Abstract
Background
Strategies to treat inflammatory skin conditions require identifying new targets involved in interactions between overlying epithelial and underlying dermal immune cells. Scavenger receptor class B type 1 (SR-B1) is a cell surface receptor that binds high-density lipoproteins (HDL) and mediates inflammatory responses in immune and endothelial cells. The SR-B1 receptor is also expressed in keratinocytes, but its role in inflammatory skin diseases remains unexplored.
Methods
To investigate keratinocyte SR-B1 in the setting of inflammation, we measured its expression in skin biopsy samples obtained from patients with psoriasis; human skin explants exposed to the inflammatory cytokine, interleukin-17A (IL-17A); and mouse skin exposed to the pro-inflammatory agent, imiquimod (IMQ). We also evaluated the effects of SR-B1 knockdown on primary keratinocyte responses to IL-17A. Finally, we employed a synthetic HDL-nanoparticle (HDL NP) to investigate the therapeutic potential of targeting SR-B1 in IL-17A-stimulated keratinocytes and in male C57BL/6 mice with IMQ-induced skin inflammation.
Results
Our data show SR-B1 expression is increased in diseased human skin and in both human and mouse models of skin inflammation. SR-B1 knockdown in keratinocytes exacerbates the inflammatory response to IL-17A, whereas targeting SR-B1 with HDL NP attenuates this response. In the IMQ murine model, topical application of HDL NPs improves the skin phenotype, normalizes SR-B1 expression, and reduces molecular and cellular markers of inflammation.
Conclusions
Overall, SR-B1 plays a role in skin inflammation and HDL NP-mediated targeting of SR-B1 in keratinocytes may offer a targeted new therapy for inflammatory skin disease.
Plain Language Summary
Inflammatory skin diseases, like psoriasis, are common and often lack effective treatment options. A protein, called scavenger receptor class B type 1 (SR-B1), known to regulate immune responses, is present in skin cells but its role in skin inflammation has not been well studied. We show that SR-B1 expression in the skin increases in psoriasis and regulates cellular inflammatory responses. We developed a molecule that targets SR-B1 and showed that it reduces disease markers in skin cells and improves outcomes in a mouse model of psoriasis-like skin inflammation. These findings suggest that SR-B1 plays a role in skin inflammation and could be targeted with topical agents to develop new treatments for inflammatory skin disease.
Similar content being viewed by others
Introduction
Epithelial and surveilling innate immune cells are the first line of defense against chemical, mechanical, microbial, and other insults at biological barriers1,2,3. Pathologic changes resulting from acute and chronic inflammation can affect barrier organs and tissues including the skin (e.g., psoriasis and atopic dermatitis), vasculature (e.g., atherosclerosis), lung (e.g., asthma) and intestine (e.g., inflammatory bowel disease)4,5,6. In many cases, a re-enforcing cycle of communication between epithelial cells and immune cells perpetuates disease resulting in progression and relapse. Local and infiltrating immune cells produce inflammatory mediators that activate epithelial cells and alter differentiation leading to cell proliferation and tissue damage7,8,9. Epithelial cells also drive pathogenic processes by secreting chemokines, cytokines, and reactive oxygen species (ROS) that amplify inflammatory signaling and immune cell recruitment5,8,9. As such, understanding new mechanisms of targeting epithelial cells to inhibit inflammatory cycles that perpetuate disease is a promising opportunity for developing novel therapeutics that can be topically delivered.
Targeting epithelial cells requires identification of molecules that participate in the maintenance of epithelial cell integrity and that become dysregulated in disease. The lipoprotein receptor, scavenger receptor class B type 1 (SR-B1), is expressed in epithelial tissues10,11,12,13 and is a known therapeutic target for modulating immune and vascular endothelial cells in the context of immunometabolic disease (e.g., atherosclerosis)14,15,16. Moreover, SR-B1 is upregulated in numerous malignancies where it supports cell survival and proliferation under metabolic and redox stress17. In the skin, the SR-B1 receptor has been shown to directly contribute to epithelial cell differentiation and wound repair13,18,19. Interestingly, cigarette smoke was found to induce time-dependent changes in SR-B1 expression and localization in keratinocytes20, which potentially reflects its dynamic role in skin injury response mechanisms. Therefore, better understanding the role of SR-B1 in epithelial inflammatory responses could uncover mechanistic insights in the study of inflammatory diseases affecting barrier organs. Further, it provides the opportunity for SR-B1 ligands to be used as novel diagnostics and targeted therapeutics.
High-density lipoprotein (HDL) endowed with the defining protein, apolipoprotein A-I (apo A-I), is the high-affinity ligand of SR-B116. Native HDL binding to SR-B1 can result in the exchange of HDL-associated lipids (e.g., phospholipids, cholesterol, cholesterol ester, lipophilic antioxidants, and biologically active lipids), proteins, and nucleic acids between the HDL particle and the cell16,21,22. As such, HDLs can directly modulate cellular lipid metabolism, the fluidity and integrity of the cell membrane, and cell-membrane anchored second messenger signaling pathways. Moreover, HDLs binding SR-B1 has been shown to reduce inflammation, oxidation, and promote wound healing21,23,24,25. It has been shown that HDLs dampen lipopolysaccharide (LPS)-induced inflammation in monocytes, stimulate endothelial nitric oxide synthase (eNOS)26, and can inhibit antigen presentation and proliferation in lymphocytes27,28.
The function(s) of HDL binding SR-B1 is dependent upon the physicochemical properties of HDL particles23,29. HDLs are dynamic, varying in size ( ~ 7-18 nm), shape, composition, and their ability to bind certain lipoprotein receptors21,30. Assigning specific structure-function relationships to natural HDLs is complicated by their heterogeneity. To overcome this challenge and more precisely study the relationship between HDL, SR-B1, and target cells and tissues, we developed functional synthetic HDL-like nanoparticles (HDL NPs) which have well-defined size, shape, and composition as well as a high binding affinity to SR-B131,32,33. Previous studies verify that HDL NPs with defined composition exert anti-inflammatory effects both in LPS-induced monocytes and macrophages and in mouse models of mechanically and chemically injured and inflamed corneas34,35. These data reinforce that SR-B1 is a target in inflammation and that HDL NPs can act as tools to study SR-B1 immunomodulation in the epithelium.
Psoriasis is a prevalent inflammatory disease of the skin with characteristic changes in keratinocyte proliferation, differentiation, and expression of inflammatory mediators9. In particular, interleukin 17 A (IL-17A) is a predominant cytokine involved in inflammatory skin conditions, particularly in psoriasis36,37. Here, we evaluate the role of SR-B1 in the setting of skin inflammation. Data demonstrate an increase in the expression of SR-B1 in skin biopsies obtained from patients with psoriasis, as well as in human skin explants and keratinocytes exposed to IL-17A. Additionally, knockdown of SR-B1 in keratinocytes amplifies inflammatory signaling in response to IL-17A. To probe potential therapeutic effects in psoriasis-like model systems, we utilize synthetic HDL NPs. Data demonstrate that HDL NPs target SR-B1 in keratinocytes and attenuate inflammatory signaling in vitro. Mechanistic studies reveal that SR-B1 functions with IL-17A/IL-17RA signaling in keratinocytes, and synthetic HDL NP binding SR-B1 reduces downstream IL-17RA-signaling. Finally, HDL NP ameliorates the inflammatory phenotype in psoriasis-like imiquimod (IMQ)-induced mouse models.
Methods
HDL NP Synthesis
HDL NPs were synthesized using an established protocol34,35. Briefly, 5 nm diameter citrate stabilized colloidal gold nanoparticles (Au NP) were incubated with a 5-fold molar excess of purified human apoA-1 (MyBioSource, San Diego, CA) for 1 hour at room temperature (RT) using a flat bottom shaker. The inner lipid, 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[3-(2-pyridyldithio)propionate] (PDP-PE) (Avanti Polar Lipids, Alabaster, AL) dissolved in dichloromethane (CH2Cl2, 1 mM) was added at a 250-fold molar excess relative to [Au NP] and the solution was briefly vortexed. Next, outer lipids, cardiolipin (heart, bovine) (CL) (Avanti Polar Lipids, Alabaster, AL) and 1,2-dilinoleoyl-sn-glycero-3-phospho-(1’-rac-glycerol) (18:2 PG) (Avanti Polar Lipids, Alabaster, AL) dissolved in CH2Cl2 (1 mM) were added to the Au NP/apoA-I/PDP-PE solution each at 125-fold molar excess to the Au NP and the solution was briefly vortexed. The NP mixture was subjected to alternating vigorous vortexing and sonication (20 s each) until the solution became opaque and pink in color. The mixture was added to a magnetic stir plate and subject to continuous mixing at room temperature for 16 hours to fully remove CH2Cl2. The solution went from opaque and pink in color to a clear solution that was colored deep red, which is consistent with the starting solution of colloidal AuNPs. The resulting HDL NPs were purified using tangential flow filtration (TFF) (KrosFlo Research Iiii TFF System, Repligen, Waltham, MA). The size, charge and concentration of HDL NPs was measured by dynamic light scattering (DLS) and UV-Vis spectroscopy (Agilent, Santa Clara, CA) where the extinction coefficient (ε) for 5 nm diameter gold nanoparticles (AuNP) is 9.696 ×106 M-1 cm-1 at λmax = 520 nm.
To synthesize fluorescently labeled HDL NPs, the intercalating dye, DiI (1,1’-Dioctadecyl-3,3,3’,3’-tetramethylindocarbocyanine perchlorate) was incorporated. DiI-HDL NPs were synthesized by adding DiI (ThermoFisher, Waltham, MA) at a 1 mM final concentration during the phospholipid addition step. Purification and quantification of DiI-HDL NPs were conducted as described above. Fluorescent labeling was confirmed using the Cytation™3 Multi-Mode plate reader (BioTek, Winooski, VT) using excitation and emission wavelengths of 545 and 570 nm, respectively.
Human Psoriasis Samples and Ex Vivo Explants
Skin tissue from five patients suffering from chronic plaque-type psoriasis were obtained through the Northwestern University Skin Biology & Diseases Resource-based Center (NU SBDRC) tissue bank. Four millimeter (mm) skin punch biopsy samples were obtained and fixed in formalin and embedded in paraffin. All patients signed informed consent, and the study was approved by the Northwestern University Internal Review Board (NU IRB; #STU00024696). Patient demographic information is reported in Supplementary Table S1. Skin specimens for full-thickness ex vivo skin explants were obtained by the NU SBDRC following abdominoplasty surgeries performed at Northwestern Memorial Hospital. The deidentified skin specimens were considered discarded materials according to NU IRB policy, thus patient consent was not required. Tissue was subsequently obtained from the NU SBDRC-Skin Tissue Engineering and Morphology (STEM) Core. All tissues were collected in compliance with NU IRB (#STU00009443). Explant experiments were performed in biological duplicates using the same donor tissue, with a total of three different donors included across independent experiments. Ten mm biopsy samples were individually mounted on metal grids. The grids were placed in 60 mm dishes and maintained in high glucose Dulbecco’s Modified Eagle’s Medium (DMEM) (Sigma-Alrich, St. Louis, MO) supplemented with 10% Fetal Bovine Serum (FBS), 0.50 µg/mL amphotericin B and 20 µg/mL gentamicin at an air-liquid interface at 37 °C and 5% CO2. After 24 hours of culture, skin explants were maintained with or without IL-17A (10 ng/mL), and the medium was changed every other day. After 4 days, the explants were harvested for histological analysis38. Formalin-fixed and paraffin-embedded human skin explants were sectioned (4 µm) and mounted on glass slides. For immunohistochemistry (IHC), slides were deparaffinized before heat-induced epitope retrieval using BOND Epitope Retrieval Solution 1 (Leica Biosystems, Deerk Park, IL). Slides were then processed using the automated Leica BOND Polymer Refine IHC protocol which consisted of a 5-minute peroxide block, 15-minute staining for SR-B1 (1:250; Novus Biologicals, Centennial, CO), an 8-minute post-primary polymer application, an 8-minute secondary antibody detection, and visualization with the Mixed DAB Refine reagent for 10 minutes. Finally, slides were counterstained with hematoxylin for 5 minutes. All IHC processing was conducted by the NU Robert H. Lurie Comprehensive Cancer Center Pathology Core Facility. SR-B1 staining was quantified using ImageJ software by counting the number of positively stained cells in the epidermis relative to the length of the epidermis (µm).
Primary Cell Culture
Human neonatal foreskin tissue was used to isolate primary keratinocytes. The use of human skin tissue is determined as “Not Human Research” by NU IRB as no patient information is linked to the samples and identifiable. Ultimately, human epidermal keratinocytes for this study were isolated from fresh, deidentified foreskin collected from the NU SBDRC with IRB approval (#STU00009443). The deidentified foreskin specimens were considered discarded materials according to IRB policy, thus legal guardian/parental consent was not required.
Normal human epidermal keratinocytes (NHEKs) were isolated from the samples following established protocols from the NU SBDRC. Foreskin samples were washed twice with 1X PBS (5 mL per wash) to remove debris. The dermal side was carefully trimmed using sterile forceps and scissors to remove residual fat and blood vessels. The trimmed foreskin was placed epidermis-side up in a 60 mm dish containing 2-3 mL Dispase II (Roche, Indianapolis, IN) and incubated at 4 °C overnight. The following day, the epidermis was gently separated from the dermis and transferred to a 100 mm dish containing TrypLE (ThermoFisher Scientific, Waltham, MA). After 10-15 minutes of incubation at 37 °C, normal calf serum (500 μL; ThermoFisher Scientific, Waltham, MA) was added to deactivate the trypsin. To release keratinocytes into solution, the epidermal tissue was scraped against the dish using sterile forceps. The cell suspension was diluted with PBS and filtered through a 40 μm strainer. Cells were pelleted by centrifugation at 220 x g for 5 minutes, resuspended, and cultured. Cells were maintained in M154CF media (Cascade Biologics, Portland, OR) supplemented with 1% Human Keratinocyte Growth Supplements (ThermoFisher Scientific, Waltham, MA), 0.07 mM CaCl2, 0.25 µg/mL amphotericin B (CellGro, Lincoln, NE) and 10 µg/mL gentamicin (Sigma Aldrich, St. Louis, MO). To induce an undifferentiated cell state that resembles the basal layer of the epidermis, the culture medium was supplemented with 0.03 mM CaCl2. Cells were co-treated with or without IL-17A (R&D Systems, Minneapolis, MN), or HDL NPs (40 nM) for 3-24 hours. In a subset of experiments, confluent monolayers were maintained for 4 days in culture media supplemented with 1.2 mM CaCl2 to induce keratinocyte differentiation.
Transfection with siRNA against SR-B1
siRNA against SR-B1 (SR-B1; AM16708) and control siRNA (ctrl siRNA; AM4611) consisting of a scrambled sequence were purchased from Invitrogen (ThermoFisher Scientific, Waltham, MA). NHEKs (350,000 cells / well) were incubated with 100 nM of siRNA oligo at the time of seeding using M154 Complete medium without antibiotics. The following day, the culture medium was replenished with fresh medium containing 0.03 mM CaCl2. After an additional incubation for 48 hours, siRNA-transfected NHEKs were co-treated with or without IL-17A (R&D Systems, Minneapolis, MN) or HDL NPs (10 nM) for 24 hours. Experiments were performed independently using cells from three different donors.
3D Human Skin Equivalents
Organotypic 3-dimensional human skin equivalents (HSEs) were constructed according to protocols provided by the NU SBDRC and consistent with previously published methods39. Collagen plugs containing 3T3-J2 fibroblasts were supplied by the NU SBDRC. NHEKs were resuspended in E medium (DMEM F12; 18 µM adenine; 0.5 µg/ml human recombinant insulin; 0.5 µg/ml human apotransferrin; 0.5 µg/ml triiodothyronine; 10 µg/ml gentamicin; 0.25 µg/ml amphotericin B; 4 mM L-glutamine; 0.4 µg/ml hydrocortisone; 10 ng/ml cholera toxin; and 5% fetal bovine serum) containing 5 ng/mL epidermal growth factor (EGF). A total of 600,000 cells suspended in 1 mL of medium were seeded onto each collagen plug. The following day, the E-medium + EGF was replaced. After 48 hours, organotypic cultures were lifted onto wire mesh and maintained at the air-liquid interface. One day after lifting to the air-liquid interface, IL-17A (25 ng/mL) was added to the culture media. Beginning 7 days after cytokine initiation, PBS or HDL NPs (40 nM) were added to the media every other day. The cultures were harvested on day 13 and either lysed for RNA analysis, fixed in 10% neutral-buffered formalin for histological analyses, or embedded in OCT media for immunofluorescent studies. Experiments were performed in biological duplicates and independently repeated using HSEs from two different donors.
Western Blotting
To assess changes in signaling pathways downstream of IL-17RA, undifferentiated cultures were co-treated with IL-17A (100 ng/mL) and HDL NPs (40 nM) for 3 hours. NHEKs were lysed in urea-based buffer containing 10 M urea, 1% w/v sodium dodecyl sulfate (SDS), 10% v/v glycerol, 60 mM Tris (pH 6.8), 0.01% w/v pyronin Y (Abcam, Waltham, MA), and protease and phosphatase inhibitor cocktails (ThermoFisher Scientific, Waltham, MA)40. Protein concentration was determined by the Amido Black method40. Lysates were loaded on SDS-PAGE gels (Mini-PROTEAN TGX Stain Free Fels, Bio-Rad, Hercules, CA) per standard protocol. For the detection of different proteins, specific antibodies were purchased from Abcam (SR-B1, ab52629, 1:1000), Santa Cruz Biotechnology [GAPDH (sc-365062, 1:2000), JNK (sc-7345, 1:200)], and Cell Signaling [phospho-SAPK/JNK (9251, 1:1000), total IκBα (4814, 1:1000)]. Bound antibody was detected by chemiluminescence (ECL, Bio-Rad, Hercules, CA). All western blots were performed three independent times using NHEKs from three different donors.
IMQ-Induced Psoriasis-Like Inflammation Model in Mice and HDL Treatment
Animal studies were conducted in accordance with the Guide for the Care and Use of Laboratory Animals and were approved by the Animal Care and Use Committee (IACUC) at Northwestern University. Male C57BL/6 mice (Jackson Laboratory, Bar Harbor ME) at 6 weeks of age were shaved and depilated under general anesthesia (n = 5/6 per condition). Each mouse was housed in an individual cage. Starting on Day 1, 62.5 mg imiquimod (IMQ) cream (5%) (Taro, Hawthorne, NY) was applied to the exposed dorsal skin for 4 consecutive days. Experimental groups received equal volumes of HDL NPs (750 nM in dH2O) mixed into a commercially available ointment. Control treatments included the vehicle, composed of equal volumes of dH2O mixed in the same ointment base. HDL NP and vehicle applications (50 µL) were topically administrated twice a day, at least 3 hours after IMQ application. A subset of mice received no IMQ or other treatments (untreated control). All mice were included in the downstream analyses. Disease severity was evaluated daily according to a modified clinical Psoriasis Area and Severity Index (PASI)41. Erythema, scaling, and thickening parameters were scored on a scale from 0 (no incidence of disease) to 4 (severe) and totaled to generate the daily cumulative score for each mouse (0-12). On day 5, mouse skins were harvested and either fixed in 10% neutral-buffered formalin for histological analyses, embedded in OCT media for immunofluorescent staining, or snap-frozen for qRT-PCR analyses. Histological processing and hematoxylin and eosin (H&E) staining were prepared by NU SBDRC-STEM. Epidermal thickness was measured on 3-4 non-overlapping images of each H&E-stained skin strip using ImageJ software (NIH). Briefly, 4-5 straight lines were drawn uniformly throughout each image from the basal layer of the epidermis to the granular layer, excluding the stratum corneum. The length of each line was measured and averaged across each image for each group.
Immunofluorescence (IF)
OCT-embedded mouse skin samples were cryosectioned at 5 µm thickness. Sections were fixed with 4% paraformaldehyde (PFA) in PBS for 10 min at RT, permeabilized with 0.1% Tween, and blocked with 10% normal goat serum for 1 hour at RT. Sections were incubated with the selected primary antibody overnight at 4 °C [rabbit anti-LOR (1:200; BioLegend, San Diego, CA), rabbit anti-SR-B1 (1:200; Novus Biologics, Centennial, CO) and mouse anti-CD3 (1:100; ThermoFisher Scientific)] followed by secondary antibody [Cy3-conjugated goat anti-mouse or AlexaFluor488-conjugated goat anti-rabbit (Jackson ImmunoResearch Laboratories, West Grove Pennsylvania)] for 1 hour at RT. Nuclei were stained with DAPI and the slides were viewed by confocal microscopy on a Nikon W1 Dual CAM Spinning Disk. LOR and CD3 signal were analyzed on 3-4 non-overlapping images using ImageJ software (NIH). For LOR analysis, regions of interest (ROIs) were drawn to include the epidermis across the image and mean fluorescent signal intensity was measured. For CD3 analysis, ROIs were drawn to isolate only the dermis, and CD3-positive cells were counted using ImageJ’s Cell Counter tool. The mean epidermal LOR intensity and number of CD3-positive dermal cells were divided by the epidermal length in each section and averaged across all images per sample. SR-B1 signal was analyzed on 1-5 non-overlapping images using QuPath42. Briefly, epidermal regions of interest were manually selected with QuPath’s polygon annotation tool, excluding the stratum corneum to avoid non-specific staining. Individual cells were identified by applying a threshold to the DAPI channel using QuPath’s Cell Detection command. The mean signal intensity of DAPI and GFP was measured for each detected cell, and the average GFP/DAPI signal per cell was then calculated.
To assess p65 nuclear translocation, undifferentiated cultures were grown on Nunc Lab-Tek II Chamber Slide System (ThermoFisher Scientific, Waltham, MA) and co-treated with IL-17A (100 ng/mL) and either HDL NPs for 3 hours. IF was conducted as described above, using a mouse anti-p65 primary antibody (1:100; SantaCruz Biotechnology, Dallas, TX).
BrdU
Keratinocyte proliferation was detected by BrdU incorporation. Mice were intraperitoneally injected with a single dose of 0.5 g/kg body weight of BrdU (Sigma-Aldrich, St. Louis, MO) 1 hour prior to mice sacrifice. For in vitro proliferation assays, BrdU (10 µM) was added to cultures left untreated or treated with IL-17A in the presence or absence of HDL NPs (40 nM) for 24 hours. Experiments were conducted in duplicate and repeated independently using keratinocytes from two different donors. OCT-embedded mouse sections or culture wells were fixed in 10% methanol for 20 minutes at -20 °C and then air-dried. Samples were washed three times for 5 minutes each with 1X PBS, followed by antigen retrieval in 2 N HCl for 30 minutes at RT. After retrieval, samples were washed three times for 5 minutes each with 0.5X TBE (tris-borate-EDTA) buffer to remove HCl, followed by sequential washes with 1X PBS, PBS-Tween (PBST), and PBS for 5 minutes each. Samples were then blocked with 10% normal goat serum in PBS at RT for 1 hour. Sections were stained with mouse-anti-BrdU (1:10; Creative BioLabs, Shirley, NY) and Cy3-labeled goat-anti-mouse as described above. Nuclei were stained with DAPI, and the slides were visualized by confocal microscopy. BrdU signal was analyzed on 3-4 non-overlapping images using ImageJ software (NIH). ROIs were drawn to isolate only the epidermis, and BrdU-positive cells were counted using ImageJ’s Cell Counter tool. The number of BrdU-positive cells were divided by the epidermal length in each section and averaged across all images per sample.
qRT-PCR
To assess chemokine expression in NHEKs under non-siRNA conditions, undifferentiated cultures were co-treated with IL-17A (100 ng/mL) and either PBS or HDL NPs (40 nM) for 24 hours. All experiments were performed in biological triplicates and repeated three independent times using NHEKs from three different donors. Total RNA was extracted using the RNeasy purification system (Qiagen, Valencia, CA). Reverse transcription of 1 µg total RNA was conducted using High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific, Waltham, MA). Real-time PCR was performed using SYBR Green PCR Master Mix (ThermoFisher Scientific, Waltham, MA) or TaqMan Fast Advanced Master Mix (ThermoFisher Scientific, Waltham, MA) on a CFX Connect Real-Time PCR Detection System (BioRad, Hercules, CA). The SYBR cycling protocol was 5 °C for 2 min; 95 °C for 10 min; then forty cycles of amplification involving melting at 95 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 45 s. The TaqMan cycling protocol was 50 °C for 2 min; 95 °C for 20 seconds; then forty cycles of amplification involving melting at 95 °C for 3 s, and annealing / extension at 60 °C for 30 s. Gene expression levels were normalized to the level of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and fold differences in expression were calculated using the Comparative CT method. RNA analysis of NHEKs under siRNA conditions, 3D HSEs and mouse skin followed the same protocol, with mouse skin first homogenized with the Qiagen Powerlyzer 24. PCR primers or TaqMan catalog numbers are listed in Supplementary Table S2.
PhenoCycler
OCT-embedded mouse skin samples were cryosectioned at 5 µm thickness and submitted to the Northwestern University’s Immunotherapy Assessment Core. The slides from -80 °C were dried in Drierite beads for 5 minutes and fixed in Acetone for 10 minutes. Slides were washed twice in hydration buffer and fixed with 1.6% PFA (Electron Microscopy Sciences, Hatfield, PA) for 10 minutes followed by two washes with hydration buffer. For antibody staining, an antibody cocktail was prepared with optimal dilutions of each antibody in a buffer containing N, J, S and G blockers (SKU 7000017; Akoya Biosciences, Marlborough, MA). Samples were first allowed to equilibrate to room temperature in Staining Buffer for 20–30 minutes, followed by incubation in a pre-blocking solution made of N, J and S blockers in Staining Buffer. The antibody cocktail containing barcoded antibodies from Akoya Biosciences: CD3-BX021 (AKYP0035) — Alexa Fluor 647, CD11b-BX025 (AKYP0040) — Alexa Fluor 488, CD11c-BX030 (AKYP0045) — Alexa Fluor 647, CD45-BX007 (AKYP0005) — Alexa Fluor 488, Ly6G-BX024 (AKYP0039) — Alexa Fluor 647, MHC II–BX014 (AKYP0006) — Atto 550 were subsequently added to the slide, and slides were incubated at room temperature for 3 hours. Slides were then rinsed with Staining buffer, fixed with 1.6% PFA in Storage buffer solution, and fixed in ice-cold methanol for 5 minutes at 4 °C. A third round of fixation with PhenoCycler Fixative reagent was applied, and slides were stored in Storage Buffer at 4 °C until ready to image. Reporters were added in 96WP plate by diluting in Reporter Stock solution as described in the PhenoCycler-Fusion User Guide with 1X Buffer, Assay Reagent, and Nuclear Stain from Akoya Biosciences. Antibody-stained slides were next equilibrated at room temperature in 1X PBS for at least 10 minutes. The flow cell was assembled onto the slide as described in the user guide. The slides were incubated in 1X additive Buffer again for 10 minutes to ensure secure sealing of the flow cell to the slide. The slide was then transferred to the flow cell carrier and imaged on a PhenoCycler-Fusion (Akoya Biosciences, Marlborough, MA). After images of all cycles had been acquired, the final QPTIFF file containing a composite image of all markers was viewed using the QuPath software.To quantify each marker, dermal ROIs were manually selected with QuPath’s polygon annotation tool. Individual cells were identified by applying a threshold to the DAPI channel using QuPath’s Cell Detection command. The QuPath Single Measurement Classifier was then applied to all DAPI-positive cells to classify cells according to each channel. The total number of cells per channel was divided by the epidermal length in each ROI and averaged across all ROIs per sample.
ICP-MS Analysis
Fresh full-thickness skin samples (n = 5 per group) were stored immediately at −80 °C until prepped for ICP-MS analysis. ICP-MS analysis was conducted at the Chemistry of Life Processes Core Facility at Northwestern University after digestion of the tissues and Au NPs using nitric acid and hydrogen peroxide (4:1 v/v), diluted with 1% hydrochloric acid. The amount of Au NPs was quantified with reference to calibrated additional standards.
HDL Binding Assay
Undifferentiated NHEKs were incubated with DiI-HDL NPs for 3 hours at 37 °C, with or without 1:100 SR-B1 blocking antibody (Novus Biologicals, Centennial CO), or 1:100 rabbit IgG isotype control antibody (Novus Biologicals, Centennial CO). Cells were washed with 1X PBS, collected, and resuspended in 300 µl 1X PBS. DAPI stain (1:100) was added to each sample prior to flow cytometric analysis. DiI fluorescence was quantified using the Cy3-channel of the BD LSR II Fortessa located at the NU Robert H. Lurie Comprehensive Cancer Center Flow Cytometry Core Facility (Chicago, IL). The experiment was performed in biological triplicates, with data analyzed using FlowJo software.
Transmission Electron Microscopy (TEM)
To visualize HDL NP binding to keratinocyte cell membranes, undifferentiated NHEKs were co-treated with IL-17A (100 ng/mL) and HDL NPs (40 nM) for 24 hours. Cells were fixed in 2% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer and processed for thin sectioning43. Ultrathin sections were made with a Leica Ultracut ultramicrotome at the NU Center for Advanced Microscopy, mounted on grids, and the grids were imaged with a FEI Tecnai Spirit G2 transmission electron microscope operating at 68 kV. Experiments were performed in both technical and biological duplicates.
Cholesterol Efflux Assay
Cholesterol efflux assays were performed using tritium-labeled cholesterol ([3H]-chol) following an adapted protocol44. Briefly, NHEKs were seeded at 250,000 cells per well in 24 well plates in M154 Complete medium on Day 1. On Day 2, labeling medium was prepared in M154 Complete medium containing [3H]-chol (2 µCi/mL) and Sandoz (2 µg/mL) (Sigma-Aldrich, St. Louis, MO). Note that Sandoz is an ACAT inhibitor used to prevent esterification of [3H]-chol. Labeling media was added and incubated for 24 hours. On Day 3, cells were washed twice in PBS and efflux samples containing either IL-17A and HDL NPs (40 nM) or IL-17A and PBS were incubated in triplicate for 3 hours at 37 °C. Efflux media was then removed, vacuum filtered, and added to 3 mL UltimaGold (PerkinElmer, Waltham, MA) scintillation fluid for scintillation counting. A separate triplicate cohort of [3H]-chol-loaded NHEKs stimulated with IL-17A was washed, air dried, and incubated at room temperature in isopropanol to extract [3H]-chol as a measure of total cholesterol at t = 0 at the beginning of efflux. Percent cholesterol efflux was calculated as the ratio of counts in the media to counts for t = 0 NHEKs. Experiments were performed in biological triplicates and repeated twice using NHEKs from two different donors.
Membrane Fluidity Assay
Membrane fluidity was assessed by using the Membrane Fluidity Kit (ab189819) (Abcam, Waltham, MA), which measures the changes in fluorescence spectral properties of lipid analog probes upon spatial interaction with the cell membrane. Undifferentiated NHEKs were left untreated or co-treated with IL-17A (100 ng/mL) and either PBS or HDL NPs (40 nM) for 3 hours. Isopropanol (0.5%), which increases the membrane fluidity, was used as a positive control. Cells were then incubated with a lipid analog probe for 1 hour at RT in the dark. Fluorescence shifts were monitored by exciting cells at 350 nm and taking emission values at 400 nm and 470 nm using the Cytation™3 Multi-Mode plate reader (BioTek, Winooski, VT). Membrane fluidity was quantified by the ratio of emission at 470 nm to emission at 400 nm. Experimental groups were conducted in triplicate and normalized to that of unlabeled cell conditions.
Statistical Analysis
Data were represented as mean ± SD. Statistical analysis of significance was calculated using either Student’s unpaired t test (when comparing two variables), one-way ANOVA (when comparing multiple variables) followed by post hoc analysis using Tukey’s test, or two-way ANOVA (when comparing interactions between two independent variables) using GraphPad Prism 10 (GraphPad Software, San Diego, CA). Statistical significance was set at *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
SR-B1 in Psoriasis and Skin Explants and Human Keratinocytes Exposed to IL-17A
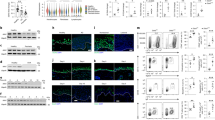
To investigate the expression of SR-B1 in the setting of skin inflammation, IHC was performed on tissue sections obtained from psoriatic skin lesions (n = 5) and human ex vivo skin exposed to IL-17A, which was confirmed to induce effective inflammatory stimulation through increased production of key antimicrobial peptides (Supplementary Table S1, Supplementary Fig. S1)38,45. Non-treated ex vivo skin samples obtained from the same individuals were used as controls (NTC)46. Imaging revealed that SR-B1 mostly localized to the basal epidermal layer in normal tissue (Fig. 1a, Supplementary Fig. S2), which is consistent with previous reports20. In psoriatic lesions and explanted human skin tissue treated with IL-17A, SR-B1 expression in the basal layers increased and SR-B1 expression extended into suprabasal layers (Fig. 1a, Supplementary Fig. S2). Analysis revealed a 3.3-fold increase (mean ± SD = 0.445 ± 0.061) in the total number of SR-B1 positive cells per length of epidermis (µm) in psoriatic skin compared to NTC (mean ± SD = 0.137 ± 0.043, t-test, p < 0.0001, 95% CI of the difference [0.232, 0.385]), and a 1.5-fold increase (mean ± SD = 0.158 ± 0.017) in IL-17A stimulated samples compared to NTC (mean ± SD = 0.106 ± 0.006, t-test, p = 0.0072, 95% CI of the difference [0.023, 0.081]) (Fig. 1b). To confirm these data in keratinocytes, western blotting was conducted on cultured, primary normal human epidermal keratinocytes (NHEKs) subjected to calcium-induced differentiation and treated with IL-17A (Fig. 1c, Supplementary Fig. S3). The terminal differentiation marker loricrin (LOR) was used to verify keratinocyte differentiation. Consistent with prior studies13, SR-B1 expression was decreased upon keratinocyte differentiation induced by high calcium. The IL-17A treatment promoted an increase in SR-B1 in differentiated NHEKs. To further understand the role of SR-B1 in modulating the IL-17A response in keratinocytes, we assessed the expression of IL-17A-driven chemokines and antimicrobial peptides following siRNA-mediated reduction of SR-B136,47. Specifically, we measured the expression of chemokine, CXCL1, and the antimicrobial peptide, S100A7, which are prominent chemoattractants for T cells, macrophages, and neutrophils, as well as markers of psoriasis36,48,49. NHEKs were transfected with either control siRNA (ctrl siRNA) or SR-B1 siRNA (SR-B1 siRNA) for 72 hours, followed by IL-17A treatment (100 ng/mL) for 24 hours (Supplementary Fig. S4). RT-PCR analyses revealed that IL-17A stimulation in SR-B1 siRNA-treated NHEKs increased the expression of CXCL1 (mean ± SD = 5.38 ± 1.28, two-way ANOVA, p = 0.0035, 95% CI of the difference [1.14, 4.74]) and S100A7 (mean ± SD = 15.1 ± 0.654, two-way ANOVA, p = 0.0060, 95% CI of the difference [2.58, 12.23]) compared to IL-17A stimulation in NHEKs treated with ctrl siRNA (Fig. 1d). These data further support that SR-B1 functions to reduce IL-17A-medieated inflammation in keratinocytes.
a Representative images following immunohistochemical (IHC) staining for SR-B1 in the epidermis from biopsy specimens obtained from patients diagnosed with psoriasis (n = 5), non-treated control human skin explant tissue (NTC) (n = 5), and IL-17A-treated human skin explant tissue (n = 3) (scale bar, 50 µm). The area above the dashed line represents the epidermis. Arrows indicate example SR-B1 positive cells (brown). b Mean number ± SD of SR-B1 positive cells relative to the length of epidermis (µm) (t-test, **p < 0.01, ***p < 0.0001). IL-17A-treated human skin explant tissue was compared to matched NTC human skin explant tissue (n = 3). c NHEKs were grown in either low calcium (0.03 mM) or high calcium (1.2 mM) to induce differentiation. Cultures were stimulated with IL-17A (100 ng/mL) for 48 hours prior to harvesting on day 2 or 4. SR-B1 and LOR expression were assessed by immunoblotting. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the loading control. Blots are representative of three independent experiments. Densitometric quantification of the ratio of SR-B1 to GAPDH and LOR to GAPDH normalized to undifferentiated, untreated NHEKs is shown below the blot. d NHEKs were transfected with control siRNA (ctrl siRNA) and siRNAagainst SR-B1 (SR-B1 siRNA) and subsequently treated with IL-17 (100 ng/mL) for 24 hours. CXCL1 and S100A7 expression were assessed by RT-PCR. Gene expression was normalized to GAPDH and shown as fold changes relative to untreated ctrl siRNA cells. Five independent experiments were performed; representative data from one experiment are shown, with values expressed as mean ± SD (two-way ANOVA, **p < 0.01).
HDL NPs Modulate Keratinocytes Stimulated with IL-17A
We sought to determine whether targeting SR-B1 by HDL NPs would modulate the IL-17A response in cultured NHEKs. We chose to study undifferentiated NHEKs as this phenotype demonstrates the highest expression of SR-B1 (Fig. 1c) and is consistent with our findings in the human psoriatic skin samples and ex vivo human skin equivalents exposed to IL-17A (Fig. 1a, b). Keratinocytes were exposed to IL-17A for 24 hours and the expression of IL-17A-driven chemokines, CCL20 and CXCL1, and antimicrobial peptides, S100A7 and S100A9, were measured36,47,48,49. IL-17A resulted in expected increases in relative gene expression of all markers as measured by RT-PCR (Fig. 2a). Conversely, HDL NP treatment of NHEKs stimulated with IL-17A significantly decreased gene expression of CCL20 by 54% (mean ± SD = 3.48 ± 0.440, one-way ANOVA, p = 0.012, 95% CI of the difference [-7.09, -1.19]), CXCL1 by 78% (mean ± SD = 0.911 ± 0.118, one-way ANOVA, p < 0.0001, 95% CI of the difference [-3.95, -2.69]), S100A7 by 52% (mean ± SD = 2.93 ± 0.897, one-way ANOVA, p = 0.0046, 95% CI of the difference [-4.99, -1.31]), and S100A9 by 55% (mean ± SD = 3.71 ± 0.661, one-way ANOVA, p = 0.0012, 95% CI of the difference [-6.66, -2.53]) as compared to IL-17A alone (Fig. 2a). To demonstrate that this effect was SR-B1-dependent, NHEKs were treated with IL-17A and HDL NP following knockdown of SR-B1 via siRNA transfection. Following SR-B1 siRNA treatment, HDL NPs no longer reduced the expression of CXCL1 (mean ± SD = 1.27 ± 0.001) compared to IL-17A alone (mean ± SD = 1.21 ± 0.034, two-way ANOVA, p = 0.413, 95% CI of the difference [-0.082, 0.204]; Fig. 2b). Similarly, in SR-B1 siRNA conditions, HDL NPs reduced S100A7 expression by only 41.9% (mean ± SD = 1.69 ± 0.077) relative to IL-17A alone (mean ± SD = 2.91 ± 0.143, two-way ANOVA, p = 0.0012, 95% CI of the difference [-1.66, -0.786]) compared to a 77.3% reduction in the ctrl siRNA conditions (IL-17A alone: mean ± SD = 1.00 ± 0.140, IL-17A + HDL NP: mean ± SD = 0.227 ± 0.011, two-way ANOVA, p = 0.010, 95% CI of the difference [-1.13, -0.259]; Fig. 2b).
a NHEKs were left untreated or stimulated with IL-17A (100 ng/mL) in the presence or absence of HDL NPs (40 nM) for 24 hours. RT-PCR of immune biomarkers associated with IL-17A stimulation in NHEKs. Gene expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and reported as fold changes relative to untreated cells. Representative data are shown, with values expressed as mean ± SD (n = 3) from three independent studies. b NHEKs were transfected with control siRNA (ctrl siRNA) and siRNA against SR-B1 (SR-B1 siRNA) and subsequently treated with IL-17 (100 ng/mL) in the presence or absence of HDL NPs (10 nM) for 24 hours. CXCL1 and S100A7 expression were assessed by RT-PCR. Gene expression was normalized to GAPDH and shown as fold changes relative to IL-17A-treated ctrl siRNA cells. Representative data are shown, with values expressed as mean ± SD from three independent studies (two-way ANOVA, **p < 0.01). c, d Representative immunofluorescent images of NHEKs stained for BrdU. The mean ± SD number of BrdU positive cells per high-powered field (HPF) were measured across three images and performed in duplicate. e, f 3D human skin equivalents (HSEs) were stimulated with or without IL-17A (25 ng/mL) for 13 days. Starting on day 7, PBS or HDL NPs were added to the HSE growth medium. e RT-PCR of the indicated genes in HSEs. The mean values ± SD of two independent experiments measured in duplicates are shown. (f) Tissue sections were stained for loricrin (green) and DAPI (blue) and imaged (scale bar, 50 µm). a, d, e Data sets were analyzed by one-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
The anti-proliferative effects of HDL NPs in cultured keratinocytes were additionally investigated using BrdU labeling. Undifferentiated NHEKs maintain a proliferative state (mean ± SD = 517 ± 51.7), which was not significantly increased by exposure to IL-17A (mean ± SD = 524.5 ± 47.24, one-way ANOVA, p = 0.959, 95% CI of the difference [-64.55, 79.88]; Fig. 2c, d). However, the addition of HDL NP led to a significant decrease (31-32%) in BrdU incorporation (mean ± SD = 356 ± 45.4) when compared to untreated (one-way ANOVA, p < 0.0001, 95% CI of the difference [-234, -89.1]) or IL-17A (one-way ANOVA, p < 0.0001, 95% CI of the difference [-241, -96.8]; Fig. 2c, d). Next, we confirmed data obtained using cultured NHEKs in the organotypic 3D human skin equivalent (HSEs) model. Similar to cultured NHEK cells, we added IL-17A to the HSEs one day after lifting the cultures to the air-liquid interface45. Beginning on day seven, either PBS (vehicle) or HDL NPs were added to the HSE growth medium. Following treatment, changes in gene expression were quantified. HDL NPs significantly reduced the levels of CCL20 (mean ± SD = 3.19 ± 0.178, one-way ANOVA, p = 0.040, 95% CI of the difference [-10.3, -0.257]), CXCL1 (mean ± SD = 4.50 ± 1.78, one-way ANOVA, p = 0.043, 95% CI of the difference [-8.68, -0.154]), and S100A7 (mean ± SD = 3.21 ± 0.349, one-way ANOVA, p = 0.033, 95% CI of the difference [-4.50, -0.205]) compared to IL-17A + PBS (Fig. 2e). Similarly, HDL NPs increased the expression of LOR, which was strongly inhibited by the addition of IL-17A (Fig. 2f).
HDL NP Targets SR-B1 and Suppresses IL-17R Signaling Pathways in Keratinocytes
Our group showed that HDL NP are functional ligands of SR-B150 that can be molecularly tuned to reduce inflammation34,35, modulate the uptake of exogenous lipid vesicles (e.g., exosomes)51, and induce cancer cell death52,53. Like native HDLs54,55, the function of HDL NPs are driven, in part, by the ability to modulate cell cholesterol flux and change cell membrane dynamics33,50,52. Changes in the cell membrane can consequently induce changes in receptor distribution and signaling51,56,57,58. Accordingly, we hypothesized that HDL NP binding SR-B1 alters IL-17R signaling in IL-17A-stimulated NHEKs. We first confirmed that HDL NPs functionally engage SR-B1 in keratinocytes. Data demonstrated that HDL NP exhibits site-specific binding to SR-B1, induced cholesterol efflux, and modulated membrane fluidity (Supplementary Fig. S5, S6). We next investigated if HDL NP binding SR-B1 modulated known cell-membrane anchored 1L-17RA signal transduction pathways. Accordingly, we examined the activation of JNK and NF-κB signaling17 in NHEKs left untreated or treated with IL-17A in the presence or absence of HDL NPs (40 nM). The HDL NPs inhibited the activation of both the JNK and NF-κB pathways induced by IL-17A, as shown by decreased phosphorylation of JNK and reduced degradation of total IκBα (Fig. 3a, Supplementary Fig. S7). Similarly, IF analysis of the NF-κB subunit p65 revealed that HDL NP treatment inhibited nuclear translocation of p65, which is required for NF-κB mediated transcription of downstream inflammatory target genes (Fig. 3b).
NHEKs were left untreated or stimulated with IL-17A (100 ng/mL) in the presence or absence of HDL NPs (40 nM) for 3 hours. a Western blots for mediators of signaling pathways downstream of IL-17RA. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the loading control. Data are representative of three independent experiments. Densitometric quantification of the ratio of p-JNK to t-JNK and t-IκBα to GAPDH is shown below the respective blot. b Immunofluorescent images from cells stained for NFκB p65 (red) and DAPI (blue) (scale bar, 25 µm).
HDL NPs Attenuate the Psoriasis-Like Phenotype in Mice Exposed to Topical Imiquimod
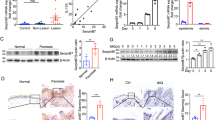
To validate our in vitro findings, we investigated the effects of topically applied HDL NPs on the psoriasis-like phenotype observed in mice topically treated with IMQ59,60. Treatments were applied daily at separate times on the shaved back skin of mice for 4 consecutive days to most effectively capture the early inflammatory responses (Fig. 4a)61,62. No adverse reactions or events were noted over the course of the study. HDL NP delivery and penetration into the epidermal layers of mouse skin was confirmed by inductively couple plasma mass spectrometry (ICP-MS) and confocal microscopy, respectively (Supplementary Fig. S8). The psoriasis-like phenotype was evaluated by comparing the skin of untreated mice with those treated with IMQ + Vehicle (IMQ + Veh) and IMQ + HDL NP. Psoriasis Area and Severity Index (PASI) scores (0-12) that subjectively quantify the severity of skin erythema, thickness, and scaling were assigned daily. By day 5, data demonstrate that HDL NP treatment resulted in a significantly improved PASI score (mean ± SD = 5.0 ± 1.0) when compared to IMQ + Veh (mean ± SD = 7.2 ± 1.6; one-way ANOVA, p = 0.022, 95% CI of the difference [-4.07, -0.326]; Fig. 4b, c). Histopathological changes were analyzed by H&E staining. Results demonstrated that HDL NPs attenuated the development of characteristic psoriatic pathology including the loss of the epidermal granular layer, parakeratosis, acanthosis, and inflammatory cell infiltration (Fig. 4d). Mice treated with HDL NPs demonstrated a significant reduction in mean epidermal thickness (39.5 ± 3.59 µm) as compared to mice treated with IMQ + Veh (49.7 ± 2.89 µm, one-way ANOVA, p = 0.007, 95% CI of the difference [-9.05, -1.58]; Fig. 4e). Topical application of IMQ in mice additionally induces cellular changes including increased keratinocyte proliferation and reduced keratinocyte differentiation59. To investigate if HDL NP mitigated these processes, keratinocyte proliferation and differentiation were quantified using confocal microscopy. The HDL NP treatment significantly decreased IMQ-induced hyperproliferation of keratinocytes, as shown by a reduction in the incorporation of BrdU in basal cell keratinocytes (mean ± SD = 0.034 ± 0.011) as compared to IMQ + Veh (mean ± SD = 0.056 ± 0.009, one-way ANOVA, p = 0.013, 95% CI of the difference [-0.039, -0.005]; Fig. 4f, g). Additionally, HDL NP treatment restored the expression of the terminal differentiation marker, LOR (mean ± SD = 1.30 ×104 ± 2.39 ×103), which is diminished in IMQ-induced mice (mean ± SD = 8.56 ×103 ± 2.62 ×103, one-way ANOVA, p = 0.042, 95% CI of the difference [172, 8710]; Fig. 4h, i)59,63. As SR-B1 levels increased in psoriatic skin and cytokine-stimulated keratinocytes (Fig. 1), we aimed to assess how SR-B1 expression changes following the observed improvement in the IMQ-induced phenotype (Fig. 4j, k). Consistent with findings from Fig. 1, immunofluorescent analysis revealed an increase in the expression and distribution of SR-B1 in mouse skin treated with IMQ + Veh (mean ± SD = 5.40 ± 0.446) versus untreated controls (mean ± SD = 3.33 ± 0.760, one-way ANOVA, p = 0.0038, 95% CI of the differences [0.763, 3.37]). Importantly, treatment with HDL NPs significantly reduced SR-B1 levels (mean ± SD = 4.19 ± 0.758, one-way ANOVA, p = 0.036, 95% CI of the differences [-2.34, -0.083]) and normalized SR-B1 expression patterns, with higher expression localized primarily to the basal layers.
a Mice were left untreated or treated daily with topical IMQ for 5 days. Control ointment (vehicle) or HDL NPs were administered topically in a subset of mice four hours after IMQ application (n = 5 per group). b, c Representative clinical pictures of mice on day 5. Psoriasis Area Severity Index (PASI) score measuring erythema, scaling, and skin thickness was assigned daily for each parameter. The line graph depicts the mean totaled score ± SD over the treatment course. d, e Representative hematoxylin and eosin (H&E) staining of mouse skin (scale bar, 50 µm). The epidermal thickness of the H&E sections was measured by ImageJ software. Data are represented as the mean ± SD. f, g Immunofluorescent (IF) staining for BrdU was conducted. Arrows indicate example BrdU positive cells. The bar graph depicts the mean number of BrdU positive cells per length of epidermis (µm) ± SD. h, i Representative IF images of mice skin sections stained for loricrin (LOR). The bar graph depicts the mean signal intensity ± SD of LOR in the epidermis. j, k Representative IF images of mice skin sections stained for scavenger receptor class B type I (SR-B1). The bar graph depicts the mean cellular signal intensity of SR-B1 over the mean signal intensity of DAPI ± SD. c, e, g, i, k The area between white dashed lines represents the epidermis. Scale bar, 50 µm. Data sets were analyzed by one-way ANOVA (*p < 0.05, **p < 0.01, ***p < 0.001).
HDL NPs Attenuate Inflammation Observed in Mice Exposed to Topical Imiquimod
Next, we sought to evaluate the impact of HDL NP treatment on keratinocyte activation and the inflammatory response by quantifying the expression of key cytokines and chemokines involved in IMQ-induced inflammation (Fig. 5a). We observed significant reductions in the expression of IL-17A (mean ± SD = 31.1 ± 14.8, t-test, p = 0.040, 95% CI of the difference [-36.4, -1.08]), IL-1β (mean ± SD = 1.18 ± 0.397, t-test, p = 0.005, 95% CI of the difference [-6.76, -1.60]), and CXCL1 (mean ± SD = 3.11 ± 1.23, t-test, p = 0.0006, 95% CI of the difference [-5.89, -2.23]) in the IMQ + HDL NP-treated mice compared to the IMQ + Veh group. Additionally, expression levels of CCL20, S100A7 and S100A9 were lower in the HDL NP-treated group, though these differences did not quite reach statistical significance. Moreover, we investigated if reductions in cytokines and chemokines were associated with changes in immune cell recruitment using immunofluorescence. Dermal expression of the general T-cell marker, CD364, significantly decreased in mice treated with HDL NPs (mean ± SD = 0.025 ± 0.014) as compared to the IMQ + Veh control (mean ± SD = 0.046 ± 0.012, one-way ANOVA p = 0.037, 95% CI of the difference [-0.041, -0.001]; Fig. 5a, b). To further evaluate innate immune responses, PhenoCycler multiplexed immunofluorescence imaging was conducted on a subset of mice (n = 2 per group; Fig. 5c, Supplementary Fig. S9). As compared to IMQ + Veh-treated skin, IMQ + HDL NP-treated skin led to a reduction in markers for general immune cells (CD45 + ), T cells (CD3 + ), macrophages (CD11b + ), neutrophils (Ly6g+ and CD11b + ), antigen presenting cells (APCs, MHC-II), and dendritic cells (CD11c + ) in the dermis. Overall, our data suggests a role for SR-B1 in inflammatory skin disease and introduces a novel therapeutic approach using synthetic HDL NPs that target SR-B1 and, subsequently, reduces pro-inflammatory signaling.
a RT-PCR of inflammatory biomarkers associated with IMQ-induced inflammation (untreated, n = 3; IMQ + Veh, n = 6; IMQ + HDL NP, n = 6). Gene expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and shown as fold changes relative to untreated mouse skin. Data sets are presented as mean ± SD and analyzed by unpaired t-test (*p < 0.05, **p < 0.01, ***p < 0.001). b, c Representative immunofluorescent images of mice skin sections stained for CD3 (scale bar, 50 µm). The bar graph depicts the number of CD3 positive cells present in the dermis (below white dashed line) per length of epidermis (µm) ± SD. Data sets (n = 5 mice per group) were analyzed by one-way ANOVA (*p < 0.05, **p < 0.01). d A subset of samples underwent PhenoCycler multiplex imaging (n = 2 per group). The left panel shows representative images of all markers (scale bar, 200 µm). To the right, each panel includes two markers indicated in the bottom left corner (scale bar, 50 µm). DAPI is shown in blue.
Discussion
The majority of individuals with mild-to-moderate inflammatory skin disease receive treatment with topical agents that are not specific to diseased cells and can cause a myriad of adverse effects65,66. Furthermore, patients with severe disease often resort to systemic biologics that, despite their effectiveness, are costly and carry the risk of severe side effects. Here, we report a novel marker, SR-B1, whose expression is increased in inflamed human and mouse skin. Further, we demonstrate that a high-affinity ligand of SR-B1 reduces inflammatory signaling in keratinocytes. As such, targeting SR-B1 with high-affinity functional SR-B1 ligands, like HDL NP, may be a novel approach to reduce inflammatory cycles between epithelial and underlying immune cells and improve healing of inflammatory skin conditions.
Our work was motivated by our findings, and those of others, that SR-B1 is differentially expressed in cells that are exposed to inflammatory mediators67,68,69 and cells under increased metabolic and redox stress70. In keratinocytes, SR-B1 expression is highest in undifferentiated versus differentiated keratinocytes13. In the current study, we observed an increase in the expression and distribution of SR-B1 in i) psoriatic skin samples, ii) human skin explants stimulated with IL-17A, and iii) mice skin treated with the pro-inflammatory agent, IMQ. Using western blotting, we confirmed increased SR-B1 expression in cultured normal human epidermal keratinocytes (NHEKs) exposed to IL-17A. While changes in the expression of SR-B1 have been reported in select cases of skin injury20, this is the first report that suggests SR-B1 may play a more general role in inflammatory diseases of the skin.
To further investigate the role of SR-B1 in keratinocytes, we silenced SR-B1 in IL-17A-stimulated NHEKs and observed a marked enhancement in pro-inflammatory chemokine production. Moreover, targeting SR-B1 using HDL NPs significantly reduced both chemokine expression and cell proliferation, as well as enhanced keratinocyte differentiation. Synthetic HDL NPs were selected due to the challenges of targeting SR-B1 with native HDLs, which, despite their anti-inflammatory and antioxidative functions, present considerable heterogeneity in their physicochemical composition23. The HDL NP platform enables precise control over particle form and composition to more accurately measure and then tune HDL NP function(s)31,33,35,50. Further, the HDL NP specifically targets SR-B1 and retains some functions of native HDLs35,51,52. To confirm the SR-B1-dependent effects by HDL NPs, we demonstrated that the anti-inflammatory response was absent or diminished following SR-B1 knockdown.
We demonstrate that SR-B1 is an immunomodulatory receptor in keratinocytes. Similar findings have been reported in macrophages, where SR-B1 deletion in primary bone marrow derived macrophages (BMMs) led to an elevation in the production of pro-inflammatory cytokines upon LPS stimulation15,71. Gain-of-function studies further demonstrated that SR-B1 overexpression attenuated the pro-inflammatory response15. As LPS activates toll-like-receptor 4 (TLR4), studies have indicated that SR-B1 also dampens downstream TLR4 signaling, particularly the NF-κB and MAP kinase pathways where JNK and p38 were found to be key mediators responsible for the heightened LPS-induced cytokine response between wild-type and SR-B1-null cells15. Regarding mechanism, these findings prompted us to investigate whether targeting SR-B1 by HDL NPs might similarly suppress downstream IL-17RA signaling. Interestingly, our results show that HDL NPs reduced the activation of JNK and NF-κB mediators, suggesting that, like macrophages, SR-B1 may serve a similar regulatory role in inflammation within keratinocytes.
Building on our in vitro findings, we were motivated to test the function of HDL NPs in vivo. To elucidate the therapeutic effects of HDL NPs, we used the well-studied IMQ mouse model where topical application of IMQ induces a significant, IL-17A-dependent inflammatory response in the treated mouse skin59,64,72. Data demonstrate that HDL NPs mitigate the IMQ-induced inflammatory phenotype. Notably, we observed improvements in key pathologies driven by epithelial processes including the proliferation and differentiation of keratinocytes and the recruitment of leukocytes. Based upon our data, we anticipate that the observed therapeutic effect was the result of HDL NP targeting keratinocyte SR-B1; however, we cannot completely rule out that HDL NP may also target infiltrating SR-B1+ immune cells where the HDL NPs have been shown to reduce inflammation54. Future studies can be conducted to map targeted cell types within both normal and inflamed skin. From a translational perspective, work is required to optimize the topical delivery of HDL NP to ensure active targeting of the intended cell populations. Additionally, it should be noted that numerous other co-factors are involved in IMQ-induced injury (and psoriasis) beyond IL-17A, which was the focus of study in the in vitro studies. As such, our current findings may not fully capture the broader impact of HDL NP-SR-B1 interactions within complex inflammatory environments. Future work will focus on exploring how HDL NP targeting SR-B1 functions across a wider range of inflammatory signals to better understand the generality and robustness of HDL NP’s anti-inflammatory effects.
In summary, this work highlights the immunomodulatory role of SR-B1 in keratinocytes and the therapeutic potential of HDL NPs in treating inflammatory skin diseases. Future research should focus on expanding the mechanisms through which SR-B1 engagement regulates inflammation, preclinical studies to optimize drug formulations and delivery, and evaluate long-term safety and efficacy. Eventually, comparative studies with existing therapies, such as topical steroids, could establish HDL NPs as a viable alternative with fewer side effects and may broaden applications extending beyond the skin to address other acute and chronic immuno-epithelial inflammatory diseases.
Data availability
Source data, datasets generated and/or analyzed during the current study, are available in the paper or are appended as Supplementary Data. The data supporting the findings of this study are available from the corresponding author upon request. Requests for the data should be submitted to: cthaxton003@northwestern.edu.
References
Schleimer, R. P., Kato, A., Kern, R., Kuperman, D. & Avila, P. C. Epithelium: at the interface of innate and adaptive immune responses. J. Allergy Clin. Immunol. 120, 1279–1284 (2007).
Larsen, S. B., Cowley, C. J. & Fuchs, E. Epithelial cells: liaisons of immunity. Curr. Opin. Immunol. 62, 45–53 (2020).
Hewitt, R. J. & Lloyd, C. M. Regulation of immune responses by the airway epithelial cell landscape. Nat. Rev. Immunol. 21, 347–362 (2021).
Hiemstra, P. S., McCray, P. B. Jr. & Bals, R. The innate immune function of airway epithelial cells in inflammatory lung disease. Eur. Respir. J. 45, 1150–1162 (2015).
Dainichi, T. & Iwata, M. Inflammatory loops in the epithelial-immune microenvironment of the skin and skin appendages in chronic inflammatory diseases. Front Immunol. 14, 1274270 (2023).
Coskun, M. Intestinal epithelium in inflammatory bowel disease. Front Med (Lausanne) 1, 24 (2014).
Martini, E., Krug, S. M., Siegmund, B., Neurath, M. F. & Becker, C. Mend Your Fences: The Epithelial Barrier and its Relationship With Mucosal Immunity in Inflammatory Bowel Disease. Cell. Mol. Gastroenterol. Hepatol. 4, 33–46 (2017).
Piipponen, M., Li, D. & Landén, N. X. The Immune Functions of Keratinocytes in Skin Wound Healing. Int. J. Mol. Sci 21 (2020).
Zhou, X., Chen, Y., Cui, L., Shi, Y. & Guo, C. Advances in the pathogenesis of psoriasis: from keratinocyte perspective. Cell Death Dis. 13, 81 (2022).
Shen, W. J., Hu, J., Hu, Z., Kraemer, F. B. & Azhar, S. Scavenger receptor class B type I (SR-BI): a versatile receptor with multiple functions and actions. Metabolism 63, 875–886 (2014).
Duncan, K. G., Bailey, K. R., Kane, J. P. & Schwartz, D. M. Human retinal pigment epithelial cells express scavenger receptors BI and BII. Biochem Biophys. Res Commun. 292, 1017–1022 (2002).
Cai, S. F., Kirby, R. J., Howles, P. N. & Hui, D. Y. Differentiation-dependent expression and localization of the class B type I scavenger receptor in intestine. J. Lipid Res 42, 902–909 (2001).
Tsuruoka, H. et al. Scavenger receptor class B type I is expressed in cultured keratinocytes and epidermis. Regulation in response to changes in cholesterol homeostasis and barrier requirements. J. Biol. Chem. 277, 2916–2922 (2002).
Gracia-Rubio, I., Martín, C., Civeira, F. & Cenarro, A. SR-B1, a Key Receptor Involved in the Progression of Cardiovascular Disease: A Perspective from Mice and Human Genetic Studies. Biomedicines 9 (2021).
Cai, L., Wang, Z., Meyer, J. M., Ji, A. & van der Westhuyzen, D. R. Macrophage SR-BI regulates LPS-induced pro-inflammatory signaling in mice and isolated macrophages. J. Lipid Res 53, 1472–1481 (2012).
Powers, H. R. & Sahoo, D. SR-B1’s Next Top Model: Structural Perspectives on the Functions of the HDL Receptor. Curr. Atheroscler. Rep. 24, 277–288 (2022).
Gaffen, S. L. Structure and signalling in the IL-17 receptor family. Nat. Rev. Immunol. 9, 556–567 (2009).
Muresan, X. M. et al. Involvement of cutaneous SR-B1 in skin lipid homeostasis. Arch. Biochem. Biophys. 666, 1–7 (2019).
Muresan, X. M. et al. SR-B1 involvement in keratinocytes in vitro wound closure. Arch. Biochem. Biophys. 658, 1–6 (2018).
Sticozzi, C. et al. Cigarette smoke affects keratinocytes SRB1 expression and localization via H2O2 production and HNE protein adducts formation. PLoS One 7, e33592 (2012).
Jomard, A. & Osto, E. High Density Lipoproteins: Metabolism, Function, and Therapeutic Potential. Front Cardiovasc Med 7, 39 (2020).
Shen, W. J., Azhar, S. & Kraemer, F. B. SR-B1: A Unique Multifunctional Receptor for Cholesterol Influx and Efflux. Annu Rev. Physiol. 80, 95–116 (2018).
Nazir, S. et al. Interaction between high-density lipoproteins and inflammation: Function matters more than concentration! Adv. Drug Deliv. Rev. 159, 94–119 (2020).
Trakaki, A. & Marsche, G. Current Understanding of the Immunomodulatory Activities of High-Density Lipoproteins. Biomedicines 9 (2021).
Yu, B.-l, Wang, S.-h, Peng, D.-q & Zhao, S.-pH. D. L. and immunomodulation: an emerging role of HDL against atherosclerosis. Immunol. Cell Biol. 88, 285–290 (2010).
Tran-Dinh, A. et al. HDL and endothelial protection. Br. J. Pharm. 169, 493–511 (2013).
Wang, S. H., Yuan, S. G., Peng, D. Q. & Zhao, S. P. HDL and ApoA-I inhibit antigen presentation-mediated T cell activation by disrupting lipid rafts in antigen presenting cells. Atherosclerosis 225, 105–114 (2012).
Grao-Cruces, E., Lopez-Enriquez, S. & Martin, M. E. & Montserrat-de la Paz, S. High-density lipoproteins and immune response: A review. Int. J. Biol. Macromol. 195, 117–123 (2022).
Mulder, W. J. M. et al. High-Density Lipoprotein Nanobiologics for Precision Medicine. Acc. Chem. Res 51, 127–137 (2018).
Fazio, S. & Pamir, N. HDL Particle Size and Functional Heterogeneity. Circ. Res 119, 704–707 (2016).
Thaxton, C. S., Daniel, W. L., Giljohann, D. A., Thomas, A. D. & Mirkin, C. A. Templated spherical high density lipoprotein nanoparticles. J. Am. Chem. Soc. 131, 1384–1385 (2009).
Luthi, A. J. et al. Nanotechnology for synthetic high-density lipoproteins. Trends Mol. Med 16, 553–560 (2010).
Luthi, A. J. et al. Tailoring of biomimetic high-density lipoprotein nanostructures changes cholesterol binding and efflux. ACS Nano 6, 276–285 (2012).
Wang, J. et al. HDL nanoparticles have wound healing and anti-inflammatory properties and can topically deliver miRNAs. Adv Ther (Weinh) 3 (2020).
Foit, L. & Thaxton, C. S. Synthetic high-density lipoprotein-like nanoparticles potently inhibit cell signaling and production of inflammatory mediators induced by lipopolysaccharide binding Toll-like receptor 4. Biomaterials 100, 67–75 (2016).
Furue, M., Furue, K., Tsuji, G. & Nakahara, T. Interleukin-17A and Keratinocytes in Psoriasis. Int J Mol Sci 21 (2020).
Mosca, M. et al. The Role of IL-17 Cytokines in Psoriasis. Immunotargets Ther. 10, 409–418 (2021).
Pfaff, C. M., Marquardt, Y., Fietkau, K., Baron, J. M. & Lüscher, B. The psoriasis-associated IL-17A induces and cooperates with IL-36 cytokines to control keratinocyte differentiation and function. Sci. Rep. 7, 15631 (2017).
Lewandowski, K. T. et al. Topically Delivered Tumor Necrosis Factor-α-Targeted Gene Regulation for Psoriasis. J. Invest Dermatol 137, 2027–2030 (2017).
Cable, C. J., Kaplan, N., Getsios, S., Thomas, P. M. & Perez White, B. E. Biotin Identification Proteomics in Three-Dimensional Organotypic Human Skin Cultures. Methods Mol. Biol. 2109, 185–197 (2020).
Neu, S. D. et al. Myeloperoxidase Inhibition Ameliorates Plaque Psoriasis in Mice. Antioxidants (Basel) 10 (2021).
Bankhead, P. et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Park, J. K. et al. MicroRNAs-103/107 coordinately regulate macropinocytosis and autophagy. J. Cell Biol. 215, 667–685 (2016).
Henrich, S. E., Hong, B. J., Rink, J. S., Nguyen, S. T. & Thaxton, C. S. Supramolecular Assembly of High-Density Lipoprotein Mimetic Nanoparticles Using Lipid-Conjugated Core Scaffolds. J. Am. Chem. Soc. 141, 9753–9757 (2019).
Singh, S. et al. Long-Term and Clinically Relevant Full-Thickness Human Skin Equivalent for Psoriasis. ACS Appl. Bio Mater. 3, 6639–6647 (2020).
Senra, L. et al. Keratinocyte-Derived IL-17E Contributes to Inflammation in Psoriasis. J. Invest Dermatol. 136, 1970–1980 (2016).
Varshney, P. et al. Transcriptome profiling unveils the role of cholesterol in IL-17A signaling in psoriasis. Sci. Rep. 6, 19295 (2016).
Zdanowska, N., Kasprowicz-Furmańczyk, M., Placek, W. & Owczarczyk-Saczonek, A. The Role of Chemokines in Psoriasis-An Overview. Medicina (Kaunas) 57 (2021).
Liang, H., Li, J. & Zhang, K. Pathogenic role of S100 proteins in psoriasis. Front. Immunol. 14 (2023).
Luthi, A. J. et al. Robust passive and active efflux of cellular cholesterol to a designer functional mimic of high density lipoprotein. J. Lipid Res 56, 972–985 (2015).
Plebanek, M. P. et al. Nanoparticle Targeting and Cholesterol Flux Through Scavenger Receptor Type B-1 Inhibits Cellular Exosome Uptake. Sci. Rep. 5, 15724 (2015).
Yang, S. et al. Biomimetic, synthetic HDL nanostructures for lymphoma. Proc. Natl Acad. Sci. USA 110, 2511–2516 (2013).
Rink, J. S. et al. Rational Targeting of Cellular Cholesterol in Diffuse Large B-Cell Lymphoma (DLBCL) Enabled by Functional Lipoprotein Nanoparticles: A Therapeutic Strategy Dependent on Cell of Origin. Mol. Pharm. 14, 4042–4051 (2017).
Shen, W. J., Asthana, S., Kraemer, F. B. & Azhar, S. Scavenger receptor B type 1: expression, molecular regulation, and cholesterol transport function. J. Lipid Res 59, 1114–1131 (2018).
Marques, P. E. et al. Multimerization and Retention of the Scavenger Receptor SR-B1 in the Plasma Membrane. Dev. Cell 50, 283–295.e285 (2019).
Desai, A. J. & Miller, L. J. Changes in the plasma membrane in metabolic disease: impact of the membrane environment on G protein-coupled receptor structure and function. Br. J. Pharm. 175, 4009–4025 (2018).
Sunshine, H. & Iruela-Arispe, M. L. Membrane lipids and cell signaling. Curr. Opin. Lipido. 28, 408–413 (2017).
Zhang, T., Hu, W. & Chen, W. Plasma Membrane Integrates Biophysical and Biochemical Regulation to Trigger Immune Receptor Functions. Front. Immunol. 12 (2021).
Jabeen, M. et al. Advanced Characterization of Imiquimod-Induced Psoriasis-Like Mouse Model. Pharmaceutics 12 (2020).
van der Fits, L. et al. Imiquimod-Induced Psoriasis-Like Skin Inflammation in Mice Is Mediated via the IL-23/IL-17 Axis1. J. Immunol. 182, 5836–5845 (2009).
Flutter, B. & Nestle, F. O. TLRs to cytokines: Mechanistic insights from the imiquimod mouse model of psoriasis. Eur. J. Immunol. 43, 3138–3146 (2013).
Sumida, H. et al. Interplay between CXCR2 and BLT1 Facilitates Neutrophil Infiltration and Resultant Keratinocyte Activation in a Murine Model of Imiquimod-Induced Psoriasis. J. Immunol. 192, 4361–4369 (2014).
Xu, Q. et al. Topical astilbin ameliorates imiquimod-induced psoriasis-like skin lesions in SKH-1 mice via suppression dendritic cell-Th17 inflammation axis. J. Cell Mol. Med 26, 1281–1292 (2022).
van der Fits, L. et al. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J. Immunol. 182, 5836–5845 (2009).
Boehncke, W. H. & Schön, M. P. Psoriasis. Lancet 386, 983–994 (2015).
Ahmed, S. S., Manchanda, Y., De, A., Das, S. & Kumar, R. Topical Therapy in Psoriasis. Indian J. Dermatol 68, 437–445 (2023).
Dieudonné, A. et al. Scavenger receptors in human airway epithelial cells: role in response to double-stranded RNA. PLoS One 7, e41952 (2012).
Lenahan, C., Huang, L., Travis, Z. D. & Zhang, J. H. Scavenger Receptor Class B type 1 (SR-B1) and the modifiable risk factors of stroke. Chin. Neurosurg. J. 5, 30 (2019).
Mullan, R. H. et al. A role for the high-density lipoprotein receptor SR-B1 in synovial inflammation via serum amyloid-A. Am. J. Pathol. 176, 1999–2008 (2010).
Gordon, J. A. et al. Upregulation of Scavenger Receptor B1 Is Required for Steroidogenic and Nonsteroidogenic Cholesterol Metabolism in Prostate Cancer. Cancer Res 79, 3320–3331 (2019).
Guo, L. et al. Scavenger Receptor BI Protects against Septic Death through Its Role in Modulating Inflammatory Response. J. Biol. Chem. 284, 19826–19834 (2009).
Gangwar, R. S., Gudjonsson, J. E. & Ward, N. L. Mouse Models of Psoriasis: A Comprehensive Review. J. Invest Dermatol 142, 884–897 (2022).
Acknowledgements
Research reported in this publication was supported by Northwestern University SBDRC of the National Institutes of Health under award number P30AR075049. Immunohistochemistry services were supported by the Northwestern University Pathology Core Facility and a Cancer Center Support Grant (NCI CA060553). Western blot analysis was performed in the Analytical bioNanoTechnology Equipment Core Facility of the Simpson Querrey Institute for BioNanotechnology at Northwestern University. ANTEC receives partial support from the Soft and Hybrid Nanotechnology Experimental (ShyNE) Resource (NSF ECCS-2025633) and Feinberg School of Medicine, Northwestern University. Imaging work was performed at the Northwestern University Center for Advanced Microscopy generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. Flow cytometry was supported by the Northwestern University RHLCCC Flow Cytometry Facility and a Cancer Center Support Grant (NCI CA060553). PhenoCycler services were provided by Northwestern University’s Immunotherapy Assessment Core (IAC). K.Q.L., C.S.T., R.M.L, and H.P discloses support for the research of this work from the Chemical Countermeasures Research Program (CCRP) executed by the National Institute of Allergy and Infectious Diseases (NIAID), National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), and the National Institutes of Health Office of the Director (NIH OD) (U54AR079795). J.T. discloses support for the research of this work from NIAMS (F31AR081685). R.M.L discloses support for the research of this work from NIH (EY019463 and EY028560). H.P. discloses support for the research of this work from NIH (EY032922), a Dermatology Foundation Research Grant and Career Development Award, Northwestern University SBDRC Pilot & Feasibility award, and an Eversight research grant.
Author information
Authors and Affiliations
Contributions
J.T. and C.S.T. conceived and designed the study and experiments. J.T. performed all experiments and data analyses. J.T. and H.P performed the animal study. A.E.C. assisted with cell culture and HDL NP synthesis. J.S.R. assisted with HDL NP synthesis and characterization and with the western blot experiments. B.E.P.W. provided guidance on keratinocyte culture experiments and experiments on human skin equivalents. F.S. obtained demographic data for the patient samples. D.B. provided control human skin explant material. K.Q.L., R.M.L., and H.P. provided dermatologic expertise and guidance regarding experimental approaches. J.T. and C.S.T. drafted the initial manuscript. All authors reviewed, edited, and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare the following competing interests: C.S.T. and A.E.C. are involved in a start-up biotechnology company that licensed technology from Northwestern University. All other authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Sankha Bhattacharya and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Trujillo, J., Calvert, A.E., Rink, J.S. et al. Keratinocyte SR-B1 expression and targeting in cytokine-driven skin inflammation. Commun Med 5, 100 (2025). https://doi.org/10.1038/s43856-025-00804-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43856-025-00804-y