Abstract
Background
The progress of artificial intelligence (AI) research in dental medicine is hindered by data acquisition challenges and imbalanced distributions. These problems are especially apparent when planning to develop AI-based diagnostic or analytic tools for various lesions, such as maxillary sinus lesions (MSL) including mucosal thickening and polypoid lesions. Traditional unsupervised generative models struggle to simultaneously control the image realism, diversity, and lesion-type specificity. This study establishes an expert-guided framework to overcome these limitations to elevate AI-based diagnostic accuracy.
Methods
A StyleGAN2 framework was developed for generating clinically relevant MSL images (such as mucosal thickening and polypoid lesion) under expert control. The generated images were then integrated into training datasets to evaluate their effect on ResNet50’s diagnostic performance.
Results
Here we show: 1) Both lesion subtypes achieve satisfactory fidelity metrics, with structural similarity indices (SSIM > 0.996) and maximum mean discrepancy values (MMD < 0.032), and clinical validation scores close to those of real images; 2) Integrating baseline datasets with synthetic images significantly enhances diagnostic accuracy for both internal and external test sets, particularly improving area under the precision-recall curve (AUPRC) by approximately 8% and 14% for mucosal thickening and polypoid lesions in the internal test set, respectively.
Conclusions
The StyleGAN2-based image generation tool effectively addressed data scarcity and imbalance through high-quality MSL image synthesis, consequently boosting diagnostic model performance. This work not only facilitates AI-assisted preoperative assessment for maxillary sinus lift procedures but also establishes a methodological framework for overcoming data limitations in medical image analysis.
Plain language summary
Images of people with dental issues can be difficult to collect and may not be representative. One example is images of maxillary sinus lesions (MSL), which refer to abnormal tissue growths within the sinus cavity. We developed a computational framework to generate high-quality MSL images and used those images to train a computational program to diagnose MSL. Results showed that the generated images were realistic and could be used to improve the diagnostic accuracy of our computational model. Our approach could be used to improve diagnosis of MSL and also could be applied to images of other parts of the body to improve development of computational diagnostic tools for diseases in those areas.
Similar content being viewed by others
Introduction
The challenges of medical data, specifically the high acquisition and annotation costs, and the class imbalance in lesion datasets due to specialized characteristics, often hinder the development of AI-based diagnostic tools, particularly in multi-lesion analysis scenarios1,2,3. For example, in the field of oral implantology, maxillary sinus lesions (MSL), which determine surgical eligibility for sinus lift, constitute a minority4,5. This is reflected in our previous study where we constructed a database of 2000 maxillary sinus images, in which pathological anomalies like mucosal thickening and polypoid lesions accounted for only 363 cases (20.2%) and 165 cases (9.17%), respectively, limiting the development of lesion-specific AI diagnostics6. These challenges underscore the need for exploring effective methods to address the medical data issues pertinent to AI in this field7,8.
The unique characteristics of medical imaging data impact data optimization strategies and AI tool performance. First, accurate lesion labeling in medical images is essential for ensuring the reliability of AI diagnostic tools, as these images must reflect clinically representative pathological features9. Second, the imaging features of the same lesion-type exhibit natural variations, such as the size and location of polypoid lesions in the maxillary sinus, necessitating models to cover diverse lesion features to enhance generalization10. Third, multi-type lesion coexistence demands integrated evaluation for accurate medical decisions11. For example, dentists must analyze concurrent MSL, including maxillary sinus mucosal thickening and polypoid lesions, to determine appropriate surgical plans for sinus elevation4. Aside from inefficiently continuing to collect and label real medical data, identifying alternative approaches to obtain realistic, diverse, and multi-type medical images becomes crucial.
Researchers typically employ data augmentation (DA) techniques to expand training datasets12. Traditional DA methods, such as geometric transformations, cannot create image features beyond those present in the original dataset, limiting their utility7,13. This limitation has driven the development of generative adversarial networks (GANs) to randomly and automatically produce lesion-specific images and optimize dataset balance, thereby enhancing the AI-based diagnosis of minor lesion categories14,15,16. For example, Maayan et al. constructed DCGANs to generate cysts, metastases, and hemangiomas in portal-phase 2D CT scans17; Zhang et al. built PG-ACGANs to generate hyperthyroidism, hypothyroidism, and normal thyroid SPECT images18; Muhammed et al. developed 9 CycleGANs to generate brain tumor MR images across different 2D slices19; Marut et al. created a series of GANs to generate skin lesion photos20; and Chou et al. constructed a GAN to generate colon polyp images21. The lesion images generated in these studies were used to enhance lesion detection and diagnostic performance. However, when dealing with multiple types of lesions, it is often necessary to train separate GANs for each17,18,19, and the automatic generation of images introduces challenges in controlling the quality of the generated images17,20,21.
Building upon traditional GANs, the StyleGAN series introduces a unique mapping network that encodes different training data features into latent space feature vectors22,23. The GANSpace algorithm, a latent space exploration method based on principal component analysis (PCA), performs dimensionality reduction and feature disentanglement, enabling linear manipulation of distinct image attributes24. This human-in-the-loop approach introduces controllability into GAN-based image generation: it not only allows flexible synthesis of realistic, diverse images through feature vector manipulation without predefined class constraints, but its class-conditional framework also eliminates the need for per-class model training, effectively addressing traditional GAN limitations25,26.
Therefore, using MSL image generation as a paradigm, this study proposes a StyleGAN2-based framework to optimize the dataset distribution and enhance the AI-based diagnosis of specific lesions. Unlike traditional GANs, this study introduces feature vector disentanglement and expert-controlled adjustments, enabling mucosal thickening and polypoid lesions image generation with regulation of anatomical realism, morphological diversity, and pathological subtypes—validated by high structural similarity indices (SSIM >0.996), low maximum mean discrepancy values (MMD <0.032), and clinician-assigned clinical scores. Incorporating the synthetic images into the diagnostic model training significantly improves its performance for both lesion types across internal and external datasets. As the early exploration of controllable image generation in dental medicine, this work provides a practical solution to data scarcity and advances diagnostic support, particularly valuable for resource-constrained settings, thereby improving clinical decision-making directly translates to enhanced patient outcomes.
Methods
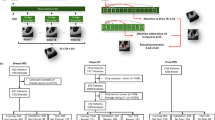
The study design flowchart is shown in Fig. 1A.
A Expert-controlled StyleGAN2-generated lesion images were incorporated into the baseline training set to improve data distribution, followed by evaluating ResNet50’s diagnostic performance. B After training StyleGAN2, high-quality MSL images, like mucosal thickening, polypoid lesions, and ostium obstruction, could be generated through an expert-controlled GANSpace interface.
Dataset development
This study adhered to the Helsinki Declaration and was approved by the Ethics Committee of the Sun Yat-sen University Affiliated Hospital (KQEC-2020-29-06). Informed consent for the use of imaging data in analysis and testing was obtained from all participants by the research team of one of the authors (Chen ZT), with all participants being fully informed. In our previous research, we had collected 2000 coronal images of the maxillary sinus at the first molar sites from CBCT scans of 1000 patients at the Hospital of Stomatology, Sun Yat-sen University, and employed a YOLOv5 object detection network to crop the maxillary sinus regions from these coronal images and standardized the annotation of MSL6. These 2000 maxillary sinus images served as training data for subsequent StyleGAN2 and ReACGAN development.
Of the 2000 maxillary sinus CBCT images, mucosal thickening accounted for 20.2%, polypoid lesions for 9.17%, and nasal obstruction for 60.45%. Therefore, this study focuses on optimizing the diagnosis of the first two lesion types exhibiting data distribution issues. To enhance the reliability of the diagnostic optimization experiment, a stochastic sampling strategy was implemented: baseline training sets were randomly sampled from the original dataset (excluding the internal test sets) to form ten groups, each containing 400 images. The lesion proportions in each group matched those in the original dataset, guiding generation quantity determination. Two independent static test sets were created, each containing 100 images: Internal Test Set 1 (50 images with mucosal thickening and 50 normal images) and Internal Test Set 2 (50 images with polypoid lesions and 50 normal images). These static test sets were used for comparing the performance of ResNet50 before and after integrating the generated MSL images into the training process.
To test the diagnostic model’s generalizability, external test sets were additionally employed under ethical approval (KQEC-2020-29-06). CBCT images from 260 patients at Guangzhou Chunzhihua Dental Clinic were collected, with imaging parameters specified in Supplementary Table 1. Using the same image processing methods, we obtained 520 annotated maxillary sinus images of the left and right maxillary sinuses. Two external test sets were randomly created, each containing 100 images: External Test Set 1 (50 images with mucosal thickening and 50 normal images) and External Test Set 2 (50 images with polypoid lesions and 50 normal images).
Construction and image generation of StyleGAN2 and ReACGAN
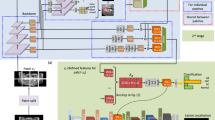
GANs consist of a generator and a discriminator and fundamentally rely on the latent space—a high-dimensional manifold composed of randomly distributed latent vectors14. During the unsupervised GAN training, the generator samples random noise from the latent space to produce initial outputs, while the discriminator assesses their realism. Through this adversarial training process, the generator progressively refines synthetic images to mimic real data distributions14. In the StyleGAN2 architecture, the mapping network discovers the relationship between training data features and latent space vectors, known as feature disentanglement22,23.
During training, the StyleGAN2 model employed an architecture in which input vectors were processed through 8-layer fully-connected networks to generate intermediate latent vectors. These vectors modulated convolutional kernels across multiple scales via modulation/demodulation modules, controlling image features at different resolutions. The generation process began with a fixed 4 × 4 × 512 tensor as the initial feature map, progressively refined through convolution, upsampling, and style injection to produce 512 × 512 images. A style injection branch controlled global appearance, while a parallel detail branch enhanced local features, effectively reducing artifacts and improving diversity/quality. Training used logistic loss with a base learning rate 0.002 for both networks, optimized via Adam optimizer27. The progressive growing strategy trained from 8 × 8 to 512 × 512 resolution over 1.2 million images, with a minibatch size of 32 distributed across GPUs (4 samples/GPU). Generator weights were smoothed using EMA (exponential moving average, half-life=10k images) with lazy regularization (generator: every 4 iterations; discriminator: every 16). Total training spanned 2700 thousand images (kimg), with checkpoints saved every 50 iterations. The lowest discriminator loss checkpoint was selected for inference. After training, GANSpace applied PCA to reduce the dimensionality of the disentangled latent space vectors, converting them into an actionable interface (Supplementary Fig. 1)24. Three well-trained implantologists with over 5 years of clinical experience manipulated and combined different feature vectors via this interface to obtain realistic, diverse, and multi-type MSL images (normal maxillary sinuses and those with mucosal thickening, polypoid lesions, and ostium obstruction) with 512 × 512 pixels. The annotation process was completed simultaneously with image generation.
ReACGAN, an improved ACGAN variant proposed by Kang et al. with demonstrated superior performance on large-scale datasets (e.g., ImageNet), was trained additionally28. The network was implemented using the open-source StudioGAN framework and employs Big ResNet as its backbone architecture29. Both generator and discriminator adopted spectral normalization and attention mechanisms (generator: layer 4; discriminator: layer 1) to enhance feature learning. The generator used a latent vector dimension of 140, a shared conditional vector dimension of 128, a convolutional channel size of 80, and employed an EMA strategy with a decay factor of 0.9999, activated after 4000 training steps. The discriminator incorporated an auxiliary classifier with an embedding dimension of 1024. The training utilized hinge loss (conditional loss coefficient = 0.25). The generator and discriminator learning rates were set to 5e-5 and 2e-4, respectively. The model employed the Adam optimizer30, with the discriminator updated twice per iteration. Each iteration included 2 discriminator updates over 40,000 total steps (batch size=16) with Adaptive Discriminator Augmentation. As a comparison to StyleGAN2, to generate two types of lesion images, ReACGAN was independently trained on mucosal thickening and polypoid lesion images from the original dataset. The autonomously generated 512 × 512 pixel images are shown in Supplementary Fig. 2.
The entire model construction and training process were performed on the local server with GPU of NVIDIA Tesla V100. The code for this study is open source and has been released at https://github.com/xhli-code/stylegan2-GANSpace.
Evaluation of generated MSL images
We evaluated the generated MSL images against their corresponding lesion images in the original dataset using SSIM and MMD31,32. SSIM measures attributes like contrast, brightness, and structural similarity, with values closer to 1 indicating higher quality. MMD evaluates feature distribution between generated and original images in high-dimensional space, where lower values (closer to 0) reflect better GAN performance and greater realism.
To guarantee the realism of generated images, a clinical validation experiment was designed. Using mucosal thickening as a prototype, 100 images with mucosal thickening labels from the original dataset, the StyleGAN2-generated images, and the ReACGAN-generated images were randomly selected, forming three blind sub-datasets for evaluation. After undergoing standardized training, three experienced implantologists with radiological expertise (referred to as Observer 1, 2, and 3), independently scored the realism of the lesion features in sub-datasets using the following scale: (1) the lesion area is entirely unrealistic or absent; (2) the lesion area exhibits obvious synthetic characteristics; (3) the lesion area approaches realism but contains notable doubts; (4) The lesion area is mostly realistic, with minor uncertainties; (5) The lesion area is convincingly realistic. For polypoid lesions, the clinical validation experiment was conducted by the same implantologists on sub-datasets containing the corresponding labels.
Diagnostic optimization experiments based on ResNet50
In our previous research, ResNet50 exhibited superior performance in the intelligent screening of maxillary sinus pathological abnormalities compared to other CNNs6. Therefore, this study continued to use this model for optimizing the diagnosis of mucosal thickening and polypoid lesions, maintaining the hyperparameters determined in previous research (epoch=600, learning rate = 0.0001, batch size = 32). Taking mucosal thickening as an example, the model was first trained on a baseline training set and obtained baseline predictions on Internal Test Set 1. Subsequently, the baseline dataset was optimized by integrating StyleGAN2- and ReACGAN-generated mucosal thickening images, followed by model retraining and post-optimization predictions. By repeating experiments across 10 different baseline training sets, the results were reported as ranges. A parallel approach was applied for polypoid lesion diagnosis, with Internal Test Set 2 used instead. In addition, this study compared the predictive effects of traditional DA (flipping and rotation) on ResNet50. To strengthen the study’s findings, external validation was performed on External Test Sets 1 and 2.
To evaluate the predictive performance, F1 score and precision–recall curves (PRC) focusing on positive class prediction efficacy33,34, and overall predictive evaluation metric receiver operating characteristic (ROC) curves35, were utilized as comparative metrics for diagnostic optimization experiments. In addition, precision and recall values at the optimal F1-score threshold were reported for ResNet50 before and after integrating StyleGAN2-generated MSL images6. Python (version 3.7) was employed for analyzing these metrics.
Visualization of the diagnostic model ResNet50
Grad-CAM, which utilizes gradients as weights to aggregate feature maps, highlights regions of interest (ROI) in deep learning models36. In this study, Grad-CAM was employed to visualize the ROI of ResNet50 on internal test images from one of the diagnostic optimization experiments, providing intuitive insights of the model’s performance before and after optimizing the baseline training set.
Statistics and reproducibility
The Shapiro–Wilk test was used to evaluate data normality. For the clinical validation experiment, the Mann–Whitney U test was used to compare the statistical differences in average scores between StyleGAN2-generated images and the other two groups (real images and ReACGAN-generated images). The Mann–Whitney U test was also employed to compare the statistical differences between various optimization techniques and the baseline data in diagnostic experiments. A P value of <0.05 was considered indicative of statistical significance.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Construction and image generation of StyleGAN2 and ReACGAN
The architecture of the StyleGAN2-based MSL image generator is shown in Fig. 1B. Within the specified iteration, the StyleGAN2 generator and discriminator completed the training, and the losses remained consistent near the end of training (Fig. 2). As data distribution issues were identified, the implantologists generated 200 images of mucosal thickening and 400 images of polypoid lesion using GANSpace to address the imbalance in both categories within the baseline training sets. Other types of images not involved in the subsequent diagnostic optimization experiments, such as normal and ostium obstruction images, can also be generated (Fig. 3). As a comparison, the trained ReACGANs autonomously generates the same number of mucosal thickening and polypoid lesion images as StyleGAN2 without implantologists intervention (Supplementary Fig. 2).
Box A contained real maxillary sinus images, while box B contained generated images. Columns a, b, c, and d represented normal maxillary sinus, mucosal thickening, polypoid lesion, and ostium obstruction, respectively (with the yellow circles indicating obstruction location). The first two lesion types were included in diagnostic optimization experiments. These results demonstrate successful generation of realistic, diverse, and multi-type maxillary sinus images.
Evaluation of generated MSL images
The StyleGAN2-generated mucosal thickening and polypoid lesion images achieved SSIM31 values of 0.9966 and 0.9962, respectively, confirming superior image fidelity. The MMD32 values were 0.0155 and 0.0317, indicating feature distribution alignment with real images. In contrast, the mucosal thickening and polypoid lesion images generated by ReACGAN exhibited image bias or artifacts and even the absence of real-world features, demonstrating lower SSIM scores of 0.4781 and 0.4564, and MMD values of 0.1994 and 0.1895, respectively, both inferior to those of StyleGAN2 (Table 1).
The clinical validation of image realism, conducted by three experienced implantologists, showed that the StyleGAN2-generated images of mucosal thickening and polypoid lesions received scores comparable to those of real images, with an average score of 4.33 ± 0.38 and 4.02 ± 0.44. In contrast, the ReACGAN-generated images were rated with an average score close to 1 for both lesion images by all three observers. The average score of StyleGAN2-generated mucosal thickening images differed significantly from that of real images (P = 0.001). In addition, statistical analyses showed significant differences between the average scores of StyleGAN2-generated images of both lesions and ReACGAN-generated images (both P < 0.001) (Tables 2 and 3).
Diagnostic optimization experiments based on ResNet50
By bootstrapping different baseline training sets for repeated diagnostic optimization experiments, the range of prediction performance for mucosal thickening and polypoid lesion is displayed in Table 4. For mucosal thickening, dataset optimization with StyleGAN2-generated images led to significant improvements in the area under the precision–recall curve (AUPRC) (P < 0.001), the area under the receiver operating characteristic curve (AUROC) (P = 0.001), and F1 score (P = 0.011) compared to the baseline. For polypoid lesions, the improvements in AUPRC, AUROC, and F1 score were more significant (all P < 0.001), with AUPRC showing an increase of nearly 14%. Conversely, ReACGAN-optimized methods showed no substantial differences from baseline in mucosal thickening or polypoid lesion detection. Except for the AUPRC in diagnosing mucosal thickening (P = 0.043), all other evaluation metrics demonstrated no statistically significant differences. Notably, traditional DA (flipping/rotation) significantly degraded performance for both lesions.
The results on the external test set are presented in Table 5. After integrating the StyleGAN2-generated images to optimize the dataset distribution, ResNet50 showed a slight improvement in diagnosing mucosal thickening compared to the baseline. In contrast, the prediction performance for polypoid lesions improved markedly, with statistically significant differences (all P of metrics = 0.001).
Visualization of the diagnostic model ResNet50
Grad-CAM visualization demonstrated that integrating the StyleGAN2-generated images into the baseline training set allowed ResNet50 to focus its ROI more precisely on the locations of mucosal thickening and polypoid lesions, thereby aiding the model in making more accurate predictions (Fig. 4).
For the two types of lesions, three images were randomly selected from Test Sets 1 and 2 for demonstration. From left to right, columns showed: original test images, ROI before dataset optimization, and ROI after dataset optimization (yellow circles indicated the approximate lesion locations). It was evident that incorporating generated images to optimize the baseline training set enhanced model focus more on lesion areas, thereby improving the reliability of the prediction results.
Discussion
This study successfully developed a StyleGAN2-based MSL image generation tool, which produced mucosal thickening and polypoid lesion images with satisfactory SSIM and MMD, indicating high image quality, realism, and diversity. After integrating StyleGAN2-generated images to optimize the baseline training sets, the diagnostic performance of the ResNet50 improved for both types of lesions, reflected in increased F1 scores, AUPRC, AUROC, and enhanced Grad-CAM visualizations.
In this study, StyleGAN2 enabled the generation of realistic and diverse MSL images under expert control, as confirmed by quantitative metrics (SSIM and MMD) and a blinded clinical validation experiment. It even exhibited hyper-realism, as the average score of StyleGAN2-generated mucosal thickening images differed significantly from that of real images (P = 0.001). In contrast, ReACGAN-generated images exhibited worse performance, indicating lower image quality that experts could easily distinguish. This performance gap originates from StyleGAN2’s feature disentanglement capability and GANSpace-driven latent space manipulation, which allow fine-tuned adjustments of feature vectors, enabling controllable image quality, data cleaning, and data annotation22,24,37. In comparison, traditional GANs like ReACGAN generate images in a fully automatic and black-box manner, making it difficult to ensure image realism and diversity, potentially leading to a substantial amount of unusable or easily identifiable fake data20,21. Such image generation poses potential risks, particularly for medical applications that require high levels of professionalism and reliability, while the lack of diversity may hinder diagnostic models from fully capturing different lesion features to improve generalization17,27.
Clinicians could generate multi-class MSL images through a unified StyleGAN2 framework through different feature vectors combination, whereas ReACGAN necessitates lesion-specific model training24. This make StyleGAN2 more suitable for medical image generation tasks, highlighting the advantages: a single network can generate multiple lesion types simultaneously, reducing unnecessary computational costs while mitigating the challenges of medical data collection that hinder the progress of computer-aided diagnosis37. Reviewing existing studies, multiple works have employed GANs to generate various types of lesion images. For instance, Frid-Adar et al. used three DCGANs to generate synthetic samples for three classes of liver lesions (cysts, metastases, and hemangiomas)17; Salehinejad et al. trained five different GANs to generate five different classes of chest diseases’ images38. Therefore, this study establishes an effective methodological framework, where the predefined class-free training approach and class-conditional nature of controllable image generation collectively hold promise for widespread application in medical DA to enhance computer-aided diagnosis in medicine.
Despite the advantages of controlling feature vectors for image generation in a visualized manner, researchers have pointed out that identifying which vectors correspond to specific image features remains a challenge22,23,24. Empirically, vectors toward the beginning typically govern large, global image attributes (such as the shape and size of the maxillary sinus), while later latent vectors tend to focus on fine, localized image details (such as the thickness of the local mucosa)24. By effectively leveraging this pattern, different vectors can be controlled and combined to generate realistic and diverse images of various types of MSL, including mucosal thickening, polypoid lesions, and ostium obstructions.
In the diagnostic optimization experiment, the baseline prediction performance for mucosal thickening using ResNet50 was relatively robust, likely due to the uniform characteristics of mucosal thickening, which are typically located at the sinus floor39. Notably, dataset optimization with StyleGAN2-generated images still led to a significant improvement in prediction performance. Grad-CAM visualizations further indicated that ResNet50’s ROI was more concentrated in areas of mucosal thickening, enhancing the model’s interpretability and credibility. For polypoid lesions, the improvement in AI-based diagnosis was more pronounced. This is likely because polypoid lesions exhibit greater variability in location, size, and presentation4, making the increased sample size particularly beneficial for model performance18. While the overall diagnostic performance improved significantly, a more nuanced evaluation revealed specific trends: for mucosal thickening, precision at the optimal F1 score increased, whereas recall remained relatively unchanged. Conversely, for polypoid lesions, recall improved, but precision decreased, suggesting that there is still room for further refinement ((Supplementary Table 2).
When evaluating the impact of ReACGAN-generated lesion images on the baseline diagnostic model, the performance for mucosal thickening showed some improvement, though only the AUPRC demonstrated a statistically significant difference (P = 0.043). In contrast, the diagnostic performance for polypoid lesions showed no improvement and even a slight decline, confirming this study’s viewpoint: images generated without expert supervision lack sufficient quality, realism, and diversity, posing greater risks in optimizing medical diagnostic tasks, which may be attributed to the relatively limited size of the dataset used in this study17. This study also compared the effect of traditional DA, such as flipping and rotation, on ResNet50’s diagnostic performance. For both mucosal thickening and polypoid lesions, traditional DA actually exerted a detrimental effect, possibly due to alterations in the original distribution of lesion features, which confused the diagnostic model6. This implies the potential instability of traditional DA methods in enhancing diagnostic performance. However, when combining the baseline training set with traditional DA and StyleGAN2-based dataset optimization methods, positive effects were observed in the prediction of polypoid lesions compared to using traditional DA alone, though these results only restored performance to baseline levels. This suggests that optimizing the dataset using StyleGAN2-generated images might mitigate the negative impact of traditional DA methods, which underscored the effect of StyleGAN2-based dataset optimization on the model.
Beyond the internal test set, the diagnostic results on the external test sets were consistent with the trends observed in the internal test set, confirming that the strategy of using StyleGAN2-generated images to optimize the baseline dataset demonstrates a certain level of generalizability, particularly in the diagnosis of polypoid lesions. For mucosal thickening, although the optimized model showed some improvement, the extent of enhancement was less satisfactory compared to polypoid lesions. This underscores the uniqueness of datasets from different sources and highlights the importance of incorporating multi-center data beyond the internal dataset for model training and evaluation40,41. Consistent with the internal test set, the diagnostic improvement for mucosal thickening was lower than that for polypoid lesions, indicating that classes with more imbalanced distributions like polypoid lesions in the original dataset benefit more significantly from dataset optimization18.
One of the limitations of this study is the moderate sample size, which necessitates the inclusion of larger-scale, multi-center, high-quality datasets to enhance the performance, reliability and generalizability of the results, and to minimize the risk of overfitting. Furthermore, exploring diverse generative models and traditional DA techniques across various classification architectures, imaging modalities, and lesion classification tasks—such as maxillary sinus tumors or tumor-like lesions—would broaden the applicability of this study. Moreover, the methodology in this study relies on expert access for data generation and annotation, which limits its application in lower-resource settings. Exploring strategies like semi-supervised learning to achieve minimal labor-intensive annotation processes warrants further consideration.
In summary, this study developed a StyleGAN2-based image generation tool for MSL, effectively addressing the challenges of medical imaging data acquisition and imbalanced data distribution for conditions such as mucosal thickening and polypoid lesions, thereby significantly improving the diagnostic performance for these conditions. This strategy can be extended to other similar applications, enabling the efficient generation of high-quality, diverse and multi-type medical images with relatively lower privacy and data risk. This advancement opens the door to the development of virtual public databases for oral medical imaging, significantly advancing the frontier of AI-driven research in dental medicine.
Data availability
Representative images generated by StyleGAN2 were included as Supplementary Data 1. The source data for the figures can be found in Supplementary Data 2. To protect study participant privacy, other data used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Code availability
The code for this study is open source and has been released at https://github.com/xhli-code/stylegan2-GANSpace42.
References
Lee, D. W., Kim, S. Y., Jeong, S. N. & Lee, J. H. Artificial intelligence in fractured dental implant detection and classification: evaluation using dataset from two dental hospitals. Diagnostics https://doi.org/10.3390/diagnostics11020233 (2021).
Wang, S. et al. Annotation-efficient deep learning for automatic medical image segmentation. Nat. Commun. 12, 5915 (2021).
Buda, M., Maki, A. & Mazurowski, M. A. A systematic study of the class imbalance problem in convolutional neural networks. Neural Netw. 106, 249–259 (2018).
Shanbhag, S., Karnik, P., Shirke, P. & Shanbhag, V. Cone-beam computed tomographic analysis of sinus membrane thickness, ostium patency, and residual ridge heights in the posterior maxilla: implications for sinus floor elevation. Clin. Oral Implants Res. 25, 755–760 (2014).
Kim, Y. et al. Deep learning in diagnosis of maxillary sinusitis using conventional radiography. Investig. Radiol. 54, 7–15 (2019).
Zeng, P. et al. Abnormal maxillary sinus diagnosing on CBCT images via object detection and ‘straight-forward’ classification deep learning strategy. J. Oral Rehabil. 50, 1465–1480 (2023).
Yi, X., Walia, E. & Babyn, P. Generative adversarial network in medical imaging: a review. Med. Image Anal. 58, 101552 (2019).
Topol, E. J. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 25, 44–56 (2019).
Litjens, G. et al. A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017).
Cossio, M. Augmenting medical imaging: a comprehensive catalogue of 65 techniques for enhanced data analysis. Preprint at https://doi.org/10.48550/arXiv.2303.01178 (2023).
Steigmann, L., Di Gianfilippo, R., Steigmann, M. & Wang, H. L. Classification based on extraction socket buccal bone morphology and related treatment decision tree. Materials https://doi.org/10.3390/ma15030733 (2022).
Simard, P. Y., Steinkraus, D. & Platt, J. C. Best practices for convolutional neural networks applied to visual document analysis. In Seventh International Conference on Document Analysis and Recognition 958–963 (IEEE, 2003).
Song, B. et al. Classification of imbalanced oral cancer image data from high-risk population. J. Biomed. Opt. https://doi.org/10.1117/1.Jbo.26.10.105001 (2021).
Goodfellow, I. J. et al. Generative adversarial nets. In Proceedings of the 27th International Conference on Neural Information Processing Systems—Volume 2. 2672–2680 (MIT Press, 2014).
Lin, Y. et al. DHI-GAN: improving dental-based human identification using generative adversarial networks. IEEE Trans. Neural Netw. Learn. Syst. 34, 9700–9712 (2023).
Bermudez, C. et al. Learning implicit brain MRI manifolds with deep learning. In Proceedings of SPIE–the International Society for Optical Engineering 10574 (SPIE, 2018).
Frid-Adar, M., Klang, E., Amitai, M., Goldberger, J. & Greenspan, H. Synthetic data augmentation using GAN for improved liver lesion classification. In 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) 289–293 (IEEE, 2018).
Zhang, H., Huang, Z. & Lv, Z. Medical Image Synthetic Data Augmentation Using GAN. In Proceedings of the 4th International Conference on Computer Science and Application Engineering. 133 (Association for Computing Machinery, 2020).
Yapici, M. M., Karakis, R. & Gurkahraman, K. Improving brain tumor classification with deep learning using synthetic data. Comput. Mater. Contin. 74, 5049–5067 (2022).
Jindal, M. & Singh, B. Bias inheritance and its amplification in GAN-based synthetic data augmentation for skin lesion classification. In 2024 3rd International Conference on Artificial Intelligence For Internet of Things (AIIoT). 1–6 (IEEE, 2024).
Chou, Y.-C. & Chen, C.-C. Improving deep learning-based polyp detection using feature extraction and data augmentation. Multimed. Tools Appl. 82, 16817–16837 (2023).
Karras, T., Laine, S. & Aila, T. A style-based generator architecture for generative adversarial networks. IEEE Trans. pattern Anal. Mach. Intell. 43, 4217–4228 (2021).
Karras, T. et al. Analyzing and improving the image quality of StyleGAN. In 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). 8107–8116 (IEEE, 2020).
Härkönen, E., Hertzmann, A., Lehtinen, J. & Paris, S. GANSpace: discovering interpretable GAN controls. In Proceedings of the 34th International Conference on Neural Information Processing Systems. 825 (Curran Associates Inc., 2020).
Deng, B., Zheng, X., Chen, X. & Zhang, M. A Swin transformer encoder-based StyleGAN for unbalanced endoscopic image enhancement. Comput. Biol. Med. 175, 108472 (2024).
Ahn, G. et al. High-resolution knee plain radiography image synthesis using style generative adversarial network adaptive discriminator augmentation. J. Orthop. Res. 41, 84–93 (2023).
Xue, Y. et al. Selective synthetic augmentation with HistoGAN for improved histopathology image classification. Med. Image Anal. 67, 101816 (2021).
Kang, M., Shim, W., Cho, M. & Park, J. Rebooting ACGAN: auxiliary classifier GANs with stable training. In Neural Information Processing Systems. (Curran Associates, Inc., 2021).
Kang, M., Shin, J. & Park, J. StudioGAN: a taxonomy and benchmark of GANs for image synthesis. IEEE Trans. Pattern Anal. Mach. Intell. 45, 15725–15742 (2022).
Kingma, D. & Ba, J. Adam: a method for stochastic optimization. In International Conference on Learning Representations (2014).
Zhou, W., Bovik, A. C., Sheikh, H. R. & Simoncelli, E. P. Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 13, 600–612 (2004).
Bernhard, S., John, P. & Thomas, H. Correcting sample selection bias by unlabeled data. In Advances in Neural Information Processing Systems 19: Proceedings of the 2006 Conference 601–608 (MIT Press, 2007).
Sabanayagam, C. et al. A deep learning algorithm to detect chronic kidney disease from retinal photographs in community-based populations. Lancet Digit. Health 2, e295–e302 (2020).
Haixiang, G. et al. Learning from class-imbalanced data: review of methods and applications. Expert Syst. Appl. 73, 220–239 (2017).
Ozenne, B., Subtil, F. & Maucort-Boulch, D. The precision–recall curve overcame the optimism of the receiver operating characteristic curve in rare diseases. J. Clin. Epidemiol. 68, 855–859 (2015).
Selvaraju, R. R. et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. in 2017 IEEE International Conference on Computer Vision (ICCV) 618–626 (IEEE, 2017).
Niehues, J. M. et al. Using histopathology latent diffusion models as privacy-preserving dataset augmenters improves downstream classification performance. Comput. Biol. Med. 175, 108410 (2024).
Salehinejad, H., Valaee, S., Dowdell, T., Colak, E. & Barfett, J. Generalization of deep neural networks for chest pathology classification in X-rays using generative adversarial networks. In 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) 990–994 (IEEE, 2018).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Kelly, C. J., Karthikesalingam, A., Suleyman, M., Corrado, G. & King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 17, 195 (2019).
Koh, P. W. et al. WILDS: a benchmark of in-the-wild distribution shifts. In Proceedings of the 38th International Conference on Machine Learning (eds Meila, M. & Zhang, T.) 5637–5664 (PMLR, 2021).
Li, X. xhli-code/stylegan2-ganspace. https://doi.org/10.5281/zenodo.15374277 (2025).
Acknowledgements
The authors thank Chenghao Zhang, Yuqing Liang, and Diallo Mariama, Guanghua School of Stomatology, Sun Yat-sen University for their assistance in data collection. This work was financially supported by National Natural Science Foundation of China (82402380), Young Science and Technology Talent Support Program of Guangdong Precision Medicine Application Association (YSTTGDPMAA202502), Guangzhou Science and Technology Program key projects (2023B03J1232), Special Funds for the Cultivation of Guangdong College Students’ Scientific and Technological Innovation (“Climbing Program” Special Funds, pdjh2023b0013), and Undergraduate Training Program for Innovation of Sun Yat-sen University (202310810).
Author information
Authors and Affiliations
Contributions
Peisheng Zeng contributed to conception, design, data acquisition, analysis and interpretation, and drafted and critically revised the manuscript. Rihui Song contributed to conception, design, algorithm writing, data analysis and interpretation, and drafted and critically revised the manuscript. Shijie Chen contributed to conception, data analysis, and interpretation, and drafted and critically revised the manuscript. Xiaohang Li contributed to design, algorithm writing, data analysis, and interpretation. Haopeng Li contributed to conception, design, algorithm writing, and data analysis. Yue Chen contributed to algorithm writing, algorithm testing, data analysis, and interpretation. Zhuohong Gong contributed to conception and data analysis and drafted the manuscript. Gengbin Cai contributed to data acquisition, analysis, and drafted the manuscript. Yixiong Lin contributed to data acquisition, analysis, and drafted the manuscript. Mengru Shi contributed to conception, design, data acquisition, analysis, and interpretation, and drafted and critically revised the manuscript. Kai Huang contributed to conception, design, algorithm guidance, data analysis and interpretation, and drafted and critically revised the manuscript. Zetao Chen contributed to conception, design, data acquisition, analysis, and interpretation, and critically revised the manuscript. All authors gave final approval and agreed to be accountable for all aspects of the work. All authors agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks McKell Woodland and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zeng, P., Song, R., Chen, S. et al. Expert-guided StyleGAN2 image generation elevates AI diagnostic accuracy for maxillary sinus lesions. Commun Med 5, 185 (2025). https://doi.org/10.1038/s43856-025-00907-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43856-025-00907-6