immutably stored on the blockchain after verification.
Key switching
After verifying the data-access policy, the masked identifiers are switched from the collective public key to the querier’s public key. The process is the same as the key-switching at the end of the aggregate-data analysis operation.
Decryption of individual identifiers
The querier decrypts the identifiers with their private key and separates those of real individuals from those of dummy individuals, which are null.
Data-access request
With the identifiers, the data querier requests access to raw-data files containing clinical or genomic information (for example, a VCF file). The data-access request generates a transaction on the blockchain that contains the cryptographic hash-based signature of the file and information about the data querier such as their name, affiliation and the description of the conducted study requiring access to individual-level data.
Access policy verification
When data providers register on the system, they write an access policy that is stored on the blockchain (Supplementary Fig. 8). Two basic options are available: broad consent and dynamic consent. Raw files’ hash signatures and access policies are linked to a data provider identity (their public key), which is used to generate transactions for registration and data upload. Thus, by reading the blockchain, computing nodes can link data providers’ files to their correspondent access policies. If broad consent is verified, the computing nodes proceed to make the data accessible. If a dynamic consent policy was chosen, the system front-end used by the data provider generates a notification when it synchronizes with the blockchain state. The data provider can review the information that is included in the data-access request and then approve or reject the request by generating another transaction that is verified by the computing nodes.
Key switching
After access policy verification, the encryption of the symmetric key used to encrypt the raw files is switched from the collective public key to the querier’s public key. The process is the same as the key-switching at the end of the data-discovery phase.
Symmetric key decryption
The data querier decrypts the symmetric key with their private key.
File download and decryption
Finally, the data querier uses the decrypted symmetric key to decrypt the raw files downloaded from the storage unit.
System implementation
We implemented the proposed system by combining different components of several open-source and widespread software platforms. For the core back-end components (database and user management) we used the state-of-the-art open-source tool for clinical data discovery, Informatics for Integrating Biology and the Bedside (i2b2). For the secure privacy-preserving component, we relied on the MedCo open-source framework, which itself relies on the distributed cryptographic library UnLynx. We extended the MedCo secure protocols by implementing, from scratch, the new dummy data-generation algorithm, the retrieval and masking of individuals’ identifiers matching the query, and the distributed obfuscation protocols. We integrated all these updates directly into the MedCo codebase. For the blockchain part of our system, we implemented our own permissioned blockchain by using the open-source Exonum framework. The Exonum framework enables the implementation of high-performance, permissioned blockchains that offer transparency and security comparable to one of the public (permissionless) blockchains.
The full prototype implementation of our platform and the data used for the experiments are publicly available on GitHub for reproducibility purposes. We note that the open-source components that we used in our implementation have been chosen for their convenience and proven reliability. However, we emphasize that our approach is tool-agnostic and that each component can be replaced by better and more efficient implementations if available.
Experimental set-up
The initial testing environment comprises three computing nodes interconnected by 10-Gbps links. The nodes were running servers that were using two Intel Xeon E5-2680 v3 CPUs @2.5 GHz that support 24 threads on 12 cores, and 256 GB of RAM. We used these servers for running the core back-end components, the secure privacy-preserving component and for storing the encrypted genetic variants and clinical attributes in a PostgreSQL database.
To test the scalability and parallelization of our system, we increased the number of computing nodes from 3 to 12. To set up our system and facilitate its deployment, we used Docker.
The blockchain was benchmarked using up to 16 computing nodes running on machines each with two Core Intel Xeon Platinum CPUs @3.4 GHz and 3.75 GB of RAM. The blockchain database was stored on a connected Elastic Block Store (EBS) drive. The nodes were distributed among 14 different locations around the globe to realistically simulate network lag.
Surveys
The survey of citizen concerns in relation to genetic testing and genetic data sharing (Fig. 1) was conducted by sending an email to 1,991 individuals on a mailing list of Nebula Genomics, a company offering personal genomics services to consumers. The recipients were requested to share their views on personal genomics by answering two questions. The first question was a multiple-choice, multi-select question that enquired about factors that deterred the participants from genetic testing. The second question asked the participants to score on a scale from 1 to 5 the importance of three different privacy guarantees that could possibly be provided. A total of 442 individuals participated in the survey.
A similar survey was conducted later after Nebula Genomics deployed the blockchain component of our system. An email was sent to 4,865 individuals on a Nebula Genomics mail list who purchased the genetic tests. A total of 407 individuals participated in the survey and answered three multiple-choice, single-select questions that reflect their views on the offered data privacy protections (Supplementary Fig. 7).
Data availability
To evaluate the performance of our system, we used open-access genomic and clinical data from The Cancer Genome Atlas (TCGA). The initial dataset contained one million genetic variants distributed across 8,000 individuals that were obtained by downloading and merging genomic data for the 32 studies of the TCGA PanCancer Atlas from https://www.cbioportal.org/, and are made available at https://github.com/ldsec/projects-data/tree/master/medco/datasets/genomic/tcga_cbio. We synthetically augmented this initial dataset to obtain the final test dataset of 28 billion genetic variants distributed across 150,000 individuals (each individual has a number of genetic variants, ranging from 15,000 to 200,000). This dataset is very large (in the order of tens of terabytes), so we cannot host it online. Instructions for reproducing the dataset augmentation are provided in the GitHub repository. It also includes a small demo dataset (https://github.com/ldsec/ccgd-platform) that is sufficient to test the code. If access to the large dataset that was used for the performance benchmarks is required, contact juan.troncoso-pastoriza@epfl.ch.
Code availability
The code is available on GitHub at https://github.com/ldsec/ccgd-platform. The code is archived on Zenodo at https://doi.org/10.5281/zenodo.455116513.
References
Regalado, A. More than 26 million people have taken an at-home ancestry test. MIT Technology Review (11 February 2019).
Farr, C. 23andMe lays off 100 people as DNA test sales decline, CEO says she was ‘surprised’ to see market turn. CNBC https://www.cnbc.com/2020/01/23/23andme-lays-off-100-people-ceo-anne-wojcicki-explains-why.html (2020).
Farr, C. Ancestry to lay off 6% of workforce because of a slowdown in the consumer DNA-testing market. CNBC https://www.cnbc.com/2020/02/05/ancestry-layoffs-of-6percent-100-people-amid-dna-test-slowdown.html (2020).
Huang, Z., Ayday, E., Fellay, J., Hubaux, J. & Juels, A. GenoGuard: protecting genomic data against brute-force attacks. In Proc. 2015 IEEE Symposium on Security and Privacy 447–462 (IEEE, 2015).
Cho, H., Wu, D. J. & Berger, B. Secure genome-wide association analysis using multiparty computation. Nat. Biotechnol. 36, 547–551 (2018).
Jagadeesh, K. A., Wu, D. J., Birgmeier, J. A., Boneh, D. & Bejerano, G. Deriving genomic diagnoses without revealing patient genomes. Science 357, 692–695 (2017).
Bogdanov, D., Laur, S. & Willemson, J. Sharemind: a framework for fast privacy-preserving computations. In Proc. Computer Security ESORICS 2008 192–206 (Springer, 2008).
Boura, C. et al. in Financial Cryptography and Data Security 183–202 (Springer, 2018).
Raisaro, J. L. et al. MedCo: enabling secure and privacy-preserving exploration of distributed clinical and genomic data. IEEE/ACM Trans. Comput. Biol. Bioinform. 16, 1328–1341 (2018).
Grishin, D., Obbad, K. & Church, G. M. Data privacy in the age of personal genomics. Nat. Biotechnol. 37, 1115–1117 (2019).
Froelicher, D. et al. UnLynx: a decentralized system for privacy-conscious data sharing. Proc. Priv. Enhancing Technol. 2017, 232–250 (2017).
Luterbacher, C. EPFL software to enable secure data-sharing for hospitals. EPFL (2 April 2002).
de Sa Sousa, J. A. G., Misbach, M., Quinn, K., Pastoriza, J. R. T. & Grishin, D. ldsec/ccgd-platform: Citizen-Centered Genomic Discovery Platform (Zenodo, 2021); https://doi.org/10.5281/zenodo.4551165
Acknowledgements
We are grateful to the Bitfury Exonum engineering team for running part of the performance measurements described in this paper. We also thank C. Redin from Lausanne University Hospital, J. Aach from Harvard Medical School and A. Pyrgelis from EPFL for their useful feedback and constructive comments on the manuscript. This work was supported in part by grant no. 2017-201 (DPPH) of the Swiss strategic focus area Personalized Health and Related Technologies (PHRT) and by grant no. 2018-522 (MedCo) of the PHRT and the Swiss Personalized Health Network (SPHN). The work was also supported by Nebula Genomics.
Author information
Authors and Affiliations
Contributions
D.G., J.L.R. and J.T.P. conceived the study. D.G., J.L.R., J.T.P. and J.P.H. wrote the manuscript. D.G., J.L.R., J.T.P., K.O., J.G., M.M., K.Q. and J.S. implemented the algorithms described in the Methods and performed the benchmark experiments. All authors discussed and reviewed the manuscript at all stages. J.F., G.M.C. and J.P.H. supervised the work.
Corresponding authors
Ethics declarations
Competing interests
D.G., K.O., K.Q. and G.M.C. are affiliated with Nebula Genomics. J.L.R., J.T.P., M.M., J.S., J.F. and J.P.H. declare no competing interests.
Additional information
Peer review information Nature Computational Science thanks Fida Dankar and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Ananya Rastogi was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Security and privacy background, example of secure distributed aggregate-data analysis, Supplementary Results (Figs. 1–9) and Supplementary Methods (Figs. 10–18).
Source data
Source Data Fig. 1
Survey responses.
Source Data Fig. 3
Code benchmarks.
Rights and permissions
About this article
Cite this article
Grishin, D., Raisaro, J.L., Troncoso-Pastoriza, J.R. et al. Citizen-centered, auditable and privacy-preserving population genomics. Nat Comput Sci 1, 192–198 (2021). https://doi.org/10.1038/s43588-021-00044-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43588-021-00044-9
This article is cited by
-
A systematic study on blockchain technology for genomic data management in healthcare: approaches, challenges, and research opportunities
Medical & Biological Engineering & Computing (2025)
-
Data breaches in healthcare: security mechanisms for attack mitigation
Cluster Computing (2024)
-
Sociotechnical safeguards for genomic data privacy
Nature Reviews Genetics (2022)
-
Data privacy through participant empowerment
Nature Computational Science (2021)