Abstract
Data generated in perturbation experiments link perturbations to the changes they elicit and therefore contain information relevant to numerous biological discovery tasks—from understanding the relationships between biological entities to developing therapeutics. However, these data encompass diverse perturbations and readouts, and the complex dependence of experimental outcomes on their biological context makes it challenging to integrate insights across experiments. Here we present the large perturbation model (LPM), a deep-learning model that integrates multiple, heterogeneous perturbation experiments by representing perturbation, readout and context as disentangled dimensions. LPM outperforms existing methods across multiple biological discovery tasks, including in predicting post-perturbation transcriptomes of unseen experiments, identifying shared molecular mechanisms of action between chemical and genetic perturbations, and facilitating the inference of gene–gene interaction networks. LPM learns meaningful joint representations of perturbations, readouts and contexts, enables the study of biological relationships in silico and could considerably accelerate the derivation of insights from pooled perturbation experiments.
Similar content being viewed by others
Main
Perturbation experiments play a central role in elucidating the underlying causal mechanisms that govern the behaviors of biological systems1,2,3. Controlled perturbation experiments measure changes in experimental readouts, such as the number of specific transcripts observed, resulting from introducing perturbations to biological systems, such as in vitro cell lines, compared with unperturbed references. Researchers use controlled perturbations in relevant biological model systems to establish causal relationships between molecular mechanisms, genes, chemical compounds and disease phenotypes. This causal understanding of foundational biological relationships has the potential to positively impact numerous important societal goals4, including the production of climate-friendly foods and materials and the development of novel therapeutics that address unmet health needs.
The path to understanding complex biological systems and developing targeted therapeutics hinges on unraveling how cells respond to perturbations. High-throughput experiments have generated an unprecedented volume of perturbation data spanning thousands of perturbations across diverse readout modalities and biological contexts, from single-cell to in vivo settings5,6,7,8,9. However, these experiments, while rich in indispensable information, vary dramatically in their protocols, readouts and model systems, often with minimal overlap. The vast scale and heterogeneity of this data, compounded by context-specific effects, make it extremely challenging to derive generalizable biological insights that drive scientific discovery. A core challenge in integrating evidence collected across heterogenous experiments is that it is difficult to disentangle effects stemming from differences in experimental context from those of the perturbation itself.
This fundamental challenge of extracting meaningful biological insights from perturbation data has spurred the development of diverse computational approaches10,11,12,13. Most existing approaches focus specifically on predicting the effects of unobserved perturbations14,15,16,17,18,19,20,21. This addresses a fundamental limitation of experimental methods: it is physically impossible to perform all possible configurations of perturbation experiments owing to the effectively infinite number of potential experimental designs (considering the time of measurement can be arbitrarily long, the number of experiments that may be conducted is already unbounded based on this dimension alone). For example, the graph-enhanced gene activation and repression simulator (GEARS)15 leverages gene representations based on domain knowledge22 to predict the effects of unseen genetic perturbations while also providing a means of identifying genetic interaction subtypes. The compositional perturbation autoencoder (CPA)19 predicts the effects of unseen perturbation combinations, including drugs as perturbagens and their dosages. Beyond perturbation effect prediction, some methods focus on other critical biological discovery tasks, such as estimating gene–gene relationships23, learning transferable cell representations24,25, modeling relationships among different types of readout26,27,28 or aiding experimental design29,30.
More recently, foundation models31,32,33,34 have emerged that are pretrained on large collections of transcriptomics data to address multiple biological discovery tasks through task-specific fine-tuning pipelines. These models, exemplified by Geneformer31 and scGPT32, use Transformer-based encoders35 to infer gene and cell representations from gene expression measurements. While their encoder-based approach offers a compelling advantage—the ability to make predictions for previously unseen contexts by extracting contextual information from gene expression profiles—it faces two substantial limitations. First, the low signal-to-noise ratio in high-throughput screens can pose a challenge to the encoder’s ability to extract reliable contextual information, which may result in limited prediction performance. Second, these models are primarily designed for transcriptomics data and are not inherently structured to accommodate diverse perturbation experiments that use other perturbation and readout modalities, such as chemical perturbations or low-dimensional screens measuring cell viability.
To enable in silico biological discovery from a diverse pool of perturbation experiments, we demonstrate that heterogeneous experimental data, regardless of perturbation type or readout modality, can be integrated into a large perturbation model (LPM) by representing perturbation, readout and context as disentangled dimensions. Similar to foundation models31,32, LPM is designed to support multiple biological discovery tasks, including perturbation effect prediction, molecular mechanism identification and gene interaction modeling. LPM is trained to predict outcomes of in-vocabulary combinations of perturbations, contexts and readouts. LPM introduces two architectural innovations that support its primary goal of handling heterogeneity in perturbation data. First, LPM disentangles the dimensions of perturbation (P), readout (R) and context (C), representing each dimension as a separate conditioning variable. Second, LPM adopts a decoder-only architecture, meaning it does not explicitly encode observations or covariates. The PRC-disentangled, encoder-free LPM architecture introduces key advantages:
-
Seamless integration of diverse perturbation data. By representing perturbation experiments as P–R–C dimensions, LPM effectively learns from heterogeneous experiment data across diverse readouts (for example, transcriptomics and viability), perturbations (CRISPR and chemical) and experimental contexts (single-cell and bulk) without loss of generality and regardless of dataset shape or format.
-
Contextual representation without encoder constraints. Encoder-based models assume that all relevant contextual information can be extracted from observations and covariates, which may be limiting due to high variability in measurement scales across contexts and a potentially low signal-to-noise ratio. By contrast, LPM learns perturbation-response rules disentangled from the specifics of the context in which the readouts were observed. A limitation of this approach is the inability to predict perturbation effects for out-of-vocabulary contexts.
-
Enhanced predictive accuracy across experimental settings. By leveraging its PRC-disentangled architecture and decoder-only design, LPM consistently achieves state-of-the-art predictive accuracy across experimental conditions.
When trained on a pool of experiments, we demonstrate experimentally that LPM achieves state-of-the-art performance in post-perturbation outcome prediction. In addition, LPM provides meaningful insights into the molecular mechanisms underlying perturbations, readouts and contexts. LPM enables the study of drug–target interactions for chemical and genetic perturbations in a unified latent space, accurately associates genetic perturbations with functional mechanisms and facilitates the inference of causal gene-to-gene interaction networks. To demonstrate the potential of LPM for therapeutic discovery, we used a trained LPM to identify potential therapeutics for autosomal dominant polycystic kidney disease (ADPKD). Finally, we show that the superior performance of LPM compared with existing methods is driven by its ability to leverage perturbation data at scale, achieving significantly improved performance as more data become available for training.
Results
LPM is a deep-learning model that integrates information from pooled perturbation experiments (Fig. 1). We train LPM to predict the outcome of a perturbation experiment based on the symbolic representation of the perturbation, readout and context (the P,R,C tuple). LPM features a PRC-conditioned architecture that enables learning from heterogeneous perturbation experiments that do not necessarily fully overlap in the perturbation, readout or context dimensions. By explicitly conditioning on the representation of an experimental context, LPM learns perturbation-response rules disentangled from the specifics of the context in which the readouts were observed. LPM predicts unseen perturbation outcomes, and its information-rich generalizable embeddings are directly applicable to various other biological discovery tasks (Fig. 1).
Top left: perturbation experiments originating from different studies (green, orange and purple indicate separate experiments) are pooled together. Each experiment is placed in the space spanned by perturbations (P), readouts (R) and experimental contexts (C), where multiple experiments generally only partially overlap in the three-dimensional (P,R,C) space. Central icon: a LPM is trained on pooled perturbation data and can be queried with the symbolic representation of perturbation, readout and context of experiments of interest to generate embeddings and predict outcomes even for configurations that were not observed during training. Top right: trained LPM can be queried to predict experiment outcome given symbolic representations of P, R and C (blue). Bottom: LPM embeddings and predictions carry rich information for a range of biological discovery tasks using transfer learning.
Predicting outcomes of unobserved perturbation experiments
We evaluated the performance of LPM in predicting gene expression for unseen perturbations against state-of-the-art baselines, including CPA19 and GEARS15 (Fig. 2). We also included baseline models that combined a Catboost regressor36 with existing gene embeddings derived from biological databases (STRING37, Reactome38 and Gene2Vec39), single-cell foundation models based on pooled gene expression data not under perturbations (Geneformer31 and scGPT32) and natural language descriptions of genes processed through ChatGPT (GenePT34). For scGPT and Geneformer, we either fine-tuned the models according to their respective instructions or used their embeddings with a CatBoost model (indicated as ‘emb’). In addition, we included the ‘NoPerturb’ baseline15 that assumes that the perturbation does not induce a change in expression. Note that no other baseline model supports predicting outcomes of chemical perturbations and that GEARS, CPA and scGPT (following author instructions) require single-cell-resolved data.
The performance of LPM was compared against state-of-the-art baselines across a variety of experimental settings, contexts and for different perturbation types. a, A comparison of methods for post-perturbation expression prediction using z-normalized data including all readouts comparing Pearson correlation (y axis) on held-out test data from eight experimental contexts (x axis) including single-cell (Replogle et al.9), bulk (LINCS7), genetic (CRISPRi and CRISPR-KO) and chemical compound interventions. b,c, In addition, we performed a comparison methods for post-perturbation expression prediction that replicates the preprocessing methodology from Roohani et al.15 and Cui et al.32. In this comparison, we calculated the Pearson correlation between true and predicted changes in log-normalized expression (control versus perturbed) measured on held-out test data for all genes (b) and on the subset of the top 20 differentially expressed transcripts (c) (y axis). Norman et al.76 include both single and multiperturbation data. Embedding (‘emb’ in parentheses) next to a baseline indicates that we used embeddings that were fine-tuned using Catboost. For baselines without this indication, we used author instructions for generating the post-perturbation expression predictions. Not all methods are suitable for all settings that LPM operates on and are therefore not included in all comparisons. Asterisks indicate statistical significance (one-sided Mann–Whitney, *P ≤ 0.05). Dots on top of bars represent random seeds.
To robustly evaluate the performance of LPM, we conducted a representative array of experiments that covers (1) a range of experimental contexts, (2) different perturbation types (chemical and genetic) and (3) varying preprocessing strategies. Across all studied experimental settings, LPM consistently and significantly outperformed the state-of-the-art baselines, regardless of preprocessing methodology. Further data from Horlbeck et al.40, which included viability readouts for pairwise CRISPRi perturbations, are presented in the Supplementary Information to demonstrate that LPM is effective even in low-dimensional settings with nontranscriptomic readouts. For details on the datasets and their preprocessing, see the Methods.
Mapping a compound-CRISPR shared perturbation space
To evaluate the ability of LPM to support the generation of insights across different types of perturbation, we trained an instance of LPM using all available data from Library of Integrated Network-Based Cellular Signatures (LINCS) experiments7 involving both genetic and pharmacological perturbations across a total of 25 experimental contexts with unique combinations of cellular contexts and perturbation types. LPM integrates genetic and pharmacological perturbations within the same latent space, enabling the study of drug–target interactions. When studying t-distributed stochastic neighbor embeddings (t-SNE)41 of the perturbation embedding space learned by the LPM, we found that pharmacological inhibitors of molecular targets are consistently clustered in close proximity to genetic CRISPR interventions that target the same genes (Fig. 3a). For example, genetic perturbations targeting MTOR and compounds inhibiting MTOR and also genetic perturbations targeting genes from the same pathway, for example PSMB1 and PSMB2, or HDAC2 and HDAC3, were clustered closely together. Qualitatively, we found that anomalous compounds that were placed distant from their putative target had been reported to have off-target activity (Fig. 3b), such as benfluorex (withdrawn due to cardiovascular side effects42) and pravastatin (shown to elicit expression changes with low correlation to other statins43). Intriguingly, we found that pravastatin moved toward nonsteroidal anti-inflammatory drugs that target gene PTGS1 in the perturbation space (Fig. 3a), indicating a potential additional anti-inflammatory mechanism of pravastatin. We found that this movement independently derived by LPM is indeed substantiated by clinical and preclinical observations that ascribe anti-inflammatory effects to pravastatin44,45,46. To further quantitatively validate these findings, we systematically compared known inhibitors of a genetic target with the genetic perturbation in embedding space as a reference. We evaluated the neighborhood of the reference in various embedding spaces and found that perturbation embeddings derived from LPM achieve considerably higher recall of known inhibitors of genetic targets compared with embeddings derived from post-perturbation L1000 transcriptome profiles or dimensionality reduced versions thereof (Fig. 3c).
a, The latent space of compound and CRISPR knockouts (reduced to two-dimensions via t-SNE) reflects known groupings of compound and genetic perturbations that target the same molecular mechanisms in bulk LINCS L1000 data from ref. 7. Genes targeted by corresponding CRISPR and compound inhibitors are color-coded in matching colors. b, Root mean squared error (RMSE) distances of known HMGCR inhibitors (statins) to the corresponding CRISPR-HMGCR perturbation in the embedding space of the LPM. Two bottom outliers are additionally annotated in a: benfluorex (withdrawn for cardiovascular side effects42) and pravastatin (shown to have low correlation to other statins78 and additional anti-inflammatory effects44,45,46). c, The RMSE-based distance between perturbation embeddings for CRISPR perturbations was used to measure the recall of known inhibitors of the respective genetic target, for different numbers of nearest neighbors. We compared LPM embeddings with those derived from post-perturbation L1000 transcriptome profiles. Bars represent the 95% confidence intervals across genetic targets (N = 89).
Learned embeddings reflect known biological relationships
To evaluate the degree to which LPM perturbation embeddings correspond to known biological functions, we extracted perturbation embeddings for well-characterized perturbations from an LPM trained on pooled single-cell perturbation data9 and compared genetic perturbations with gene function annotations as curated by Replogle et al.9 using the comprehensive resource of mammalian protein complexes (CORUM)47 and search tool for recurring instances of neighbouring genes (STRING)37 databases. We found that LPM implicitly organizes perturbations according to their molecular functions (Fig. 4a) and that these embeddings are significantly (P ≤ 0.01) more predictive of gene function annotations than existing state-of-the-art gene perturbation embeddings (Fig. 4b), including those derived from curated databases such as STRING37 and Reactome38, derived from co-expression datasets in Gene2Vec39 and derived from the single-cell unperturbed gene expression foundation models Geneformer31 and scGPT32 and gene embeddings based on natural language descriptions processed through ChatGPT (GenePT34).
a, LPM perturbation (P) embeddings (t-SNE embedded in two-dimensional (2D) space). Each point represents a CRISPRi perturbation color-coded by the molecular function of its respective genetic target from ref. 9. b, Performance of LPM perturbation (P) embeddings compared with existing state-of-the-art gene embeddings derived from large-scale genetic screens and public pathway and interaction databases in predicting gene function annotations from ref. 9 (P value calculated via one-sided Mann–Whitney test). Dots on the top of bars represent replicates across N = 5 random seeds. c, LPM context (C) embeddings (2D t-SNE representation) quantify similarity between experimental contexts. Intriguingly, we found that contexts are grouped with respect to the model system under study (shown in the figure) or by type of perturbation (not shown), depending on the t-SNE random seed used.
To qualitatively assess the information contained within context representations of LPM, we used the LPM model trained on combined LINCS data from the perturbation embedding experiment above to generate context embeddings. We found that—depending on the t-SNE random seeds used—either cell types tend to cluster together with matching cell types from other experiments (Fig. 4c), or the context embeddings tend to cluster based on the perturbation methodology (CRISPR versus compound screens; not depicted). The qualitative results imply that the information contained within the learned context embeddings carries information regarding biological semantics and could thus be valuable in downstream analyses, such as for quantifying the similarity of contexts.
In silico discovery of candidate therapies for ADPKD
We hypothesized that the ability of LPM to conduct perturbation experiments in silico with high accuracy while reflecting underlying biological function could be used to discover potential candidate therapeutics for diseases with known genetic causes, such as ADPKD. ADPKD is a genetic disease suspected to be caused by mutations in PKD148 that are reported to lead to a lack of functional PKD1—eventually manifesting in dose-dependent cystogenesis49,50,51,52. ADPKD affects more than 12 million people worldwide53 and may lead to severe long-term complications, such as end-stage renal disease (ESRD) and the dependence on dialysis or a kidney transplant. There are no curative treatments available for ADPKD. A potential hypothesis for a therapeutic could be to upregulate expression of the functional allele of PKD1 in heterozygous carriers of PKD1 mutations to make up for the nonfunctional allele and thereby reach a sufficient level of functional PKD1 that may inhibit further progression of ADPKD. To identify potential therapeutics that could increase PKD1 expression in individuals with ADPKD, we conducted an in silico perturbation experiment using an LPM trained on pooled LINCS compound and genetic perturbation data to predict which clinical-stage drugs may lead to upregulation in PKD1 levels in HA1E embryonic kidney cells cultured under the LINCS L1000 protocol54. We found that triptolide, simvastatin and other statins were among the top clinical-stage drugs predicted to cause increased PKD1 expression in vitro (Fig. 5a). Our findings align well with previous literature, where effects of commercially available statins were shown to increase the expression of PKD1 in pancreatic cancer cell line MiaPaCa-255. We note that Huang et al.56 found no significant change in PKD1 expression in mice exposed to atorvastatin. As simvastatin is a Food and Drug Administration (FDA)-approved medicine that is prescribed preventatively for cardiovascular indications, we conducted a retrospective, matched cohort study57,58 using a non-linear propensity score estimator59 to validate the in silico hypothesis that simvastatin may lead to reduction in ESRD progression in real-world clinical data from the Optum deidentified Electronic Health Record database. Notably, we found that—among individuals diagnosed with ADPKD60—exposure to simvastatin over 1 year or longer was associated with a significant decrease (5-year relative risk 0.86, P = 0.0405, and 10-year relative risk 0.74, P = 0.0003) in progression to ESRD61 compared with those not exposed to any statins predicted by LPM to increase expression of PKD1 (Fig. 5b). Several of the therapeutics predicted to increase PKD1 are substantiated by literature; for example, pravastatin was shown to be associated with improved kidney markers in a clinical study in young individuals62, and triptolide led to a reduction of cystogenesis in murine models63,64. PKD1 was neither measured nor perturbed in LINCS, the 5,310 chemical perturbations were not all tested in HA1E cells, and the in silico LPM experiments were therefore essential to enable this study. We note that these findings should not be considered definitive and that further research is required to validate and support them.
a, Using LPM, we conducted an in silico perturbation study in which we identified clinical-stage drugs that are predicted to upregulate PKD1 in embryonic kidney cells. A lack of functional copies of PKD1 is hypothesized to be causally involved in ADPKD pathogenesis and progression49,50,51. We found that triptolide, simvastatin (bold) and other statins, are the top predicted upregulators of PKD1 among clinical-stage drugs. For reference, we also include the predicted CRISPRi on vasopressin receptor 2 (AVPR2) to simulate the effect of the FDA-approved AVPR2 antagonist tolvaptan79,80 that is mechanistically distinct81,82. Bars represent model predictions in the form of z scores. b, Because simvastatin is commonly prescribed for cardiovascular indications, we were able to conduct a retrospective cohort study in large-scale electronic health records to further substantiate the potential efficacy of simvastatin in reducing ADPKD progression in the clinic. Most notably, we found that—among individuals diagnosed with ADPKD—1 year or longer exposure to simvastatin (blue) is associated with a significant (P ≤ 0.05, 5-year relative risk (RR) of 0.86 and 10-year RR of 0.74) reduction in progression to ESRD compared with those not exposed to statins (red). The 95% confidence intervals were estimated using the Nelson–Aalen estimator83,84.
Facilitating inference of causal gene–gene relationships
To assess to what degree the accuracy of the predictions of LPM translate to capturing mechanistic interactions between genes, we used LPM in the context of causal inference of gene interaction networks. Normally, these networks are inferred from perturbation experiments in which only a subset of all genes were perturbed. By contrast, we measured the enhancement in performance when those networks were inferred from the same experimental data enriched with missing, unmeasured CRISPRi perturbations predicted in silico using LPM. In particular, to perform network inference, we applied corresponding methods that demonstrated best-in-class performance on the recent CausalBench challenge23,65 and were designed specifically for inferring gene–gene networks from perturbational single-cell RNA sequencing data. We found that augmenting the original data with in silico perturbation outcomes, before applying network inference using above-mentioned methods, leads to a significant improvement in terms of false omission rate (FOR) in comparison with existing state-of-the-art methods for gene–gene network inference that do not have access to perturbation imputation (Fig. 6). These results underscore the utility of LPM in supporting the inference of more comprehensive and accurate causal interactions tailored to a given experimental context and the ability of LPM to learn generalizable, causal interactions between perturbations.
a, We used LPM to predict post-perturbation transcriptomes for unseen perturbations, completing a partially observed experimental space where only half (orange area) of the possible CRISPRi perturbations across the genome were experimentally observed. We hypothesized that access to the computationally completed dataset (blue plus orange area) may enable the state-of-the-art Guanlab85 gene network inference method to more accurately infer the gene–gene interactions for genes not experimentally perturbed. b, We found that the combination of LPM imputation and the Guanlab method (LPM+Guanlab) significantly outperformed existing methods for gene–gene network inference using the partially observed dataset alone in terms of FOR in a gene network inference benchmark23 using single-cell data from Replogle et al.9. Dots on top of bars represent replicates across N = 11 random seeds (P values calculated using one-sided Mann–Whitney–Wilcoxon).
LPM performance improves with more training data
In contrast to data-rich domains such as natural language processing, where scaling of model performance with additional data has been studied experimentally66,67, it is not yet clear to what degree in silico biological discovery can benefit from the availability of additional data across both contexts and perturbations for pooling. Establishing data scaling patterns in biology has historically been more difficult than in predominantly digital domains such as natural language processing and computer vision because biological perturbation data can often not be naively aggregated owing to the intricate connection between experimental context, data processing methodologies and batch effects68,69. To elucidate the potential performance benefits of additional data for LPM, we computationally evaluated the prediction performance in terms of Pearson correlation coefficient ρ for predicting unseen perturbations when varying the number of datasets covering multiple contexts and perturbations in a single context available for model training (Extended Data Fig. 1). The performance of LPM significantly (P ≤ 0.05) improves both when more datasets covering multiple contexts and when more perturbations in a single context are available for training.
Discussion
LPM demonstrates that integrative learning across heterogeneous perturbation screens can deliver accurate, in silico estimates of perturbation-, readout- and context-specific experimental outcomes. We found that the use of LPM—either independently or in combination with a causal network inference algorithm—significantly outperforms existing state-of-the-art methods, providing an experimental proof of concept for the potential to accelerate biological discovery with computationally generated evidence. The ability to generate unobserved experimental data for critical biological questions, such as what the estimated effects of unseen perturbations would be, could accelerate the generation of insights and complement experimentally generated data—particularly in settings that are difficult, time-intensive or resource-intensive to study in real-world laboratory experiments. Notably, we found that LPM implicitly learns rich latent space embeddings for perturbations, readouts and experimental contexts as is required to achieve their explicit training objective to predict yet unseen experimental outcomes. The rich latent space embeddings of LPM enables a range of downstream biological discovery tasks (only a subset of the potential use cases are investigated in this study), which demonstrates the versatility and multitask capability of LPM that captures underlying mechanistic relationships in data.
LPM still faces important limitations. First, the training data used in our study are publicly available and sufficiently standardized; however, non-immortalized cell lines, rare cell types, primary tissues and patient-derived samples remain underrepresented. Second, the model can interpolate and handle symbols within its training vocabulary but cannot yet extrapolate to unseen symbols—for instance, novel cell types or perturbations—unless suitable pretrained embeddings are explicitly supplied. Nevertheless, recent trends indicate that in the near future, as perturbational experimental data becomes more abundant, the experimental space will be sufficiently covered, rendering in-vocabulary approaches sufficient for most tasks. Third, hidden batch effects, inconsistent preprocessing and incomplete metadata can still erode performance, as in other large-scale biological models. Fourth, the ADPKD case study is retrospective and therefore vulnerable to unobserved confounders; mechanistic conclusions will remain provisional until prospective validation. As a further limitation, we considered only a single genetically validated marker in our ADPKD study but therapeutic candidates must be optimized with regard to multiple criteria, including safety, pharmacokinetics and pharmacodynamics. It is important to note that further clinical validation is needed to conclusively establish causality for the predictions of LPM in the context of ADPKD. Finally, we would like to emphasize that gene network inference is a distinct and complex field of research70, and future studies will need to explore additional datasets and benchmarks to further validate findings in this area. Our study, however, is focused on demonstrating the potency of high-quality perturbation-effect predictors, such as LPM, to complement existing network inference methods.
Several experimental directions could address these gaps, including prospective perturbation screens in primary and patient-derived cells to test whether LPM maintains accuracy outside immortalized lines and under different dosages71. Applying LPM to data derived from clinical settings could present valuable opportunities to identify novel therapies or patient cohorts that are likely to respond to specific treatments, thus advancing the field of personalized medicine. By leveraging these datasets, LPM could help to pinpoint biomarkers of response (for example, clinical covariates) and further optimize therapeutic strategies for patients based on their unique molecular profiles. In general, if curated and standardized perturbation data continue to grow, parameter-scaling results suggests that larger LPM variants could yield proportional gains in predictive accuracy and mechanistic resolution. Systematic efforts to reduce batch effects and harmonize metadata will be as important as algorithmic advances in realizing that potential.
Methods
Problem formulation
We consider every experimental system subject to a perturbation (represented symbolically) for which we observe a readout. For example, an experiment could be conducted in a single-cell in vitro system in which transcript counts are measured after CRISPRi targeting a specific gene. A biological model system is considered to be a black box, and no prior knowledge is assumed about the internal mechanism that gives rise to observed readouts.
The totality of the experimental context, including model system under study and the experimental protocol used, is represented by the variable \(C\in {\mathcal{C}}\) and is referred to as the context of the experiment. The context C is a symbolic description of the system itself and implicitly represents all the covariates that constitute the experimental conditions, for example, biological context details such as cell type, genetic background and incubation protocols. We consider a perturbation to be any input to the system that is not already included in the context, including a chemical compound, a gene knockout or a disease that has perturbed the system are examples of perturbations. Let \(P\in {\mathcal{P}}\) be the vector that describes a perturbation. Similar to the context C, P is a symbolic representation of the perturbation. For instance, CRISPRi_STAT1 would symbolically represent CRISPR interference of gene STAT1. In addition, multiperturbations that are symbolically represented as, for example, CRISPRi_STAT1+CRISPRa_FOXF1 (CRISPR interference of gene STAT1 coupled with CRISPR-mediated transcriptional activation of FOXF1), are modeled as a function of corresponding embeddings. In the experiments in this Article, we used the embedding average. The symbolic description of the measurements observed in the system that is under perturbation is represented by a readout \(R\in {\mathcal{R}}\), where \({\mathcal{R}}\) is a set of symbols that correspond to all possible discrete values that represent observed readouts. For example, R can represent the gene expression of the gene PSMA1, denoted as Transcript_PSMA1. The concrete measurement taken in context C after perturbation P using readout R is represented by \(Y\in {\mathcal{Y}}\subseteq {\mathbb{R}}\). It is notable that the experimental observation Y is distinct from the readout R in that R symbolically describes the type of measurement taken, whereas Y is a concrete instance of that measurement in the experimental context C under perturbation P.
Let O = (P, R, C, Y) be the stack of aforementioned random variables and \({\mathcal{I}}=\{1,2,\ldots \}\) be the index set of all possible potential observed samples. Therefore, the index \(i\in {\mathcal{I}}\) refers to one potential observation O(i) = (P(i), R(i), C(i), Y(i)). Let \({{\mathcal{D}}}_{{\rm{obs}}}=\{{O}^{(1)},{O}^{(2)},\ldots ,{O}^{({n}_{{\rm{obs}}})}\}\) be the set of observations that has nobs data points and \({{\mathcal{I}}}_{{\rm{obs}}}\subseteq {\mathcal{I}}\) be the set of associated indices. It is clear that Y is not independent from P, R and C. We want to learn the causal model q(Y∣do(P = p), R, C). Here, q is the probability distribution of the outcome Y in a biological system within the context C when the perturbation p is applied and the readout R is observed. We would like to leverage the structural dependence between these variables to estimate q from \({{\mathcal{I}}}_{{\rm{obs}}}\) so it can predict the outcome of unobserved (perturbation, readout and context) combinations indexed by \(j\in {{\mathcal{I}}}_{{\rm{unobs}}}={\mathcal{I}}\backslash {{\mathcal{I}}}_{{\rm{obs}}}\). Mathematically, we want to estimate
for any combination \((P,R,C)\in {\mathcal{P}}\times {\mathcal{R}}\times {\mathcal{C}}\). This is possible only if the spaces \({\mathcal{P}}\), \({\mathcal{R}}\) and \({\mathcal{C}}\) have some structure that allows the concept of distance to be defined. For example, for a system with context C(j), predicting the effect of perturbation P(j) on readout R(j) is possible if the outcome of a similar perturbation on a similar readout is already observed for a system within a similar context. Clearly, discussing similarities requires the relevant spaces to possess some structure in which a distance metric can be defined. As (P, R, C) are in essence discrete symbolic values, it is necessary to first transform them into more tractable spaces that we call embedding spaces. Let \({Z}_{P}\in {{\mathcal{Z}}}_{P}\subseteq {{\mathbb{R}}}^{{d}_{{Z}_{P}}}\), \({Z}_{R}\in {{\mathcal{Z}}}_{R}\subseteq {{\mathbb{R}}}^{{d}_{{Z}_{R}}}\) and \({Z}_{C}\in {{\mathcal{Z}}}_{C}\subseteq {{\mathbb{R}}}^{{d}_{{Z}_{C}}}\) be the random variables that represent the embeddings of P, R and C, respectively. The transformation maps \({\phi }_{p}:{\mathcal{P}}\to {{\mathcal{Z}}}_{P}\) \({\phi }_{r}:{\mathcal{R}}\to {{\mathcal{Z}}}_{R}\) and \({\phi }_{c}:{\mathcal{C}}\to {{\mathcal{Z}}}_{C}\) that induce such structure in the embedding spaces are learned from \({{\mathcal{I}}}_{{\rm{obs}}}\). In other words, the information of the observed data is learned in ϕp(⋅), ϕr(⋅) and ϕc(⋅) functions. This means that, for any unseen (P, R, C) tuples, their corresponding embeddings ZP, ZR and ZC implicitly contain some information from \({{\mathcal{I}}}_{{\rm{obs}}}\). This is indeed the reason that enables knowledge transfer to unseen perturbations, readouts and contexts. With the learned embedding space, equation (1) can be written as
where the subscript ‘emb’ emphasizes that the map is defined from the embedding spaces instead of the original spaces. Due to the learned structure in the embedding spaces, it is expected that qemb(⋅) be more accessible to learn than q(⋅).
Model architecture
Building on the problem formulation described in the ‘Problem formulation’ section, we designed the architecture of LPM as shown in Extended Data Fig. 2a. Because P, R and C are discrete random variables, it is simple to implement the corresponding embeddings ZP, ZR and ZC using symbol vocabularies and learnable look-up tables (Extended Data Fig. 2b). Symbol vocabularies map symbols to indices, while look-up tables map indices to learnable weights that we treat as embeddings. This model can nevertheless be trivially generalized to include more complex perturbations or context descriptions; for example, multiple perturbations can be implemented as a sum of individual perturbations15,19. The prediction network is a neural network that is learned end-to-end together with the embeddings, by backpropagating the error using the Adam optimizer72. We found the multilayer perceptron architecture with ReLU activation functions, implemented on top of concatenated embeddings, to work satisfactorily (Extended Data Fig. 1c). We note that an extensive architecture search was not performed and further architecture tuning could potentially further improve results.
The key property of our model that enables scaling training across heterogeneous high-throughput perturbation screens (Fig. 1) is its conditioning on the readout R. To clarify why this simple trick is effective, consider an alternative description of the causal model equation (1) that does not condition on the R, that is, \(q{\prime} (Y| P,C,{{\mathcal{I}}}_{{\rm{obs}}})\) (ref. 73). In this case, \(Y\in {\mathcal{Y}}\subseteq {{\mathbb{R}}}^{{d}_{y}}\) is a vector (not a scalar) whose dimension dy is the number of readouts. The challenge is that de facto each perturbation screen has its own subset of phenotypic readouts. Even when the same modality—such as the transcriptome—is measured in two datasets, they often capture different subsets of that modality. The problem exacerbates if different modalities are used for training (for instance, proteome along with transcriptome), or if a large number of datasets is included in the training process. Related previous works alleviate this issue by selecting only readouts that appear in all considered perturbation experiments. However, this approach is clearly suboptimal because it discards relevant information. Moreover, it becomes impractical when scaling to many datasets, because the size of the overlapping feature set shrinks as the number of datasets increases. Moreover, certain experimental measurement technologies, such as DRUG-seq74, may contain missing values. LPM is designed to be robust to missing readouts as well.
Data sources
The datasets used for benchmarking include single-cell and bulk data, genetic (CRISPRi, CRISPR activation (CRISPRa) and CRISPR-knockout (CRISPR-KO)) and chemical compound perturbations, and single- and multiperturbation settings. The full overview of all used data is presented in the Supplementary Information. Single-perturbation single-cell data contain two experimental contexts from ref. 9 : Replogle et al. (K562) and Replogle et al. (RPE1). The data are based on transcriptome measurements generated after DepMap essential genes75 have been perturbed using CRISPRi Perturb-seq technology. The data were sequenced in chronic myeloid leukemia (K562) and retinal pigment epithelial (RPE1) cell lines, respectively. In the single-cell space, we also used multiperturbation experiments of type CRISPRa from Norman et al.76, which were performed also on K562 cells using Perturb-seq. For bulk data, we used the expanded Connectivity Map Lincs 2020 screens (https://clue.io)7, on both pharmacological and CRISPR-KO perturbations. A total of 26 biological contexts from LINCS studies based on bulk data were used, encompassing different cell and perturbation types. We discarded LINCS contexts that had too few perturbations (<300), for simplicity, as they did not make a difference in our analysis. To further simplify the analysis, we used only the most commonly appearing drug doses (10 μM) and observation times (24 h).
Data preprocessing
We used two preprocessing approaches to test robustness and perform a fair comparison against competing methods. In our first set of experiments, for single-cell data from ref. 9, we used the z-normalized version of the datasets, as recommended and provided by the authors. For single-perturbation single-cell data, z normalization was performed per gemgroup (batch). Single-guide RNAs that target the same gene were aggregated to represent a single perturbation. We removed cells containing multiple knockdowns to simplify the evaluation, focusing exclusively on predicting unobserved perturbations rather than combinations of observed perturbations. For bulk data, we used the preprocessed data that included quality control as provided by Subramanian et al.7 (level 5, phase II data). We kept only 978 experimentally measured readouts and dropped inferred gene expressions. In our second set of experiments, we used data from both single-perturbation experiments Replogle et al. (K562) and Replogle et al. (RPE1), as well as multiperturbation experiments from Norman et al.76 processed as described in ref. 15 (log-transformed and filtered to 5,000 highly variable genes). This preprocessing strategy is arguably the most established in the literature for evaluating perturbation models.
Benchmarking
As a part of our benchmarking, we compared LPM against six baselines: (1) CPA19, (2) GEARS15, (3) CatBoost36 combined with precomputed gene embeddings from STRING37, Reactome38 and Gene2Vec39, (4) Geneformer31, (5) scGPT32 and (6) GenePT34. Geneformer and scGPT were either fine-tuned according to the authors’ instructions or used as frozen embedding generators (suffix ‘emb’). The NoPerturb baseline15 was included as a perturbation-agnostic control. For performance benchmarks (Fig. 2), we used cross-validation and held out a single experimental context as the target context for each fold. Within the target context, test and validation data were randomly held out (stratified by perturbation) and excluded from training, while the remaining target context data and all data from nontarget contexts were used to train LPM (experimental details provided in the Supplementary Information). For GEARS and Catboost-based models, only data from the target context were used because including additional contexts did not benefit those methods. For CPA, due to architectural constraints, we could only include single-cell data from the same experimental studies (that is, all Replogle data). For each target context, we trained models for different random seeds to quantify uncertainty. The remaining details of our experiments are given in the Supplementary Information. They include hyperparameter selection, learning details, baselines and metrics used, and details related to specific downstream tasks.
Statistics and reproducibility
No statistical method was used to predetermine sample size. The experiments were not randomized. Data collection and analysis were not performed blind to the conditions of the experiments. Source code is available in the code repository77.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Perturbation data used in this study are from publicly available sources, including Norman et al.76 (GSE133344), Replogle et al.9 (Figshare), Horlbeck et al.40 (GSE116198) and Subramanian et al.7 (https://clue.io/). The Optum deidentified Electronic Health Record database used to validate in silico findings in real-world data is available for accredited researchers from Optum, but third-party restrictions apply to the availability of these data. The data were used under license for this study with restrictions that do not allow the data to be redistributed or made publicly available. Data access to the Optum deidentified Electronic Health Record database may require a data sharing agreement and may incur data access fees. Source data are provided with this paper.
Code availability
Source code is available via GitHub at https://github.com/perturblib/perturblib and via Zenodo at https://doi.org/10.5281/zenodo.15671137 (ref. 77).
References
Meinshausen, N. et al. Methods for causal inference from gene perturbation experiments and validation. Proc. Natl Acad. Sci. USA 113, 7361–7368 (2016).
Rubin, A. J. et al. Coupled single-cell CRISPR screening and epigenomic profiling reveals causal gene regulatory networks. Cell 176, 361–376 (2019).
Tejada-Lapuerta, A. et al. Causal machine learning for single-cell genomics. Nat. Genet. 57, 797–808 (2025).
Biermann, F., Kanie, N. & Kim, R. E. Global governance by goal-setting: the novel approach of the un sustainable development goals. Curr. Opin. Environ. Sustain. 26, 26–31 (2017).
Shalem, O., Sanjana, N. E. & Zhang, F. High-throughput functional genomics using CRISPR–Cas9. Nat. Rev. Genet. 16, 299–311 (2015).
Rauscher, B., Heigwer, F., Breinig, M., Winter, J. & Boutros, M. GenomeCRISPR—a database for high-throughput CRISPR/Cas9 screens. Nucleic Acids Res. 45, gkw997 (2016).
Subramanian, A. et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171, 1437–1452 (2017).
Oughtred, R. et al. The Biogrid Interaction Database: 2019 update. Nucleic Acids Res. 47, D529–D541 (2019).
Replogle, J. M. et al. Mapping information-rich genotype–phenotype landscapes with genome-scale Perturb-seq. Cell 185, 2559–2575 (2022).
Fröhlich, F. et al. Efficient parameter estimation enables the prediction of drug response using a mechanistic pan-cancer pathway model. Cell Syst. 7, 567–579 (2018).
Dixit, A. et al. Perturb-Seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866 (2016).
Lopez, R., Gayoso, A. & Yosef, N. Enhancing scientific discoveries in molecular biology with deep generative models. Mol. Syst. Biol. 16, e9198 (2020).
Dong, M. et al. Causal identification of single-cell experimental perturbation effects with CINEMA-OT. Nat. Methods 20, 1769–1779 (2023).
Kamimoto, K. et al. Dissecting cell identity via network inference and in silico gene perturbation. Nature 614, 742–751 (2023).
Roohani, Y., Huang, K. & Leskovec, J. Predicting transcriptional outcomes of novel multigene perturbations with GEARS. Nat. Biotechnol. 42, 927–935 (2024).
Yuan, B. et al. Cellbox: interpretable machine learning for perturbation biology with application to the design of cancer combination therapy. Cell Syst. 12, 128–140 (2021).
Lotfollahi, M., Wolf, F. A. & Theis, F. J. scGen predicts single-cell perturbation responses. Nat. Methods 16, 715–721 (2019).
Hetzel, L. et al. Predicting cellular responses to novel drug perturbations at a single-cell resolution. Adv. Neural Inf. Process. Syst. 35, 26711–26722 (2022).
Lotfollahi, M. et al. Predicting cellular responses to complex perturbations in high-throughput screens. Mol. Syst. Biol. 19, e11517 (2023).
Wu, Y. et al. Predicting cellular responses with variational causal inference and refined relational information. Preprint at https://arxiv.org/abs/2210.00116arXiv (2022).
Bunne, C. et al. Learning single-cell perturbation responses using neural optimal transport. Nat. Methods 20, 1759–1768 (2023).
Consortium, GeneOntology The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32, D258–D261 (2004).
Chevalley, M. et al. A large-scale benchmark for network inference from single-cell perturbation data. Commun. Biol. 8, 412 (2025).
Lopez, R. et al. Learning causal representations of single cells via sparse mechanism shift modeling. Proc. Mach. Learn. Res. 213, 662–691 (2023).
Rosen, Y. et al. Universal cell embeddings: a foundation model for cell biology. Preprint at bioRxiv https://doi.org/10.1101/2023.11.28.568918 (2023).
Schmauch, Benoít et al. A deep learning model to predict rna-seq expression of tumours from whole slide images. Nat. Commun.11, 3877 (2020).
Arslan, S. et al. Large-scale systematic feasibility study on the pan-cancer predictability of multi-omic biomarkers from whole slide images with deep learning. Preprint at bioRxiv https://doi.org/10.1101/2022.01.21.477189 (2022).
Mehrizi, R. et al. Multi-omics prediction from high-content cellular imaging with deep learning. Preprint at https://arxiv.org/abs/2306.09391 (2023).
Mehrjou, A. et al. GeneDisco: a benchmark for experimental design in drug discovery. In International Conference on Learning Representations (2022).
Lyle, C. et al. DiscoBAX discovery of optimal intervention sets in genomic experiment design. In Proc. 40th International Conference on Machine Learning 23170–23189 (PMLR, 2023); https://proceedings.mlr.press/v202/lyle23a.html
Theodoris, C. V et al. Transfer learning enables predictions in network biology. Nature 618, 616–624 (2023).
Cui, H. et al. scGPT: toward building a foundation model for single-cell multi-omics using generative AI. Nat. Methods 21, 1470–1480 (2024).
Hao, M. et al. Large scale foundation model on single-cell transcriptomics. Nat. Methods 21, 1481–1491 (2024).
Chen, Y. & Zou, J. Simple and effective embedding model for single-cell biology built from ChatGPT. Nat. Biomed. Eng. 9, 483–493 (2025).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 30, (2017).
Prokhorenkova, L. et al. Catboost: unbiased boosting with categorical features. Adv. Neural Inf. Process. Syst. 31; https://proceedings.neurips.cc/paper_files/paper/2018/file/14491b756b3a51daac41c24863285549-Paper.pdf (2018).
Szklarczyk, D. et al. String v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Fabregat, A. et al. The reactome pathway knowledgebase. Nucleic Acids Res. 46, D649–D655 (2018).
Du, J. et al. Gene2vec: distributed representation of genes based on co-expression. BMC Genomics 20, 7–15 (2019).
Horlbeck, M. A. et al. Mapping the genetic landscape of human cells. Cell 174, 953–967 (2018).
Van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Tribouilloy, C. et al. Benfluorex and valvular heart disease. Presse Med. 40, 1008–1016 (2011).
Jiang, Jiayue-Clara, Hu, C., McIntosh, A. M. & Shah, S. Investigating the potential anti-depressive mechanisms of statins: a transcriptomic and mendelian randomization analysis. Transl. Psychiatry 13, 110 (2023).
Blake, G. J. & Ridker, P. M. Are statins anti-inflammatory? Trials 1, 161 (2000).
McGown, C. C., Brown, N. J., Hellewell, P. G., Reilly, C. S. & Brookes, Z. L. S. Beneficial microvascular and anti-inflammatory effects of pravastatin during sepsis involve nitric oxide synthase III. Br. J. Anaesth. 104, 183–190 (2010).
Sommeijer, D. W. et al. Anti-inflammatory and anticoagulant effects of pravastatin in patients with type 2 diabetes. Diabetes Care 27, 468–473 (2004).
Giurgiu, M. et al. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 47, D559–D563 (2019).
Reeders, S. T. et al. A highly polymorphic DNA marker linked to adult polycystic kidney disease on chromosome 16. Nature 317, 542–544 (1985).
Hopp, K. et al. Functional polycystin-1 dosage governs autosomal dominant polycystic kidney disease severity. J. Clin. Invest. 122, 4257–4273 (2012).
Rossetti, S. et al. Incompletely penetrant PKD1 alleles suggest a role for gene dosage in cyst initiation in polycystic kidney disease. Kidney Int. 75, 848–855 (2009).
Gainullin, V. G. et al. Polycystin-1 maturation requires polycystin-2 in a dose-dependent manner. J. Clin. Invest. 125, 607–620 (2015).
Lanktree, M. B., Haghighi, A., di Bari, I., Song, X. & Pei, Y. Insights into autosomal dominant polycystic kidney disease from genetic studies. Clin. J. Am. Soc. Nephrol. 16, 790 (2021).
Radhakrishnan, Y., Duriseti, P. & Chebib, F. T. Management of autosomal dominant polycystic kidney disease in the era of disease-modifying treatment options. Kidney Res. Clin. Pract. 41, 422 (2022).
Duan, Q. et al. L1000CDS2: LINCS L1000 characteristic direction signatures search engine. NPJ Syst. Biol. Appl. 2, 16015 (2016).
Gbelcová, H. et al. Variability in statin-induced changes in gene expression profiles of pancreatic cancer. Sci. Rep. 7, 44219 (2017).
Huang, Tong-sheng et al. Long-term statins administration exacerbates diabetic nephropathy via ectopic fat deposition in diabetic mice. Nat. Commun. 14, 390 (2023).
Hernán, M. A. & Robins, J. M. Using big data to emulate a target trial when a randomized trial is not available. Am. J. Epidemiol. 183, 758–764 (2016).
Abadie, A. & Imbens, G. W. Matching on the estimated propensity score. Econometrica 84, 781–807 (2016).
Chen, T. & Guestrin, C. XGBoost: a scalable tree boosting system. In Proc. 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD '16). 785–794 (Association for Computing Machinery, 2016); https://doi.org/10.1145/2939672.2939785
Kalatharan, V. et al. Positive predictive values of international classification of diseases, 10th revision coding algorithms to identify patients with autosomal dominant polycystic kidney disease. Can. J. Kidney Health Dis. 3, 2054358116679130 (2016).
Friberg, L., Gasparini, A. & Carrero, JuanJesus A scheme based on ICD-10 diagnoses and drug prescriptions to stage chronic kidney disease severity in healthcare administrative records. Clin. Kidney J. 11, 254–258 (2018).
Cadnapaphornchai, M. A. et al. Effect of pravastatin on total kidney volume, left ventricular mass index, and microalbuminuria in pediatric autosomal dominant polycystic kidney disease. Clin. J. Am. Soc. Nephrol. 9, 889 (2014).
Leuenroth, S. J. et al. Triptolide is a traditional Chinese medicine-derived inhibitor of polycystic kidney disease. Proc. Natl Acad. Sci. USA 104, 4389–4394 (2007).
Leuenroth, S. J., Bencivenga, N., Igarashi, P., Somlo, S. & Crews, C. M. Triptolide reduces cystogenesis in a model of ADPKD. J. Am. Soc. Nephrol. 19, 1659 (2008).
Chevalley, M. et al. The CausalBench challenge: a machine learning contest for gene network inference from single-cell perturbation data. Preprint at https://arxiv.org/abs/2308.15395 (2023).
Kaplan, J. et al. Scaling laws for neural language models. Preprint at https://arxiv.org/abs/2001.08361 (2020).
Hoffmann, J. et al. Training compute-optimal large language models. Preprint at https://arxiv.org/abs/2203.15556 (2022).
Leek, J. T. et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 11, 733–739 (2010).
Errington, T. M. et al. Investigating the replicability of preclinical cancer biology. eLife 10, e71601 (2021).
Pratapa, A., Jalihal, A. P., Law, J. N., Bharadwaj, A. & Murali, T. M. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat. Methods 17, 147–154 (2020).
Schwab, P. et al. Learning counterfactual representations for estimating individual dose–response curves. In AAAI Conference on Artificial Intelligence (2020).
Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. Preprint at https://arxiv.org/abs/1412.6980 (2014).
Wu, Y. et al. Variational causal inference. Preprint at https://arxiv.org/abs/2209.05935 (2022).
Ye, C. et al. Drug-seq for miniaturized high-throughput transcriptome profiling in drug discovery. Nat. Commun. 9, 4307 (2018).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576 (2017).
Norman, T. M. et al. Exploring genetic interaction manifolds constructed from rich single-cell phenotypes. Science 365, 786–793 (2019).
Miladinovic, D. et al. perturblib/perturblib: DOI-archived v0.1. Zenodo https://doi.org/10.5281/zenodo.15671137 (2025).
Jiang, X. & Prabhakar, A. et al. Control of ribosomal protein synthesis by the microprocessor complex. Sci. Signal. 14, eabd2639 (2021).
Gimpel, C. et al. International consensus statement on the diagnosis and management of autosomal dominant polycystic kidney disease in children and young people. Nat. Rev. Nephrol. 15, 713–726 (2019).
Torres, V. E. et al. Tolvaptan in patients with autosomal dominant polycystic kidney disease. N. Engl. J. Med. 367, 2407–2418 (2012).
Wang, X. et al. Protein kinase a downregulation delays the development and progression of polycystic kidney disease. J. Am. Soc. Nephrol. 33, 1087–1104 (2022).
Wang, X., Wu, Y., Ward, C. J., Harris, P. C. & Torres, V. E. Vasopressin directly regulates cyst growth in polycystic kidney disease. J. Am. Soc. Nephrol. 19, 102 (2008).
Colosimo, E., Ferreira, F., Oliveira, M. & Sousa, C. Empirical comparisons between Kaplan–Meier and Nelson–Aalen survival function estimators. J. Stat. Comput. Sim. 72, 299–308 (2002).
Davidson-Pilon, C. lifelines: survival analysis in Python. J. Open Source Softw. 4, 1317 (2019).
Deng, K. & Guan, Y. A supervised LightGBM-based approach to the GSK.ai CausalBench Challenge (ICLR 2023). OpenReview https://openreview.net/forum?id=nB9zUwS2gpI (2023).
Acknowledgements
We thank X. L. Zhao, C. Weis and S. Bauer for their valuable feedback.
Author information
Authors and Affiliations
Contributions
D.M. and P.S. initiated the project. D.M., T.H., A.G., M.C. and A.M. contributed to model development. D.M., A.G., T.H. and L.S. contributed to software development and conducted experiments. D.M., A.G., T.H. and L.S. contributed to writing the manuscript. P.S., M.B. and B.S. provided supervision and strategic direction.
Corresponding authors
Ethics declarations
Competing interests
D.M., M.C., A.G., L.S., A.M., M.B. and P.S. are employees and shareholders of GSK plc. T.H. is a former employee of GSK plc.
Peer review
Peer review information
Nature Computational Science thanks Fotis Psomopoulos and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Kaitlin McCardle, in collaboration with the Nature Computational Science team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Performance of LPM as a function of training data availability.
Performance comparison in terms of Pearson correlation coefficient ρ in predicting the outcomes of unseen experiments of LPM when varying a. the number of perturbations available for training in a target context and b. the number of different contexts available for training. Dots correspond to individual runs with a different random seed, and the blue line corresponds to the inferred trend (the average value with SD depicted in shaded blue). The dashed grey line denotes the performance of the ‘NoPerturb’ baseline, which does not take perturbation information into account. The performance of LPM is significantly increased (* = p ≤0.05, ** = p ≤0.01; one-sided Mann-Whitney test) when more perturbations and more contexts are available.
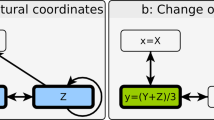
Extended Data Fig. 2 Model architecture.
a. Graphical model shows the dependencies between random variables previously described in Section 4.1. Dashed lines indicate implicit bi-directional dependencies that enable transfer learning across datasets. Symbolic perturbation, readout, and context descriptors (P,R,C) are first embedded (ZP, ZR, ZC), then used to generate output Y that represents the value of the readout R. b. Embeddings are implemented as learnable look-up tables. P, R, and C identify indices in the corresponding tables. c. Concatenated embeddings are forward propagated through a multilayer perceptron to predict the output Y
Supplementary information
Supplementary Information
Supplementary text, Supplementary Tables 1–4 and Supplementary Figs. 1 and 2.
Source data
Source Data Fig. 2
Source data accompanying the corresponding figure.
Source Data Fig. 3
Source data accompanying the corresponding figure.
Source Data Fig. 4
Source data accompanying the corresponding figure.
Source Data Fig. 6
Source data accompanying the corresponding figure.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Miladinovic, D., Höppe, T., Chevalley, M. et al. In silico biological discovery with large perturbation models. Nat Comput Sci 5, 1029–1040 (2025). https://doi.org/10.1038/s43588-025-00870-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s43588-025-00870-1
This article is cited by
-
Interpretation, extrapolation and perturbation of single cells
Nature Reviews Genetics (2026)
-
Interpolating perturbations across contexts
Nature Computational Science (2025)