Abstract
Background:
Increased spinal curvature is one of the most recognizable aging traits in the human population. However, despite high prevalence, the etiology of this condition remains poorly understood.
Methods
To gain better insight into the physiological, biochemical, and genetic risk factors involved, we developed a novel machine learning method to automatically derive thoracic kyphosis and lumbar lordosis angles from dual-energy X-ray absorptiometry (DXA) scans in the UK Biobank Imaging cohort. We carry out genome-wide association and epidemiological association studies to identify genetic and physiological risk factors for both traits.
Results
In 41,212 participants, we find that on average males and females gain 2.42° in kyphotic and 1.48° in lordotic angle per decade of life. Increased spinal curvature shows a strong association with decreased muscle mass and bone mineral density. Adiposity demonstrates opposing associations, with decreased kyphosis and increased lordosis. Using Mendelian randomization, we show that genes fundamental to the maintenance of musculoskeletal function (COL11A1, PTHLH, ETFA, TWIST1) and cellular homeostasis such as RNA transcription and DNA repair (RAD9A, MMS22L, HIF1A, RAB28) are likely involved in increased spinal curvature.
Conclusions
Our findings reveal a complex interplay between genetics, musculoskeletal health, and age-related changes in spinal curvature, suggesting potential drivers of this universal aging trait.
Plain Language Summary
In our study, we investigated how spinal curvature changes with age, and the underlying factors contributing to this phenomenon. We developed a machine learning model to precisely measure upper spine (kyphotic) and lower spine (lordotic) angles from X-ray scans. We observed a clear age-related increase in both angles, with sex-specific differences. Our analysis revealed significant associations with fat composition and musculoskeletal traits. Lower muscle mass and bone density was correlated with greater curvature. Through genetic analysis, we also identified several genes influencing spinal curvature. Our findings suggest a complex interaction between aging, physiology, and genetics in shaping spinal curvature, with potential implications for preventive and therapeutic strategies.
Similar content being viewed by others
Introduction
Increased spinal curvature is a frequently observed phenomenon during aging in humans and in other species1,2,3. Drift to higher thoracic kyphosis and lumbar lordotic angles starts in early ages, is gradual, and takes many decades to develop4,5. These physiological declines are generally considered to be non-pathological unless kyphosis becomes so severe as to exceed 40° (hyperkyphosis), or 30° in lordosis regions (hyperlordosis) on the sagittal plane4. Hyperkyphosis is known to have a significant heritable component (~ 60-70% heritability of clinical hyperkyphosis in twin studies) and affects females more than males6,7,8,9,10. On the other hand, lumbar hyperlordosis is an understudied condition11.
At the extremes, acute injuries, vertebral body fractures, and degenerative joint disorders are believed to be the main drivers of spinal hypercurvature12. In the general population, increased spine curvature develops largely in the absence of severe events. Prevalent aging conditions such as loss of bone mineral density, declines in muscle strength, and metabolic disorders have been suggested to play an important role8,13. Over time, continued biomechanical stresses on the spine and aging-related declines in the repair capacity of cartilage and bone tissues can lead to the development of vertebral fractures, increased back pain, and consequently reduced mobility5,14. Cumulative adverse consequences include an increased chance of falls, reduced health span, and increased mortality15,16.
While the negative consequences of an aging spine on health outcomes are clear, the genetic risk factors and underlying molecular mechanisms of this phenomenon remain poorly understood. Recent advances in the UK Biobank (UKBB) large-scale prospective imaging study create an opportunity to characterize changes to spine curvature with age in a general population17. We capitalized on this resource by developing machine learning models to assess spine curvature across 41,212 dual-energy X-ray absorptiometry (DXA) scans. Using this fully automated machine learning (ML) pipeline, we were able to measure Cobb angles from a sagittal view in both thoracic and lumbar regions, thus providing us with an opportunity to examine shared and unique risk factors in different regions of the spine.
Our analysis extends to the study of underlying pathological conditions, physiological, blood biochemistry, and genetic risk factors. We assessed contributions to spine angle from 415 diseases, 15 musculoskeletal traits, 59 biomarkers, and all common genetic variants. We further carry out Mendelian randomization (MR) to test for potential causal involvement of genes within these loci with changes in spinal curvature. To our knowledge, this is the first study to identify genetic risk factors involved in the development of kyphotic and lordotic angles.
Methods
Study population: the UK Biobank
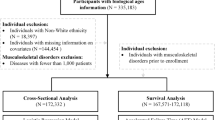
The UK Biobank (UKBB) recruited 503,000 community-based adult volunteers aged 40-69 from 2006-2010 in 22 study centers across the United Kingdom: England, Wales, and Scotland18. In the current analysis, we evaluate the UKBB imaging subgroup of 41,212 participants who returned between 2014-2020 to undergo spinal dual-energy X-ray absorptiometry (DXA) assessment (Lunar iDXA densitometer; GE Healthcare, Chicago, Illinois). At the baseline visit, extensive demographic and biometric information were collected including age, race, smoking status, height, and weight via questionnaires and physical measurements. The Townsend Deprivation Index (TDI), a composite measure for socioeconomic status calculated from unemployment, car and home ownership, and household crowding status, was evaluated from self-reported metrics19. Additionally, blood was collected for serum biomarker and genomic analyses. Information on prior disease conditions were derived from baseline data and clinical data aggregated across healthcare providers: the Health Episode Statistics database from England, Patient Episode Database for Wales, and the Scottish Morbidity Record 01 for Scotland. The International Classification of Diseases, Tenth Revision (ICD10) was used.
This study was approved by the North West Multi-Centre Research Ethics Committee (MREC) and informed consent was collected from all participants. Further information on the study design of the UKBB has been previously published18.
Machine learning
Detailed methods are available in the Supplementary Machine Learning Methods document. In brief, we constructed an image-processing pipeline that (1) establishes a trace along the center of the vertebral column, (2) identifies an appropriate thoracic or lumbar section of the spine for analysis of kyphosis or lordosis, respectively, and (3) measures the angle difference between lines tangential to the spinal traces at the top versus bottom of that section as a proxy for the Cobb angle (Supplemental Fig. 12). For kyphotic angle, the thoracic spine approximately above and including vertebra T12 was analyzed. For lordotic angle, the lumbar spine approximately between and including vertebrae T12 through L5 was analyzed. Full details can be found in the Supplementary Methods and GitHub repository (https://github.com/graham-calico/KyphoLordoDxa.git). Kyphosis and lordosis were measured using the python tool “spineCurve.py”. The following command-line options were invoked, for kyphosis: “--aug_flip --side_facing right”; and for lordosis: “--lumbar --aug_flip --side_facing right --aug_tilt 0.5”. Exact reproduction of the results in this manuscript should additionally invoke the “--legacy” flag. Further methods are described in Supplementary Machine Learning Methods.
Statistical analyses
For all analyses, two-tailed P-values were calculated. Significance was defined as a false discovery rate-corrected p-value (pFDR) < 0.05. All epidemiological analyses were performed with R version 4.3.1.
We assessed participant characteristics at the time of the imaging visit, stratified for presence and absence of hyperkyphosis (>40°) and hyperlordosis (>30°). Continuous variables were summarized as medians and interquartile ranges (IQR), and categorical variables were presented as counts and percentages. The correlation between kyphotic and lordotic angles was displayed using locally smoothed regression with span = 0.75 and the correlation coefficient (r) was calculated using Pearson product-moment correlation after confirming the linearity assumption could be reasonably met. Unadjusted Wald maximum likelihood estimation was used to calculate the risk ratio between prevalent hyperkyphosis and hyperlordosis.
To evaluate the association between age and kyphosis/lordosis angles, several analyses were conducted. First, the sex-stratified association between age and each kyphotic and lordotic angle was plotted using locally smoothed regression (span = 0.75) with 95% confidence intervals (ggplot2). Second, participants were binned by age decile (e.g. 45-54, 55-64, etc.), and the proportion of individuals in each respective age category that had high kyphosis (40° to 50°+) and lordosis (30° to 40°+) angles by 5° intervals was graphed. Last, the mean age of participants in each 10° kyphosis by lordosis angle was presented as a heatmap.
To gain further insight into factors that might contribute to increased spinal curvature, we used multivariable linear regressions to evaluate the association of Cobb angles with 414 pre-existing medical conditions with prevalence > 200 in our population, 59 clinical chemistry measures, and 15 body measure traits (adiposity, bone mineral density, muscle, lung traits) in UK Biobank. All associations were adjusted for age, age², sex, age*sex, smoking, BMI, and Townsend deprivation index. FDR-adjusted significant associations between Cobb angle (either kyphosis or lordosis) and pre-existing conditions and clinical chemistry measures were presented. All 15 adiposity and skeletal-muscular body measure associations were presented, regardless of significance since they were related to our main study interest of phenotypic body composition associations with kyphosis and lordosis angle. All these skeletal-muscular associations were adjusted for variables listed above except BMI was not included in the adjustment of measures highly collinear with BMI calculation (BMI, height, total fat mass, and visceral adipose tissue).
We expected that the associations of various musculoskeletal traits on spine curvature could be non-linear. Thus, to assess ranked independent predictors of Cobb angles (out of body measure traits, age, sex, and smoking status), we ran variable selection using a 10-fold cross-validated boosted generalized regression model with interaction depth = 2 and modeled selected features using splines in the generalized additive model (GAM)20. Variables with relative influence > 0 were selected for the model and relative variable influence percentages were presented. In secondary analyses, we reran all these linear regressions after stratifying by sex to examine potential effect modification by sex of these associations.
Genetic analyses
UKBB imputed genotypes were used in all genetic analyses17. Further details on their methods have been previously published (https://biobank.ndph.ox.ac.uk/showcase/showcase/docs/impute_ukb_v1.pdf). We excluded SNPs with (1) minor allele frequency < 1%, (2) low imputation quality with info value < 0.9, (3) genotype missingness > 10%, or (4) demonstrated deviation from Hardy-Weinberg equilibrium (HWE p < 1x10−10). Participants were excluded from the genomics analyses if they were (1) not of European ancestry (field ID 22006), (2) had heterozygosity or genotype call rate outliers (field ID 22027), (3) demonstrated sex chromosome aneuploidy (field ID 22019), (4) had a mismatch between genetic sex and self reported sex (field ID 22001), or (5) were genetically related (field ID 22011). This yielded a total of 9,911,384 SNPS and 33,413 participants in the genetic analyses. Bonferroni correction was used to evaluate significance in all the genetics analyses and a threshold of pFDR < 0.1 was used. This study was conducted on a pre-existing dataset so was not preregistered.
Genome wide association study
For the initial genetic wide association study (GWAS), a whole-genome regression model was conducted on kyphosis and lordosis separately using REGENIE, a computationally efficient, machine learning-based parallel analysis approach21. In brief, REGENIE first uses cross-validated ridge regression by SNP block for dimension reduction of the genetic data. Then, a second cross-validated ridge regression for each trait is conducted to combine predictors from the first ridge regression into an overall prediction by trait, which is then decomposed by chromosome for a leave-one-chromosome-out (LOCO) scheme. These LOCO predictors are used as a covariate where each phenotype is tested by the set of imputed SNPs, also adjusting for genotype SNP chip (Illumina vs Affymetrix), sex, age, age2, age*sex, and recruitment center. We verified that the test statistics showed no inflation compared to the expectation using the genomic control lambda coefficient (1.13 for kyphosis and 1.09 for lordosis) and the intercept (0.999, SD 0.009 for kyphosis and 1.000, SD 0.084 for lordosis) of linkage disequilibrium score regression.
Multi-trait analysis of GWAS (MTAG)
The method for conducting MTAG from GWAS statistics has been previously described22. MTAG enhances statistical power by leveraging the genetic correlation between traits to generate trait-specific estimates for each SNP. Based on linkage disequilibrium score regression (LDSC) estimates of genetic correlations between kyphosis and lordosis, a joint analysis of the two traits using the GWAS summary statistics was conducted using MTAG. Briefly, variants were restricted to those with minor allele frequency (MAF) > 0.01, and those that passed quality control (QC) in the individual trait GWAS. We verified that MTAG results were not inflated compared to the expectation using the genomic control lambda coefficient (1.14 for kyphosis and 1.11 for lordosis) and the intercept (0.998, SD 0.0085 for kyphosis and 0.9842, SD 0.0089 for lordosis) of LDSC.
Genetic correlation and heritability
To examine if the genetic variation linked to spine angles shares genetics with other traits, we calculated genetic correlations with GWAS catalog phenotypes for kyphosis and lordosis, respectively. All available summary statistics from the EBI GWAS catalog (www.ebi.ac.uk/gwas/) were downloaded on October 19, 202223. Of the 25,475 summary statistics files, 20,021 could be successfully re-formatted by the munge step of LDSC24. Of those, LDSC calculated a positive heritability for 14,843 summary statistics files. Of these traits, 10,170 and 10,138 for kyphosis and lordosis, respectively, yielded non-null SNP heritabilities and were used for downstream genetic correlation analysis.
In addition to traits available on the EBI GWAS catalog, we also estimated the heritability of and genetic correlations between kyphosis or lordosis angles with musculoskeletal traits measured in the UKBB (adiposity, bone mineral content, muscle and lung traits). LDSC was calculated using the repository at https://github.com/bulik/ldsc/, version aa33296. We estimated genetic correlation and heritability using default parameters and the –rg command and –h2 command, respectively (example: ldsc.py --rg kyphosis.sumstats.gz, lordosis.sumstats.gz --ref-ld-chr eur_w_ld_chr/ --w-ld-chr eur_w_ld_chr/ --out). Allele polarization was pre-harmonized to match the reference file w_hm3.snplist. Analyses were adjusted for sex, age, age², age*sex, and recruitment center.
Fine-mapping
GCTA was used for approximate conditional analysis25,26. We considered all variants that passed QC (above) and were within 500 kb of the index variant, except in the major histocompatibility complex (MHC) region due to the complex linkage disequilibrium (LD) structure of the region (GRCh38::6:28,510,120-33,480,577)27. For the LD calculation reference panel, we used genotypes from 5,000 randomly-selected, unrelated, Northern European UKBB participants. For individual loci, variants with locus-wide evidence of association (pjoint < 1x10⁻6) were considered conditionally independent. A threshold of p < 5x10⁻8 was used as the genome-wide association threshold for defining a locus.
Construction of genetic credible sets
For every distinct GWAS signal, credible sets were calculated at a threshold of 95% probability of containing at least one variant with a true non-zero effect size. First, the natural log approximate Bayes factor, Λj, was computed for the jth variant within the fine-mapping region:
where βj and Vj are the estimated effect size and corresponding variance, respectively28. The parameter ω denotes the prior variance in allelic effects and is estimated as (0.15σ)2, where σ is the standard deviation of the phenotype. This standard deviation (σ) was estimated using the formula below:
Here, Var(βj) is the variance of the beta coefficients, fj is minor allele frequency, nj is sample size, and σ2 is the coefficient of the regression which was used to estimate σ with \(\sigma =\sqrt{{\sigma }^{2}}\).
In genetic loci with multiple distinct association signals, we used two techniques (1) exact conditional analysis adjusting for all other index variants in the fine-mapping region and (2) marginal analysis not adjusting for other index variants within the locus. In genetic loci with only a single association signal, we used an unconditional meta-analysis.
Given l variants in the region, we then calculated the posterior probability, πj, that the jth variant was driving the association using:
where γ denotes the prior probability for no association at this locus and k indexes the variants in the region (with k = 0 indicating no association in the region). To account for the expected false discovery rate of 5%, we set γ = 0.05 since a threshold of pmarginal < 5 × 10−8 was used to identify loci for fine-mapping.
For each signal, the 95% credible set was then constructed by (1) ranking all variants by their Bayes factor and (2) including all ranked variants until their cumulative posterior probability exceeded 0.99.
PheWAS
Fine-mapped lead SNPs were annotated to potential functional genes using the Ensembl Variant Effect Predictor, Open Targets, and dbSNP databases. We also considered previous significant associations in the GWASAtlas catalog and summarized our findings in Table 1.
Colocalization
We performed colocalization tests to evaluate whether two traits (spine angle and gene expression) share the same underlying causal variants. For gene expression colocalizations, we used summary statistics from GTEx v829. For physiological and disease trait colocalizations, we used UKBB summary statistics of standardized quantitative and ICD10-categorized traits30. We identified UKBB phenotypes where the minimum p-value within the +/−500kb region around the SNP locus was <5 × 10-8. Colocalization analysis was conducted using the coloc R package with default priors and including all variants within 500 kb of the index variant31. Two genetic signals were considered to have strong colocalization evidence if PP3 + PP4 ≥ 0.99 and PP4/PP3 ≥ 5 and suggestive colocalization evidence if PP3 + PP4 ≥ 0.8 and PP4/PP3 ≥ 331.
Mendelian randomization analysis
To further evaluate causal associations between the genes identified and spinal curvature, we conducted MR analyses32. In MR analysis, we used cis-regulatory or splicing variants that affect RNA expression and function of implicated genes as instrumental variables. We evaluated whether these expression and splicing changes drive changes in a phenotype of interest. Since genetic variants are theoretically randomly assigned when passed from parents to offspring, this method is expected to minimize the effect of confounding and avoids reverse causation.
The MR method relies on three important assumptions: (i) the genetic variant is associated with the exposure, (ii) the genetic variant is not associated with any confounders, (iii) the genetic variant is not associated with the outcome through any pathway other than via the exposure (no horizontal pleiotropy). Since multiple tissues can affect spine curvature, we carried out two sample MR analyses across all 54 tissue types measured in GTEx v829. To identify independent SNP instruments for each exposure, we pruned GWAS-significant SNPs (p-value < 5 × 10-8) for each risk factor using a threshold of r2 < 0.01 and LD window of 250 kb. The construction of reference panels for LD calculations was described in the fine-mapping section. When multiple independent eQTLs were observed for the same gene in a single tissue within GTEx, we evaluated the causal effect of a gene within each genome-wide significant locus on kyphosis and lordosis using inverse variance weighting (IVW). For genes with only a single independent eQTL association in a tissue, we used the Wald ratio test. A notable limitation of eQTL-based MR analysis was that bone and connective tissues were not available in the current version of GTEx (v8), so we cannot rule out the possibility that causal effects on spine angles are not due to gene expression in missing tissues.
For sensitivity analyses, the MR analysis was also reconducted using (1) MR-Egger regression and (2) weighted median-based and mode-based tests of the ordered Wald ratio estimates33,34. Since weighted median and mode estimates assume that the estimates from pleiotropic variants are outliers, these methods tend to be less sensitive to the effect of pleiotropic variants35. On the other hand, MR-Egger involves a weighted linear regression to evaluate the marginal effect of each SNP on the outcome on the marginal effect of each SNP to the exposure. Thus, this method evaluates the overall directional pleiotropic contribution of weak instrumental SNPs on the risk estimate. As such, we used these methods as sensitivity analyses to confirm that there was a consistent directional effect between the variants and traits relative to the IVW and Wald ratio results35.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Results
Cobb angle annotation by machine learning
We developed ML models to mimic a well-established Cobb angle measurement technique used in radiological practice36. Briefly: a U-net segmentation model was trained to label the spine body, and a previously published37 object-detection model was used to identify appropriate regions for Cobb angle estimation. Training and testing data from those models were separate from the Cobb angle preliminary training and testing sets described below.
Cobb angles were estimated between tangent lines drawn through appropriate spine regions using Deming regression. In lateral DXA scan images, four human annotators (PhD spine experts) measured two sets of Cobb angles across 320 images selected randomly from the UKBB dataset, excluding randomly-chosen images that were set aside for the training or testing of upstream models (i.e. spine segmentation, bridge object detection, left/right classification). Four independent annotators measured T12-T5 kyphotic angle and 3 independent annotators measured L4-L1 lordotic angle (Fig. 1A). These images and their annotations were randomly split into a preliminary training dataset (120 images), used to calibrate parameters of the ML & regression pipeline described above, and a testing dataset (200 images), used for final evaluation of each model’s performance. There was a moderate degree of concordance across the human annotators (r = 0.66-0.91, Supplementary Fig. 1).
The ML-based estimation algorithms for both kyphosis and lordosis performed well on the 200-image testing dataset, closely matching the results of human annotators with high correlation values (r = 0.90 and 0.89; MSE = 17.39 and 35.78 for the kyphosis and lordosis models, respectively) (Fig. 1B, C). The lordosis model has slightly lower accuracy, likely due to greater variability in the ability to define the location of the L4 and L5 junctions.
Epidemiology of kyphosis and lordosis
Among the 41,212 UKBB participants included in this study, 48.3% were male. The median age of DXA imaging subjects was 64 [interquartile interval (IQI) 58, 70] years (Supplementary Data 1). A total of 3702 (9.0%) participants had hyperkyphosis (angle > 40°), and 3,109 (7.5%) had hyperlordosis (angle > 30°), of which 1,057 (2.6%) participants had both. Thoracic kyphosis angles were strongly correlated to lumbar lordosis angles (r = 0.49, p-value < 0.001, Supplementary Fig. 2). Kyphosis and lordosis angles were systematically higher in females compared to males at all ages (Fig. 2A). Both kyphotic and lordotic angles increased with age, at approximately the same rate: 2.28° (IQI 2.16°, 2.39°) of kyphotic angle and 1.32° (IQI 1.19°, 1.44°) of lordotic angle per decade of life (not adjusted for other factors).
a Kyphotic and lordotic angles as a function of age, stratified by sex. Spinal curvature increases with age in both sexes with approximately the same slope, but kyphotic and lordotic angles are greater in females. Confidence intervals represent SEs. b Prevalence of hyperkyphosis and hyperlordosis increases by age category. c Mean age increases jointly in higher kyphotic and lordotic angle categories.
Since both angles increase with age, we asked if they also increased as a function of each other. In the multivariable model, adjusted for age and sex, the kyphotic angle increased an additional ~0.4° for every degree of lordotic angle (Supplementary Fig. 4). Consequently, there was a graded increase in the prevalence of hyperkyphosis and hyperlordosis with age (Fig. 2B). By age 75+, the prevalence of hyperkyphosis and hyperlordosis was 15.4% and 10.4%, respectively. Participants with hyperkyphosis were 5.22 (4.89, 5.58) times more likely to also have hyperlordosis than those without hyperkyphosis and, relatedly, participants with hyperlordosis were 4.90 (4.61, 5.21) times more likely to also have hyperkyphosis. Those with higher observed kyphosis and lordosis angles tended to be older (Fig. 2C).
Disease associations with spine curvature
Various pre-existing pathological conditions were associated with increased spine angles (Fig. 3A). First, among the strongest associations for kyphosis and lordosis angles were previous diagnoses of intervertebral disc disorders such as disc compression disorders (M41, M47, M51, G55) and osteoporosis (M81). Prior diagnosis of multiple sclerosis (MS) was highly associated with increased kyphotic angle by 2.24° (0.83°, 3.64°), potentially due to the effect of MS on posture38. We also observed that a variety of gastric conditions including esophagitis, esophageal disease, and gastro-esophageal reflux disease (GERD) [K21, K22, K20] and, perhaps relatedly, diaphragmatic hernia (K44) were associated with increased kyphotic angles, also possibly due to the association of kyphosis with posture, reduced back muscle strength, and increased intra-abdominal pressure39. Interestingly, we observed that metabolic disorders, such as diabetes, gout, and hyperlipidemia (E11, E14, M10, E78), were associated with decreased spine curvature. This effect was generally more pronounced in females, where diabetes was linked to reduction of kyphotic angle by 1.50° (0.71°, 2.30°) versus 0.63° (0.06°, 1.19°) in males (Supplementary Fig. 3). The association with these disorders with kyphosis and lordosis angles tended to be linear (Supplementary Fig. 4). More comorbid conditions were associated with kyphotic angles than lordotic angles, likely in part due to a higher precision in our machine learning-based estimates of kyphotic angles.
Angle as a function of a diseases (a) or biomarker values (b). Beta is a degree change in kyphosis and lordosis angle with the presence of a prior condition or per standard deviation increase in biochemical measure. All analyses were adjusted by age at visit, sex, BMI, smoking status, and Townsend deprivation index.
In serum biochemistry data, we observed that elevated levels of plasma alkaline phosphatase (ALP) were associated with significant increases in both kyphosis and lordosis angles, supporting established literature that osteoporosis is an important contributing factor to increased spine curvature (Fig. 3B). Similar to the inverse associations we found between kyphosis and lordosis angles with metabolic comorbidities, in biochemistry data, elevation of metabolic biomarkers (hemoglobin A1c [HbA1c], urate, and triglycerides) were associated with significant reductions in kyphotic angles. This inverse association between biomarkers of metabolic disorders and kyphosis was consistent for both sexes (Supplementary Fig. 5).
Association of musculoskeletal traits with spine curvature
To evaluate how aging of the musculoskeletal system contributes to increased spinal curvature, we performed a phenotypic association scan with muscle, adiposity, and bone traits across the entire skeletal system in UKBB. Significant associations, adjusted for multiple hypothesis testing, are shown in Fig. 4A. We observed that bone mineral density (BMD) measures across the entire skeletal system were strongly associated with spine curvature. Both kyphotic and lordotic angles were consistently larger in those with lower BMD. For example, the kyphotic angle increased by 0.23° (0.17°, 0.31°), and the lordotic angle increased by 0.47° (0.40°, 0.54°) per standard deviation loss of spine BMD. This directional association was mostly consistent for both males and females across skeletal sites, with associations stronger in females than males (Supplementary Fig. 6).
a Association of musculoskeletal traits with spine curvature. Analyses were adjusted by age at visit, sex, BMI, smoking status, and Townsend deprivation index, except analyses examining BMI, height, total fat mass, and visceral adipose tissue were not adjusted for BMI. Beta is a degree change in angle per standard deviation increase in physiological measure. b Importance of cross-validated gradient boosting algorithm (GBM)-selected variables for predicting spine curvature.
Among muscle measurements, lower muscle mass in both the legs and the arms was also associated with increased spinal curvature, indicating the important role skeletomuscular factors play in the development of pathology (Fig. 4A). On average angle increased by 0.86° (0.79°, 0.93°) kyphosis and 0.88° (0.81°, 0.95°) lordosis per standard deviation loss of leg muscle mass (Supplementary Fig. 6). Decreased grip strength was also strongly associated with increased spine angles.
Interestingly, we observed that fat composition measures showed a differential effect between lordosis and kyphosis (Supplementary Fig. 7). Higher adiposity (total and visceral fat) was linked with increased lordotic angles but reduced kyphotic angles. Further examining regional measures of adiposity, abdominal adiposity was more strongly associated with lordotic angle, potentially resulting from increased anterior pressure on the lumbar spine, while kyphosis was more strongly and inversely associated with trunk adiposity (Supplementary Fig. 8). The same differential effect was observed with body mass index (BMI) and was consistent for males and females (Supplementary Fig. 9).
Since physiological measurements are strongly associated and can be non-linearly dependent on each other, to identify the most important predictors of spine curvature we used gradient boosting algorithm (GBM)40. While predictors of angles at both sites were similar, their relative contribution (as measured by variable importance) was different (Fig. 4B). Consistent with single trait analysis, the top independent predictors of kyphosis were age, muscle mass, visceral adiposity, grip strength, and BMD. Independent predictors of lordosis were similar to those for kyphosis, but sex, height, and BMI were stronger predictors of lordosis.
Genetic risk factors of spine curvature
Leveraging genotyping information, we next evaluated genetic risk factors associated with the development of spinal curvature. We carried out a genome wide association study (GWAS) for both kyphosis and lordosis angles within the participants of Northern European origin (n = 33,413). Both traits had significant heritability (h2 kyphosis: 0.34 [0.30, 0.38], lordosis: 0.26 [0.22, 0.30]). We found that there was a strong genetic correlation between kyphotic and lordotic angles (Rg = 0.71, p-value = 1.58x10⁻48) indicating that common genetic variation contributes to both traits.
The GWASes identified eight genome-wide significant loci associated with kyphosis and six genome-wide significant loci associated with lordosis (Supplementary Fig. 10). Since we observed a strong genetic correlation between kyphosis and lordosis, to increase the statistical power, we carried out multi-trait analysis of GWAS (MTAG) to jointly model these traits22. The results of the MTAG analysis (Fig. 5A) for each trait were generally consistent with individual trait GWAS (Supplementary Fig. 9). Given the greater power and precision of MTAG, we focus our downstream analyses on the MTAG summary statistics for each trait in the remaining sections.
a MTAG GWAS manhattan plot with fine-mapped lead SNPs annotated. b GWAS catalog phenotypic correlations with kyphosis and lordosis angle. In green are significant associations where pFDR < 0.05. c Comparison of genetic and phenotypic correlations. Displayed in the upper left triangle are genetic correlations for selected musculoskeletal traits. Displayed in the lower right triangle are phenotypic correlations of selected musculoskeletal traits. Both traits show similar patterns of genetic and phenotypic correlations, with greater kyphosis and lordosis angles anti-correlated with other traits. Significant correlations are marked with asterisks. d Genetic summary figure of GWAS, colocalization (eQTL and sQTL), and MR associations by lead SNP and gene.
Genetic correlation
To examine if the genetic variation linked to spine angles shares genetics with other traits, we calculated genetic correlations with 10,170 and 10,138 GWAS catalog phenotypes for kyphosis and lordosis, respectively (Fig. 5B, Supplementary Data 2). Top traits associated with kyphosis were measures of muscle mass (appendicular lean mass), bone mineral density (heel, total body), and diseases related to the esophagus (esophageal disorder, Barrett’s esophagus, hiatal hernia, GERD). The top genetic correlations of lordosis were similar to that of kyphosis but with stronger genetic associations with body type measures including height, BMI, waist circumference (Supplementary Data 3). In line with epidemiological association with GERD diagnosis, we observed a strong genetic correlation between both kyphosis and lordosis with esophageal diseases and omeprazole usage (Rg between 0.10 and 0.25, all p-values < 1x10⁻3), further suggesting that esophageal diseases share genetic risk factors with spine curvature traits.
To assess the similarity of demographic-adjusted genetic and phenotypic correlations, we computed an all-by-all correlation matrix for selected musculoskeletal traits and displayed it in a single heatmap (Fig. 5C). While most traits showed strong phenotypic correlation with one another, genetic correlations were less statistically powered and therefore more sparse. As expected, strong interdependence between all musculoskeletal traits can be observed with shared genetic origins. A mechanistic understanding of how these traits contribute to the development of spine curvature requires a deeper understanding of the loci involved (Fig. 6).
Loss of bone mineral density, decreased muscle mass, and decreased lung capacity are associated with increased kyphosis and lordosis angles. Adiposity has opposing effects on the thoracic and lumbar spine: decreasing kyphosis and increasing lordosis. Aging is a common risk factor for declines in physiological function and presentation of comorbid conditions such as osteoporosis, type 2 diabetes, and esophageal diseases. Both traits are strongly genetically correlated and share common risk alleles such as PTHLH → BMD, COL11A1 → connective tissue, and ETFA → muscle mass, as well as a number of genes involved in DNA repair and cellular homeostasis processes (RAD9A, MMS22L, HIF1A, RAB28).
Gene prioritization
To better understand genetic risk factors we carried out two types of analyses (i) fine-mapping followed by colocalization with Genotype-Tissue Expression (GTEx) RNA splicing and RNA expression quantitative trait loci (eQTLs) in 49 available tissues (ii) MR with GTEx RNA expression (in the 54 tissues available) as a mediator of Cobb angle outcomes29. Combining these multiple lines of evidence, we were able to prioritize genes with high likelihood of being involved in the development of kyphosis and lordosis angles.
Fine-mapping identified 12 unique variants associated with kyphosis and lordosis angles (seven with kyphosis and eight with lordosis) (Table 1, Supplementary Data 4). Of these, there were three overlapping fine-mapped lead single nucleotide polymorphisms (SNPs) shared by kyphosis and lordosis: rs881926 near SFRP4, rs9651832 near CCDC91, and rs3783820 near SLC38A6. Variation near RAB28 and MMS22L genes showed a stronger association with kyphosis, while variation near COL11A1, OTOG, RAD9A, and ETFA genes showed a stronger association with lordosis. Full fine-mapping results can be found in Supplementary Data 5 and 6. Previous GWAS studies have implicated SNPs in these loci with multiple musculoskeletal traits such as height, heel BMD, appendicular lean mass, various measures of lung function, and spinal stenosis (Supplementary Fig. 11).
Using variants identified in fine-mapping with either lordosis or kyphosis, we ran colocalization analyses of Cobb angles with RNA expression data (eQTLs and splicing quantitative trait loci [sQTL]) across multiple tissues profiled in GTEx v8 (Supplementary Data 7). (See Methods). Although we observed colocalization with both traits for some genes, we found more associations with lordosis (Fig. 5D). Some of the fine-mapped SNPs, colocalized with RNA expression of multiple genes within the same loci. For example, we observed colocalization of kyphotic and lordotic angles with both PTHLH and CCDC91 on the chromosome 12 locus, along with both H1F1A and L3HYPDH on chromosome 14.
We followed up colocalization analysis with MR. In MR, genetic variants (instruments) are used to estimate the causal effect of an exposure (RNA expression) on an outcome (spinal curvature). The detailed description of methodology and its limitations can be found in the Methods. Given correlated RNAseq expression patterns observed in colocalization analysis, we can not rule out the possibility of horizontal pleiotropy, but we did find support for the potential mediation effect of multiple genes. We found that decreased expression of RAD9A (multiple tissues), ETFA (skeletal muscle), COL11A (adipose tissue), ISL2 (esophagus), and increased expression of RNASEH2C (brain), and CCDC91 or PTHLH and SCRAPER (multiple tissues) were significantly associated with increased Cobb angles (Supplementary Data 8 and 9, Fig. 5D).
Following MR-STROBE guidelines41, we evaluated the sensitivity of our MR results. We repeated analyses using MR-Egger and median-weighted MR methods. Effect sizes were concordant across different estimation methods. MR-Egger did not find statistically significant associations, but this was expected given lower statistical power of the methodology. When multiple instruments were available, to avoid estimation bias we tested for heterogeneity of instruments. We did not find significant heterogeneity across instruments.
We summarized our findings in Fig. 6. Overall, a complex picture of genetic risk factors emerges, involving genes expressed in multiple tissues with fundamental roles in maintenance of tissues and cellular homeostasis. This included genes such as: COL11A1 (maintenance of connective tissue), RAB28 (intracellular trafficking), MMS22L (DNA double-strand break repair), TWIST1 (transcription factor involved in skeletal-vascular development), EPDR1 (glycoprotein related to neurogenesis), PLEKHA7 (key adherens junction component), RAD9A (DNA double-strand break repair), PTHLH (parathyroid like hormone), HIF1A (hypoxia response including angiogenesis and glycolysis), and ETFA (mitochondrial fatty acid beta-oxidation). Many genes also displayed non-specific tissue expression patterns.
Discussion
Increased spinal curvature is a universally recognized aging trait; however, the genetic and physiological etiology of this trait has not been well-characterized. In this study, we leveraged machine learning models to automatically quantify kyphosis and lordosis angles for 41,212 community-dwelling middle-aged to older adults in UKBB. Physiology, serum biochemistry, and genetics data were analyzed to build an integrated epidemiological and biological understanding of spinal curvature.
We observed that increased curvature in both the lumbar and thoracic spine were strongly intercorrelated and age-dependent (2.4° and 1.5° per decade of life). Previous investigations of the effect of aging on spine angles in males and females did not paint a consistent picture of sex-specific differences in kyphosis and lordosis angle and how they change with age42,43. In our analysis, we see a consistent aging effect in both males and females, but with a different baseline of higher kyphosis and lordosis angles in females by the age of 40. Although we do not have measurements at earlier ages, it is plausible that differences in spine curvature arise much earlier in life or due to developmental differences between sexes. While our cross-sectional analysis of aging effects could be biased by the characteristics of the cohort, they are consistent with previous longitudinal assessment of aging effects44.
As the population ages, both hyperkyphosis and hyperlordosis increased, reaching a substantial prevalence of 15.4% and 10.4%, respectively, in those age 75+. While the prevalence and assessment of hyperkyphosis has been well-characterized, the prevalence of hyperlordosis and its cause is limited45,46. Previous epidemiological studies of hyperkyphosis have implicated several potential pre-existing conditions, such as vertebral fractures and osteoporosis, that could contribute to the development of this pathology45. We have conducted a systematic evaluation of all common pre-existing conditions associated with kyphosis and lordosis angles. The majority of pre-existing conditions associations (multiple sclerosis, osteoporosis, and gastroesophageal diseases) were the same between kyphosis and lordosis, matching in both direction and effect sizes. In our analysis of biochemical factors, we likewise observed similar correspondence between kyphosis and lordosis. These results led us to conclude that the increase in spinal curvature at two sites is driven by a similar set of underlying factors.
Our analysis of physiological measures suggested that the loss of muscle mass (and relatedly muscle strength) and decreased bone mineral density were the factors most strongly associated with increased spinal curvature. The slope of age-dependent angle increase was approximately the same for both males and females, even though females on average had lower muscle mass, lower BMD, and higher prevalence of osteoporosis. This suggests that the aging effect was not due to baseline differences in BMD and muscle measures and further highlights the important, complex relationship between spinal curvature with muscle mass and BMD by age8,47,48,49.
Examining the effect of body adiposity measures on spine angles, we were surprised to find that increased adiposity (total fat mass, visceral fat, and BMI) was associated with decreased kyphotic angles, but increased lordotic angles. Among physiological measures considered, adiposity was the only measure that showed this opposing relationship between the two traits. This effect remained after adjusting for age, sex, and respective spine angles. The increased lumbar lordosis with adiposity may be due to the biomechanics of the spine, where the lumbar region compensates for increased adiposity in the trunk by moving the spine forward to adjust the center of gravity50. Alternatively, decreased kyphotic angles were linked to a higher prevalence of metabolic disorders (type 2 diabetes, gout, elevated glucose, and HbA1c), but less so with lordotic angles. It is possible that metabolic disorders can result in an incidental increase in BMD, perhaps strengthening the upper spine and thus decreasing kyphotic angle, while more abdominal adiposity can increase the load on the lower spine50,51,52.
Genetic analyses elucidated the complex origin of spine curvature traits. To our knowledge, this is the first GWAS of kyphosis and lordosis published to-date. We found that angles at both sites had substantial heritability (25–35%) and were strongly genetically correlated, indicating that common genetic drivers influence spine angles at both sites. This is comparable to prior studies on clinical hyperkyphosis which have similarly found high heritability in twins9,10. Genetic correlation with other traits was consistent with phenotypic associations, with appendicular lean mass and BMD among the strongest correlated traits. Interestingly, in both phenotypic and genetic associations, we observed an inverse correlation between height and spine angles (higher kyphosis/lordosis associated with smaller stature/height). It was not clear to us if this was a trivial observation. However, this seemed to indicate that there could be shared genetics that determine these skeletally-related traits.
We also observed a strong genetic correlation of increased spine curvature with the development of gastroesophageal disease (GERD and Barrett’s esophagus). This genetic correlation was consistent with increased use of proton-pump inhibitors (PPIs) such as omeprazole and GERD being among the strongest associated pre-existing conditions, as well as future GERD diagnosis events. It is plausible that a genetically driven decline in muscle mass is a common factor contributing to both conditions53. Increased incidence of GERD is also known to be associated with metabolic syndrome (eg. diabetes, obesity)54,55. However, we did not observe a significant genetic correlation between GERD and metabolic syndrome. This suggests that metabolic syndrome is unlikely a common genetic risk factor between GERD and spine curvature. The link between these gastroesophageal diseases and spine curvature may also lie in the common use of PPIs for treatment. Chronic use of PPIs which has been associated with increased fracture risk and osteoporosis which may therefore lead to increased spinal curvature56,57.
From genetic colocalization and MR, most of the alleles and implicated genes we identified were previously reported in GWASAtlas as associated with broad muscular-skeletal traits such as height, BMD, appendicular lean mass, and maintenance of connective tissues58. Molecular functions and expression patterns of implicated genes did not point to any single cell type or specific tissue involvement, rather they indicated involvement of global cellular maintenance processes. For example, we found support for the hypothesis that expression of genes involved in DNA repair processes (RAD9A), and microtubule-dependent chromosome segregation (SCAPER) are involved in spinal curvature59,60. Potentially in line with the observation that fundamental DNA repair processes are playing a role, previous studies have demonstrated that elevated p53 activity, and partial loss of mitochondrial checkpoint protein BubR1 can lead to increased senescence and dramatically increased spinal curvature61,62.
A potential causal association of parathyroid hormone-like hormone (PTHLH) with the development of kyphosis is also interesting, as it specifically points to the role of calcium/phosphate homeostasis in the pathology. Rare variants in PTHLH gene are known to produce skeletal defects63. A number of other genes involved in calcium/phosphate homeostasis, such as KLOTHO and FGF23 have been shown in animal model systems to lead to the development of kyphosis, likely through BMD loss and development of osteoporosis64. Further studies are necessary to unravel the link between hyperparathyroidism and spine curvature. As PTHrP therapy is an approved treatment for osteoporosis and is known to increase vertebral BMD, it might be possible to develop intervention strategies targeted to the kyphotic population with this genetic variation65.
Other genes such as ETFA, critical to normal mitochondria maintenance and metabolism, could be contributing to the development of kyphosis via skeletal muscle declines66,67. It is plausible that reduced capacity to repair cellular machinery in muscle and connective tissues results in damage to these tissues over the lifetime of individuals, consequently leading to increased spinal curvature68,69. These effects are potentially exacerbated in individuals who carry mutations in cellular maintenance pathways and become more apparent as their development of spine curvature accelerates with age68,70,71.
Beyond genetic predisposition, our findings suggest that bone and muscle physiology play an important role in the progression of kyphosis. As such, interventions such as resistance training have the potential to improve both muscle mass and strength, as well as increase bone mineral density72,73,74. Indeed, physical therapy in hyper-kyphotic individuals was shown to improve kyphosis by ~3–5°75,76,77. It is also plausible that, if increased spinal curvature is detected early and can be attributed to osteoporosis, medications such as bisphosphonates could potentially offer additional benefits44,78.
There are several limitations in the current study. DXA scans used to quantify Cobb angles were conducted in the supine lateral position, which could affect spine curvature as compared to an orthostatic measure79. In the analysis of lumbar lordosis, we were not able to reliably identify the L5 vertebrae due to pelvis obstruction, thus all Cobb angles were computed from L1-L4. This can lead to increased noise in this measure. Additionally, the number of hand annotated samples used to train the machine learning model was limited. Despite this, we are confident in the automated Cobb angle measures because the accuracy was strong (r = 0.906 and 0.877 for kyphosis and lordosis, respectively). The association between aging and spinal curvature was based on cross-sectional data and thus does not represent individual aging trajectories, but rather overall population trends. Participants of the UKBB imaging study are healthy volunteers recruited from the general community and therefore tend to be healthier than the general population80; however, the cohort is well-suited to answer questions about various genetic, aging, and pathological factors that contribute to increased spinal curvature. Additionally, the significant colocalization findings of a few SNPs (rs1318756, rs3783820, rs9651832, rs71457747, and rs12908334) with RNA expression (eQTL/sQTL) of multiple genes may suggest potential horizontal pleiotropy in the genetic analyses, especially for MR. Another limitation to note for the colocalization and MR analyses was that many skeletal and connective tissues were unavailable in GTEx. Thus, the possibility of associations between genetic expression in these missing tissues and spine angles cannot be dismissed.
Overall, our study identified that declines in processes involved in muscle and bone maintenance were greatly involved in age-related increases in spinal curvature. We found that increased body adiposity has distinct and opposing effects on lumbar and kyphotic angles, and increased spine curvature was associated with the development of GERD. Beyond age, maintenance of muscle mass was a key driver for kyphosis, while fat composition measures were the strongest drivers of lordosis. Analysis of genetic risk alleles revealed that molecular mechanisms were shared across multiple tissues and involve maintenance of cellular homeostasis such as DNA repair, RNA biogenesis, and mitochondrial metabolism and maintenance.
Data availability
Data from the UK Biobank is available on request (https://www.ukbiobank.ac.uk/). Summary statistics are available in the GWAS catalog under accession number (GCST90570047,GCST90570048).
Code availability
Our open-source code can be accessed at https://github.com/graham-calico/KyphoLordoDxa.git81.
References
Fon, G. T., Pitt, M. J. & Thies, A. C. Jr Thoracic kyphosis: range in normal subjects. AJR Am. J. Roentgenol. 134, 979–983 (1980).
Lin, R. M., Jou, I. M. & Yu, C. Y. Lumbar lordosis: normal adults. J. Formos. Med. Assoc. 91, 329–333 (1992).
Benoist, M. Natural history of the aging spine. Eur. Spine J. Suppl 2, S86−S89 (2005).
Ailon, T., Shaffrey, C. I., Lenke, L. G., Harrop, J. S. & Smith, J. S. Progressive spinal kyphosis in the aging population. Neurosurgery 77, S164–S172 (2015).
Sparrey, C. J. et al. Etiology of lumbar lordosis and its pathophysiology: a review of the evolution of lumbar lordosis, and the mechanics and biology of lumbar degeneration. Neurosurg. Focus 36, E1 (2014).
Roghani, T., Zavieh, M. K., Manshadi, F. D., King, N. & Katzman, W. Age-related hyperkyphosis: update of its potential causes and clinical impacts-narrative review. Aging Clin. Exp. Res. 29, 567–577 (2017).
Yau, M. S. et al. Heritability of thoracic spine curvature and genetic correlations with other spine traits: the framingham study. J. Bone Miner. Res. 31, 2077–2084 (2016).
Lorbergs, A. L. et al. A longitudinal study of trunk muscle properties and severity of thoracic kyphosis in women and men: the framingham study. J. Gerontol. A Biol. Sci. Med. Sci. 74, 420–427 (2019).
Stone, M. A. et al. Heritability of spinal curvature and its relationship to disc degeneration and bone mineral density in female adult twins. Eur. Spine J. 24, 2387–2394 (2014).
Damborg, F., Engell, V., Andersen, M., Kyvik, K. O. & Thomsen, K. Prevalence, concordance, and heritability of scheuermann kyphosis based on a study of twins. JBJS 88, 2133 (2006).
Murray, K. J., Le Grande, M. R., Ortega de Mues, A. & Azari, M. F. Characterisation of the correlation between standing lordosis and degenerative joint disease in the lower lumbar spine in women and men: a radiographic study. BMC Musculoskelet. Disord. 18, 330 (2017).
Katzman, W. et al. Association of spinal muscle composition and prevalence of hyperkyphosis in healthy community-dwelling older men and women. J. Gerontol. A Biol. Sci. Med. Sci. 67, 191–195 (2012).
Patel, R. et al. Trabecular bone score and its association with Cobb angle kyphosis in older men: a cross-sectional study for the Osteoporotic fractures in men (MrOS) Study. Osteoporos. Int. 33, 1171–1176 (2022).
Bartynski, W. S., Heller, M. T., Grahovac, S. Z., Rothfus, W. E. & Kurs-Lasky, M. Severe thoracic kyphosis in the older patient in the absence of vertebral fracture: association of extreme curve with age. AJNR Am. J. Neuroradiol. 26, 2077–2085 (2005).
Koelé, M. C. et al. The association between hyperkyphosis and fall incidence among community-dwelling older adults. Osteoporos. Int. 33, 403–411 (2022).
Kado, D. M., Huang, M.-H., Nguyen, C. B., Barrett-Connor, E. & Greendale, G. A. Hyperkyphotic posture and risk of injurious falls in older persons: the Rancho Bernardo Study. J. Gerontol. A Biol. Sci. Med. Sci. 62, 652–657 (2007).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018).
Sudlow, C. et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 12, e1001779 (2015).
Townsend, P. Deprivation*. J. Soc. Policy 16, 125–146 (1987).
Wood, S. N. Generalized Additive Models: An Introduction with R Second Edition, Vol. 496 (CRC Press, 2017).
Mbatchou, J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 53, 1097–1103 (2021).
Turley, P. et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 50, 229–237 (2018).
Sollis, E. et al. The NHGRI-EBI GWAS catalog: knowledgebase and deposition resource. Nucleic Acids Res. 51, D977–D985 (2023).
Grotzinger, A. D. et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav 3, 513–525 (2019).
Bulik-Sullivan, B. K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
MHC region of the human genome. Genome Reference Consortium. https://www.ncbi.nlm.nih.gov/grc/human/regions/MHC (2025).
Wakefield, J. Bayes factors for genome-wide association studies: comparison with P-values. Genet. Epidemiol. 33, 79–86 (2009).
GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Churchhouse, C. Rapid GWAS Of Thousands Of Phenotypes For 337,000 Samples In The Uk Biobank. http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank (2017).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet 10, e1004383 (2014).
Smith, G. D. & Ebrahim, S. Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease?. Int. J. Epidemiol. 32, 1–22 (2003).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40, 304–314 (2016).
Hartwig, F. P., Davey Smith, G. & Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46, 1985–1998 (2017).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525 (2015).
Vrtovec, T., Pernus, F. & Likar, B. A review of methods for quantitative evaluation of spinal curvature. Eur. Spine J. 18, 593–607 (2009).
Sethi, A., Ruby, J. G., Veras, M. A., Telis, N. & Melamud, E. Genetics implicates overactive osteogenesis in the development of diffuse idiopathic skeletal hyperostosis. Nat. Commun. 14, 2644 (2023).
Muhtaroglu, M. et al. Evaluation of thoracic kyphosis angle and respiratory functions in patients with multiple sclerosis. Ann. Appl. Sport Sci. 9, 0–0 (2021).
Imagama, S. et al. Influence of lumbar kyphosis and back muscle strength on the symptoms of gastroesophageal reflux disease in middle-aged and elderly people. Eur. Spine J. 21, 2149–2157 (2012).
Friedman, J. et al. glmnet: Lasso And Elastic-net Regularized Generalized Linear Models. Astrophysics Source Code Library, ascl:2308.011 (2023).
Skrivankova, V. W. et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR statement. JAMA 326, 1614–1621 (2021).
Milne, J. S. & Lauder, I. J. Age effects in kyphosis and lordosis in adults. Ann. Hum. Biol. 1, 327–337 (1974).
Mohan, M. & Huynh, L. Sex Differences in the spine. Curr. Physi. Med. Rehabi. Rep. 7, 246–252 (2019).
Kado, D. M. et al. Factors associated with kyphosis progression in older women: 15 years’ experience in the study of osteoporotic fractures. J. Bone Miner. Res. 28, 179–187 (2013).
Katzman, W. B., Wanek, L., Shepherd, J. A. & Sellmeyer, D. E. Age-related hyperkyphosis: its causes, consequences, and management. J. Orthop. Sports Phys. Ther. 40, 352–360 (2010).
Been, E. & Kalichman, L. Lumbar lordosis. Spine J 14, 87–97 (2014).
Hicks, G. E. et al. Cross-sectional associations between trunk muscle composition, back pain, and physical function in the health, aging and body composition study. J. Gerontol. A Biol. Sci. Med. Sci. 60, 882–887 (2005).
Katzman, W. B., Miller-Martinez, D., Marshall, L. M., Lane, N. E. & Kado, D. M. Kyphosis and paraspinal muscle composition in older men: a cross-sectional study for the Osteoporotic Fractures in Men (MrOS) research group. BMC Musculoskelet. Disord. 15, 19 (2014).
Kaiser, J. et al. Correspondence between bone mineral density and intervertebral disc degeneration across age and sex. Arch. Osteoporos. 13, 123 (2018).
Miranda, A. P. O. C., Penha, P. J., Pereira, L. G., Pessoa, W. C. & João, S. M. A. Influence of sex and body mass index on the thoracic Kyphosis and Lumbar lordosis. J. Manipulative Physiol. Ther. 45, 508–514 (2022).
Ma, L. et al. Association between bone mineral density and type 2 diabetes mellitus: a meta-analysis of observational studies. Eur. J. Epidemiol. 27, 319–332 (2012).
Muka, T. et al. The association between metabolic syndrome, bone mineral density, hip bone geometry and fracture risk: the rotterdam study. PLoS ONE 10, e0129116 (2015).
Kim, Y. M. et al. Association between skeletal muscle attenuation and gastroesophageal reflux disease: a health check-up cohort study. Sci. Rep. 9, 20102 (2019).
Sun, X.-M., Tan, J.-C., Zhu, Y. & Lin, L. Association between diabetes mellitus and gastroesophageal reflux disease: A meta-analysis. World J. Gastroenterol. 21, 3085–3092 (2015).
El-Serag, H. The association between obesity and GERD: a review of the epidemiological evidence. Dig. Dis. Sci. 53, 2307–2312 (2008).
Fraser, L.-A. et al. The effect of proton pump inhibitors on fracture risk: report from the Canadian multicenter Osteoporosis study. Osteoporos. Int. 24, 1161–1168 (2013).
Center for Drug Evaluation & Research. FDA Drug Safety Communication: Possible Increased Risk Of Fractures Of The Hip, Wrist, And Spine With The Use Of Proton Pump Inhibitors. http://www.fda.gov/Drugs/DrugSafety/PostmarketDrugSafetyInformationforPatientsandProviders/ucm213206.htm (2019).
Liu, X. et al. GWAS Atlas: an updated knowledgebase integrating more curated associations in plants and animals. Nucleic Acids Res. 51, D969–D976 (2023).
Höpfler, M. et al. Mechanism of ribosome-associated mRNA degradation during tubulin autoregulation. Mol. Cell 83, 2290–2302.e13 (2023).
Tong, H. L., Jiang, R. Y., Zhang, W. W. & Yan, Y. Q. MiR-2425-5p targets RAD9A and MYOG to regulate the proliferation and differentiation of bovine skeletal muscle-derived satellite cells. Sci. Rep. 7, 418 (2017).
Tyner, S. D. et al. p53 mutant mice that display early ageing-associated phenotypes. Nature 415, 45–53 (2002).
Baker, D. J. et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 479, 232–236 (2011).
Klopocki, E. et al. Deletion and point mutations of PTHLH cause brachydactyly type E. Am. J. Hum. Genet. 86, 434–439 (2010).
Kuro-o, M. et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 390, 45–51 (1997).
Miller, P. D. et al. Effect of abaloparatide vs placebo on new vertebral fractures in postmenopausal women with osteoporosis: a randomized clinical trial. JAMA 316, 722–733 (2016).
Schiff, M., Froissart, R., Olsen, R. K. J., Acquaviva, C. & Vianey-Saban, C. Electron transfer flavoprotein deficiency: functional and molecular aspects. Mol. Genet. Metab. 88, 153–158 (2006).
Fiorentino, T. V. et al. Pioglitazone corrects dysregulation of skeletal muscle mitochondrial proteins involved in ATP synthesis in type 2 diabetes. Metabolism 114, 154416 (2021).
Bertram, C. & Hass, R. Cellular responses to reactive oxygen species-induced DNA damage and aging. Biol. Chem. 389, 211–220 (2008).
Mognato, M. DNA Repair: An Update. (BoD – Books on Demand, 2019).
Almeida, M. & O’Brien, C. A. Basic biology of skeletal aging: role of stress response pathways. J. Gerontol. A Biol. Sci. Med. Sci. 68, 1197–1208 (2013).
Pignolo, R. J., Law, S. F. & Chandra, A. Bone aging, cellular senescence, and Osteoporosis. JBMR Plus 5, e10488 (2021).
Katzman, W. B., Sellmeyer, D. E., Stewart, A. L., Wanek, L. & Hamel, K. A. Changes in flexed posture, musculoskeletal impairments, and physical performance after group exercise in community-dwelling older women. Arch. Phys. Med. Rehabil. 88, 192–199 (2007).
Ball, J. M., Cagle, P., Johnson, B. E., Lucasey, C. & Lukert, B. P. Spinal extension exercises prevent natural progression of kyphosis. Osteoporos. Int. 20, 481–489 (2009).
Bansal, S., Katzman, W. B. & Giangregorio, L. M. Exercise for improving age-related hyperkyphotic posture: a systematic review. Arch. Phys. Med. Rehabil. 95, 129–140 (2014).
Itoi, E. & Sinaki, M. Effect of back-strengthening exercise on posture in healthy women 49 to 65 years of age. Mayo Clin. Proc. 69, 1054–1059 (1994).
Katzman, W. B. et al. Sex differences in response to targeted kyphosis specific exercise and posture training in community-dwelling older adults: a randomized controlled trial. BMC Musculoskelet. Disord. 18, 509 (2017).
Katzman, W. B. et al. Targeted spine strengthening exercise and posture training program to reduce hyperkyphosis in older adults: results from the study of hyperkyphosis, exercise, and function (SHEAF) randomized controlled trial. Osteoporos. Int. 28, 2831–2841 (2017).
Tucci, J. R. et al. Effect of three years of oral alendronate treatment in postmenopausal women with osteoporosis. Am. J. Med. 101, 488–501 (1996).
Kado, D. M. et al. Comparing a supine radiologic versus standing clinical measurement of kyphosis in older women: the fracture intervention trial. Spine 31, 463–467 (2006).
Keyes, K. M. & Westreich, D. UK Biobank, big data, and the consequences of non-representativeness. Lancet 393, 1297 (2019).
Ruby, J. G. Machine learning code for measurement of Kyphosis and Lordosis in DXA scans. Zenodo. https://doi.org/10.5281/ZENODO.15725218 (2025).
Acknowledgements
The authors would like to acknowledge Danny Park for his assistance with the bulk download of the EBI catalog. In addition, the authors would like to thank Elena Sorokin for downloading and processing GTEx v8 for colocalization and MR analyses. This research was supported by Calico Life Sciences LLC and conducted under UK Biobank Resource application number 18448.
Author information
Authors and Affiliations
Contributions
E.M. conceived the study. F.M.W. carried out epidemiological analyses and wrote original and revised versions of the manuscript. A.S. carried out genetic analyses. J.G.R. developed the machine learning pipeline. N.T. supported image annotation. M.V. supported image annotation, reviewed literature, and provided clinical perspective. All authors contributed to writing and editing, provided critical comments, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors are or were full-time employees of Calico Life Sciences LLC.
Ethics
All UKBB data used in this study were anonymized and were accessed through project number 18448. The National Research Ethics Service Committee of North West-Haydock (REC reference: 11/NW/0382) approved the UKBB project. Signed informed consent was obtained electronically from all participants https://biobank.ctsu.ox.ac.uk/crystal/crystal/docs/Consent.pdf.
Peer review
Peer review information
Communications Medicine thanks Roei Levy, Julia Ive and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
.Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, F.M., Ruby, J.G., Sethi, A. et al. Characterizing aging-related genetic and physiological determinants of spinal curvature. Commun Med 5, 291 (2025). https://doi.org/10.1038/s43856-025-01003-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43856-025-01003-5