Abstract
Despite being rich in natural resources and scientific talent, Africa continues to bear a staggering infectious disease burden. Historically, health innovation on the continent has relied on international funding and has been constrained by limited infrastructure and the emigration of skilled professionals. Data science tools offer a promising alternative, typically requiring fewer costly resources than traditional empirical research, with the potential to empower African scientists to generate tangible and impactful health solutions for the continent. Rapid progress in data science is expected to transform infectious disease research; thus, it is encouraging that numerous African initiatives are already applying data science tools to tackling pressing unmet medical needs, particularly in drug discovery. These efforts include identifying novel therapeutic targets, predicting drug-like molecules and their synthesis, enhancing clinical trial success rates and preparing for future disease threats. This review examines the current landscape of data science in infectious disease drug discovery across Africa.
Similar content being viewed by others
Introduction
Africa’s infectious disease burden
The WHO African Region (referred to hereafter as ‘Africa’) suffers from a disproportionately high burden of disease and is the only continent on which communicable diseases claim more lives than non-communicable diseases1. The five biggest infectious ‘killers’ in Africa are acute respiratory infections, diarrhoea, HIV/Aids, malaria, and tuberculosis, which together are responsible for 60% of Africa’s total infectious disease burden, leading to more than 2.4 million deaths per year. The impact of these numbers is compounded due to survivors of infectious disease in Africa being much more likely to suffer from debilitating complications, with the African continent further accounting for a disproportionately high number (52.5%) of global disability-adjusted life years originating from infectious diseases1. According to the United Nations, Africa’s population is projected to quadruple over the next century2. Temperatures across the continent are also expected to rise between 3 °C and 4 °C (5.5 °F and 7 °F) over the next century, potentially leading to more drought, conflict and biodiversity loss, and raising the possibility of future pandemics through increases in the habitable ranges of animal disease vectors or by increasing pathogen survival time3,4.
Consequently, reducing the burden of infectious diseases in Africa calls for a multipronged strategy that includes better sanitation, broader access to vaccines, and affordable treatment options. However, many of those diseases, such as mycetoma, leishmaniasis or pneumonia caused by drug-resistant Klebsiella, remain without effective or accessible treatments and, even when therapeutic options are available, the continent still faces significant challenges in drug distribution and primary healthcare, aggravated by the emergence of drug resistance5. Additionally, the rich genetic diversity of the continent is often associated with varying responses to medication, resulting in suboptimal treatments or unforeseen adverse reactions. This issue is exacerbated by the predominant use of cohorts of European ancestry in clinical research as well as the absence of African-centric preclinical screening models and tools6. Therefore, incorporating African data into drug discovery and development is essential for advancing a research agenda that reflects the continent’s unique needs and context7,8.
Infectious diseases prevalent in Africa
As the area for greatest potential impact, African-led drug discovery research primarily focuses on infectious diseases with local relevance but without adequate or optimal treatment options. These diseases are caused by a variety of pathogens and fall into the categories of neglected tropical diseases (NTDs, or those that have largely been overlooked by the international community) and emerging diseases (those first appearing or re-appearing in a population after a long period of absence). Importantly, there are other diseases that do not fit into either of these categories but remain major health issues for the continent, such as malaria, tuberculosis and HIV.
Parasite-borne diseases are particularly widespread in Africa and are challenging to treat as their causative agents typically possess complex life cycles, have animal host reservoirs and transmission vectors, and evade the human immune response through invasion of human cells. Malaria, leishmaniasis and schistosomiasis are three relevant examples. Africa carries 94% of the >200 million worldwide cases of malaria each year, with an associated healthcare cost of more than US$12 billion annually9. While malaria research and development (R&D) has attracted more resources from various international entities, R&D for leishmaniasis and schistosomiasis is severely limited. Leishmaniasis, a sandfly-transmitted disease, is endemic to areas inhabited by over one billion people, yet treatment options remain limited. Meanwhile, at least 90% of people at risk of being infected by schistosomiasis, mainly due to lack of adequate sanitation and potable water, live in Africa.
Bacterial diseases represent another prominent sphere of infectious disease R&D as resistance to existing antibacterials becomes increasingly pervasive; consequently, new compounds and treatment regimes are urgently needed. A primary research focus on the continent is the development of improved treatments for tuberculosis (TB), the top infectious disease killer caused by a single pathogen, Mycobacterium tuberculosis (Mtb), and a concerning example of the appearance of antimicrobial-resistant strains. Existing treatment regimens have lengthy periods of up to 6–9 months for drug-sensitive TB and 18–24 months for drug-resistant TB, often with intolerable side effects. Salmonellosis is another devastating zoonotic infection caused by Salmonella bacteria and causing almost 50,000 deaths per year in Africa. Lastly, the ESKAPE pathogens represent a set of highly infectious and multidrug-resistant bacteria that have been prioritised by the WHO for urgent development of novel therapeutic agents before all existing treatment options fail. In 2019, the sub-Saharan Africa region harboured the highest burden of antimicrobial resistance-related deaths at 23.5 deaths per 100,000 inhabitants compared to other regions10.
Challenges for drug discovery in Africa
Drug discovery is a notoriously risky, expensive, and time-consuming enterprise, contributing to the assumption that it is unsuitable or unrealistic in Africa. Indeed, after the high attrition rates of the discovery phase, the probability of successfully transitioning a compound from Phase I clinical trials to regulatory approval is only around 10%11. The total mean cost from discovery to regulatory approval has recently been estimated to be $1.6 billion12. As a result, therapeutic areas such as infectious diseases, which present their own scientific challenges for drug discovery13,14 and that do not yield the desired return on investment, often struggle to secure funding. Although Africa showcased its capabilities in disease surveillance, pathogen sequencing15,16 and clinical trials17 during the Covid-19 pandemic, continent-wide integrated drug discovery is the critical ‘missing link’ in the biomedical value chain.
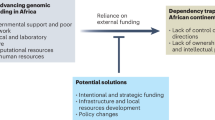
To face the formidable challenges associated with the drug discovery enterprise, Africa must first strengthen its research and higher education capabilities. Across much of the continent, universities remain constrained by poorly-equipped laboratories, limited shipping services, unstable power supply and unreliable internet connectivity. Weak procurement chains and a lack of locally-produced supplies mean that it is more difficult and expensive to bring reagents and equipment into the continent18. The few available specialised instruments are often shared between institutions from multiple countries and are limited in capabilities, as it is common for them to have been donated as retired instruments from facilities in the developed world19. With some notable exceptions, the meagre infrastructure and support for research are dismaying for African PhD-holders20, with many who return home to Africa after a stint abroad facing challenges to be competitive in the global research landscape21. Others never return, contributing to the continent’s ‘brain drain’22, the emigration of skilled nationals resulting in a depletion of human resources in their countries of origin.
Despite ambitious training programmes around the continent, Africa still lacks a critical mass of skills in science and health innovation. The number of tertiary graduates in Africa is projected to rise from 103 million to 240 million by 2040, highlighting the urgent need to provide local job opportunities to the next generation of scientists23, replete with appropriate financial compensation. Alarmingly, scientific output from the continent represents less than 1% of the global share24 and the sub-Saharan Africa region invests only 0.5% of its gross domestic product, less than a third of the world’s average (1.8%)25. Consequently, drug discovery efforts rely on international funding agencies and depend on them to establish priorities and research goals, affecting local leadership26.
The promise of data science for African drug discovery
In recent years, boosted by the outstanding progress of artificial intelligence (AI), data science has become an essential component of the biomedical research enterprise. Arguably, Africa is the region that may benefit the most from these advances, offering its citizens the tantalising possibility of technological ‘leapfrogging’ to close the gap with wealthier nations in the Global North. AI methodologies analyse existing data for patterns that can then be used for predictions and insight into novel data points, including the identification of potential drug leads prior to chemical synthesis and testing. This can be an enabler in resource-constrained settings where the cost of laboratory experiments is often prohibitive. Drug discovery—a field that is eminently interdisciplinary and relies on cumulative evidence to progress compounds to the clinic—has particularly benefited from AI methods27,28. With sustained funding, world-class infrastructure and abundant data, the field has been spearheaded by non-communicable disease research in the Global North—as demonstrated by a plethora of AI-developed drugs progressing to clinical trials29. Arguably, poorly-funded research fields such as those in infectious diseases, and NTDs in particular, have the potential of benefiting the most from these promising advances30.
However, this is dependent on African countries investing in and providing access to the three main ingredients that underpin AI—affordable and reliable power, digital infrastructure, and data. The remainder of this narrative review will highlight the major advances in the development of AI/ML tools in drug discovery projects on the continent, and discuss the recent advances in the field that will help bypass the slow adoption of these novel methods to advance the creation of an African research ecosystem powered by data science and AI/ML.
Drug discovery initiatives in Africa
Drug discovery is a highly integrated endeavour that requires cooperation amongst multiple scientific disciplines across chemistry, biology, pharmacology, and so on. There is no formal academic training to become a drug discovery scientist, which instead comes from on-the-job training after specialisation in a relevant PhD programme. However, as the African innovative pharmaceutical industry is still in a nascent phase of development, there is limited capability to foster and grow a critical mass of skills. Consequently, African contributions to drug discovery are typically limited to small, focused studies within a particular scientific discipline in the context of a postgraduate degree.
One of the most notable achievements in African drug discovery comes from the Holistic Drug Discovery and Development (H3D) Centre at the University of Cape Town in South Africa. Working in collaboration with the Medicines for Malaria Venture (MMV), H3D spearheaded an international research effort that produced the first small-molecule drug candidate to be discovered and advanced into clinical development entirely within Africa. This candidate, known as MMV390048, moved into Phase II trials with African patients, marking progress not only in malaria treatment but also in the continent’s capacity for both fundamental and translational research. Importantly, MMV390048 represented the first Plasmodium kinase inhibitor to reach clinical testing—a significant milestone given that kinases have traditionally been targeted for cancer drug discovery rather than for malaria31,32. The achievements of the H3D Centre highlight that fostering a thriving ecosystem for health innovation depends on strong partnerships at both local and global levels; such partnerships bring together governments, universities, product development partners, philanthropic bodies and industry stakeholders33. To ensure lasting impact, these collaborations should be complemented by a human-focused strategy in which research institutions actively cultivate and strengthen scientific leadership capabilities.
In 2018, the African Academy of Sciences launched a collaboration through its Grand Challenges initiative together with the H3D Centre, MMV and the Gates Foundation. The joint initiative set out to strengthen the foundations of drug discovery across the continent, giving rise to the Grand Challenges Africa Drug Discovery programme. Central to this effort was the creation of capable research teams, supported not only by funding but also by exposure to industry expertise and mentorship. Sixteen projects were awarded grants34, with the H3D Centre providing continued guidance and capacity-strengthening support for participating groups. In 2023, the Gates Foundation and LifeArc jointly invested $7.2 million towards selected projects over 3–5 years as part of the Grand Challenges African Drug Discovery Accelerator (GC ADDA) network established with the H3D Foundation35. The GC ADDA grantees represent eight different African countries with two flagship projects addressing malaria and tuberculosis drug discovery. Funding has also been provided to support an African drug metabolism and pharmacokinetics research network and the assembly of an African-derived natural products library. A portion of the funding has gone to the H3D Foundation, established in 2019 to strengthen the ability to attract, develop and retain talented African researchers in innovative R&D, thereby supporting the GC ADDA network, with the H3D Centre serving as its technical partner.
A snapshot of some of the recent work contributing to African-flavoured drug discovery and development across the continent reveals advances from fields as diverse as basic pathogen biology (probing field isolates of malaria in Mali to better understand P. malariae intra-erythrocytic development and invasion36) to the use of P. falciparum field isolates to optimise the selection and combination of dose regimens for antimalarial treatment37. Critical efforts to better understand the safe and efficacious use of medicines in populations of African ancestry is being conducted as part of the work of the Zimbabwe-based African Institute for Biomedical Science and Technology38, while elucidation of genome sequences of local strains of pathogens, for example, for S. aureus, provides a valuable data resource for the development of new vaccines39.
Although initial efforts to build and strengthen capacity have been focused on individual African researchers, new centres of research have begun to emerge. In 2022, a collaborative drug discovery hub was launched in West Africa, coordinated by the University of Ghana together with the Noguchi Memorial Institute for Medical Research. Its starting priority is malaria drug discovery. Over the longer term, the hub seeks to expand its facilities and expertise to support end-to-end drug discovery, including compound screening, optimisation and characterisation, with the ultimate goal of delivering preclinical drug candidates40. A further emerging drug discovery centre based in Cameroon (Central Africa), the University of Buea Centre for Drug Discovery, is currently investigating putative antiviral compounds against HIV and SARS-CoV-2 from natural product sources41.
Leveraging data science for drug discovery in Africa
Traditionally, drug discovery begins with an identified ‘hit’ compound that is found by phenotypic or target-based high-throughput screens of thousands or even millions of compounds. This is followed by costly and resource-intensive rounds of experiments and optimisation, including human clinical trials, until the drug reaches regulatory approval. Some of the ways in which AI and data science are proposed to accelerate the traditional drug discovery pipeline are listed in Table 1, with those of particular relevance to the African context highlighted.
A recent Wellcome Trust report on the potential of AI in drug discovery identified three major use-cases of data science in small-molecule drug discovery: (i) identifying and validating novel drug targets; (ii) small molecule design and refinement; and (iii) the evaluation of safety profiles of therapeutics42. In the African context, the potential is augmented by the fact that (i) known therapeutic targets are critically lacking for (neglected) pathogens, (ii) the intensive rounds of experimentation and throughput necessary to design new medicines are unfeasible locally, and (iii) African populations are not adequately represented in clinical trials, increasing treatment risks. Of these three points, the preclinical ones—namely, target identification and molecular design—are by far the most thoroughly explored.
A distinction ought to be made between data science for antimicrobial drug discovery globally, and data science efforts for which the scientific leadership resides in Africa. Globally, AI has already shown promise in prioritising broad-spectrum antimicrobials43, as well as compounds against Acinetobacter baumannii44, providing genuinely new chemical entities for further exploration. Alongside these advancements, generative AI tools capable of systematically exploring the antimicrobial chemical space are emerging45. The field is expected to continue benefiting from AI applications, including large language models (e.g. GPT), image analysis for phenotypic screening data, and comprehensive processing of ‘omics’ data. This approach is likely to draw from the progress made in other well-established areas, like anticancer drug discovery, in which various data types—including transcriptomics profiles and genetic screenings—are integrated to forecast drug activity seamlessly46. Additionally, we anticipate increased development of host-directed therapies, which either block pathogen infection or stimulate the human immune system.
Unfortunately, although the ultimate goal of much of this research is to meet the healthcare needs of Africa, the majority, if not all, of it has originated and been developed outside the continent. It may be unrealistic to expect that Africa will generate all the necessary data to ‘train’ AI models for drug discovery in a standalone fashion. Creating large datasets, which are crucial for the success of AI tools, is often prohibitively expensive, even for a single centre in the Global North. Indeed, initiatives like the Tuberculosis Drug Accelerator47, the Malaria Drug Accelerator48, MMV and CO-ADD49 rely on data gathered from multiple centres around the world. Thus, in the context of decolonising research, drug discovery presents unique challenges compared to other fields such as epidemiology and vector control, in which data production occurs on-site, and local strategies are necessary to leverage these data effectively. In drug discovery, and especially preclinical drug discovery, bioactivity screening data is ideally produced with high throughput in large facilities worldwide; the challenge is to make this worldwide data actionable in Africa, ideally in the form of AI tools deployed on-site or accessible via free or affordable online inference services.
In our experience, adoption of AI can be challenging and particularly slow in Africa. Often, no computational skill sets exist on premises to support the implementation of AI tools, which tend to demand advanced technical expertise and frequently require fine-tuning to a particular problem of interest. Additionally, AI tools are heavily reliant on datasets of sufficient volume and quality, which are typically not made accessible to the broader scientific community, and user-friendly proprietary implementations of AI often require prohibitively expensive licenses that prevent access in resource-constrained institutions. To further discuss these matters, we now expand on the various factors that contribute to the successful implementation of AI for global health and consider the current state of the field in Africa.
A recent bibliometric analysis shows that between 2013 and 2022, of the top ten countries publishing about ‘AI in Africa’, only two, South Africa and Nigeria, are actually located on the African continent50. This demonstrates that AI development in general is still in its infancy in Africa. Nevertheless, in the last few years, the combination of academic, non-profit and small start-ups has rendered the first promising results, with South Africa emerging as a leading hub for the application of AI to drug discovery. We recently published the first end-to-end implementation of a virtual screening cascade for malaria and tuberculosis at the H3D Centre in South Africa in collaboration with Ersilia51. Similarly, scientists at the University of Pretoria in South Africa are also developing AI/ML models to forecast the antimalarial potential of new drug candidates52,53. With the support of Collaborative Drug Discovery Inc., a US-based pharmaceutical software firm with a strong philanthropic ethos, other research groups across Africa have started establishing AI/ML virtual screening pipelines for tackling tuberculosis54. Aside from the three major disease areas (malaria, tuberculosis and HIV), modest developments towards adoption of data science and AI/ML for NTDs are also happening on the continent; a recent perspective describes the benefits, limitations, and pitfalls of AI/ML tools for antiviral drug discovery41, and AI could also accelerate the identification of therapeutic options for Ebola virus disease55.
Data science presents an avenue to circumvent the inherent challenges of infectious disease research, compounded by a lack of data, difficult model systems, and poor local scientific infrastructure in the African context. Moreover, emerging AI/ML methods have the potential to reduce overall development costs in low- and middle-income countries, paving the way for locally-d-eveloped, affordable drugs from within these regions.
Data availability
Effective application of AI/ML methods to drug discovery is highly dependent on sufficient and high-quality data, including the outcome of phenotypic assays, receptor-ligand interactions, toxicity of compounds and, whenever available, clinical data. Most laboratories across Africa do not possess the funding and infrastructure to perform high-throughput experimental assays and must resort to publicly available data as a starting point for AI/ML model training; for example, available from ChEMBL56, PubChem57, DrugBank58 and BindingDB59. This presents several challenges, starting with data curation from multiple sources, and requiring more careful interpretation of the models, ensuring the results obtained from an AI/ML model trained with external data can be confidently applied to the chemical space and experimental conditions of interest. Indeed, recent advances in AI/ML can leverage the scarce data existing for poorly-studied diseases to enhance research in low-resourced settings. Those include transfer learning techniques, in which neural networks are pre-trained on larger corpuses of data and then fine-tuned to the specific task at hand60, and few-shot and zero-shot learning techniques devised to learn from very few labelled data points61,62. In addition, the development of large language models (LLMs; including LLAMA, GPT or BLOOM) opens the door to using unstructured text as inputs for AI/ML modelling63,64. These novel methods hold promising potential to speed up research in the low-data scenario for infectious diseases.
AI-based target identification
Pathogen biology for most of the causative agents of NTDs is largely unexplored, partially due to the complexity of in vitro culture (for example, for Cryptosporidium species65) or the lack of adequate animal models, even for better-studied diseases like malaria66. Without proper parasitology studies, target identification remains a challenge, and most drug discovery pipelines necessarily need to start by phenotypic, rather than target-based, assays. Advances in AI-driven tools for predicting protein structure, such as AlphaFold67 and ESMFold68,69, are bringing the field closer to performing molecular docking experiments on proteins for which crystallographic data are lacking. This approach has already elucidated a resistance mechanism in P. falciparum linked to mutations in the PfATP4 ion channel70, uncovered new therapeutic targets in T. cruzi (the parasite responsible for Chagas disease)71, and supported investigations into a range of viral proteins72. Nevertheless, caution should be exercised when using those novel tools, as docking-based approaches demonstrate that AlphaFold predicted proteins might not be accurate enough73. Current efforts by the spin-off from the developers of AlphaFold, Isomorphic Labs, are focused on improving model performance74.
Finally, advances building on AlphaFold are accelerating the mapping of host-pathogen interactions by revealing molecular complexes75 and, when combined with network biology approaches, they offer powerful tools for identifying critical targets in both the host and the pathogen76,77. To name an example, an Mtb-human protein-protein interaction involving 34 proteins secreted by Mtb revealed a switch between the host’s antiviral and antibacterial immune responses, which can be further exploited therapeutically78.
Data science infrastructure
The development of biomedical research data science centres in Africa might seem a challenging endeavour, given that many countries still face unreliable electricity and internet supplies79, which can disrupt data collection, storage and processing. These disruptions not only compromise the integrity of health data but also limit the deployment of advanced health technologies. Ensuring widespread access to cost-effective and dependable electricity and access to the internet, particularly in rural and under-resourced areas, is therefore essential to unlocking the full potential of data science for health systems strengthening in Africa.
Fortunately, recent technology developments are providing several avenues to bridge this gap. On one hand, the rise of local AI agents, i.e. AI-based models that run entirely in the user’s hardware, is bringing LLMs to areas with poor internet connectivity and low computational infrastructure80. For example, the Mozilla-backed initiative Llamafile already provides locally executable files for many LLMs. On the other hand, the growth of cloud computing providers allows researchers to leverage scalable, graphics processing unit-accelerated computing systems from their local institutions81 and, by placing their computational pipelines in remote servers, bypasses the danger of electricity and internet cuts, and reduces the need for expensive solar or generator back-up systems. In sum, technological advances provide a great avenue for the nascent data science landscape in Africa, yet these advances must be coupled with the support of international initiatives such as the recent NIH Harnessing Data Science for Health Discovery and Innovation in Africa82, as well as the investment of local governments83.
Open-source drug discovery
Finally, the open-source paradigm, i.e. releasing data, code and results in real-time so that researchers can contribute to the project as it develops, and not only upon publication, is uniquely well-suited to facilitate the development of data science for infectious disease research in Africa. Open-source drug discovery harnesses collective contributions—by sharing data, software and methodologies—to find patent-free drugs. This approach has been pioneered by the Open Source Malaria consortium, which is actively working on several series of potential antimalarial hits84, and has expanded to other disease areas, including antibiotic-resistant bacteria (OSA) and the fungal infection mycetoma (MycetOS). Those initiatives provide engagement opportunities via computational challenges, including the DREAM Challenges, and result in a plethora of novel open AI/ML tools for malaria and other diseases85,86. When paired with open-source platforms that make these tools accessible to non-specialist researchers—such as the Ersilia Model Hub87—this approach creates opportunities for African scientists to form collaborative networks of data scientists, software developers, medicinal chemists and biologists, all guided by the principle of open source and open science. The recently published Covid Moonshot project demonstrates how a crowdsourcing approach to drug discovery can accelerate the finding of new drugs at low cost88.
Opportunities in an African context
The absence of a local innovative pharmaceutical industry on the continent became apparent during the Covid-19 pandemic, as African countries were mostly reliant on treatment and vaccine development from outside the continent89. As demonstrated by the aforementioned Covid Moonshot project and the Canadian-led prostate cancer treatment development90, computational tools can provide an advantage to resource-limited areas of research, creating a unique opportunity to expedite Africa’s drug discovery research agenda. Particularly, two avenues for leveraging those methods in Africa stand out in the context of infectious disease (Fig. 1).
On one hand, drug repurposing, in which existing drugs are investigated for use in a different therapeutic area, is an attractive strategy that aims to reduce the time and costs of pharmaceutical research by leveraging existing data to advance quickly through the early drug discovery stages. Since drug repurposing depends on extensive data from existing drugs and diseases, data science tools hold promise in allowing researchers to, for example, automate the identification of active ligands and prioritise multi-target scores for promising molecules. While this could be a boon for polypharmacological investigations, issues of selectivity and toxicity still need to be considered.
Secondly, natural products remain an untapped source of novel compounds, particularly in Africa, which boasts a vast array of plants and animals with potential medicinal properties91,92,93. Work in this field has led to several natural product databases, such as NANPDB and SANCDB, that can be used as an initial data source for virtual screening and prioritisation for further investigation94,95. Natural products, once identified, are typically complex to access synthetically and are often computationally processed through various scaffold-hopping approaches to identify a more drug-like pharmacophore for further exploration96. Finally, traditional and indigenous knowledge remains a source of undisclosed potential for finding new therapeutic options for endemic diseases in Africa. Almost 5000 plants are used in African traditional medicine97, and the first examples of successful collaboration between scientists and traditional healers are emerging98. Data science and AI/ML techniques can also be employed to systematically collect, organise and analyse the wealth of traditional medicine and indigenous knowledge to identify promising targets or compounds based on traditional uses.
While the rich genomic diversity of the continent’s population may appear to be a complicating factor in treating disease in Africa, it also represents an opportunity to deliver highly optimised therapies. To advance this goal of precision medicine, the Human Heredity and Health in Africa consortium has invested in developing biorepositories for African-specific genome data while building a pan-African bioinformatics network, H3ABioNet, to advance computational biology research on the continent99. Growing the pharmacogenetics knowledge base to better understand the interplay between African genetic diversity and treatment outcomes relies on data science approaches to identify prevalent variants of pharmacogenes in African populations relevant to drug metabolism in order to optimise drug dosage for African patients, as exemplified by Project Africa GRADIENT100. While little data are, in principle, required for this approach, it is critical that infrastructure, trained staff and secure databases are developed and maintained.
Once prospective treatments advance to the clinical stage of research, it is important for compounds to be tested in representative populations to ensure that safety and effectiveness can be accurately assessed before regulatory approval. However, like the challenges faced in early drug discovery research, Africa lacks the critical mass of infrastructure, skilled clinical research practitioners and funding to facilitate this research at scale. It comes as no surprise, therefore, that despite being home to 17% of the global population, only 3% of clinical trials take place on African soil101. To bridge this gap, the African Union established the African Medicines Agency in 2021 to advance the goal of a standardised legal and ethical framework for robust regulatory review of medical research in Africa102. As AI methodologies are broadly applicable across datasets, data science tools offer much promise at multiple points in the clinical research pipeline by assisting with trial design, participant enrolment, patient monitoring and analysis of clinical end-points103.
Outlook
While infectious diseases remain a significant burden on health in Africa, the emergence of data science tools has opened new avenues for accelerating drug discovery in the region. In this review, we have outlined the crucial role of data science tools and their diverse applications in infectious disease drug discovery on the continent. ML algorithms, data mining and computational modelling offer powerful approaches for analysing complex biological data, identifying potential drug targets, and expediting drug development. Integrating genomics, proteomics and epidemiological data will allow for a holistic understanding of pathogens and disease progression to facilitate the prioritisation of drug candidates tailored to Africa’s unique epidemiological landscape. Various drug discovery initiatives are flourishing on the continent, and these should be accompanied by data science tools and pipelines adapted to their needs.
Leveraging these data science assets will require collaborative efforts amongst researchers, clinicians, public health experts and data scientists. Initiatives that foster interdisciplinarity and data sharing will enhance the accessibility and usability of these tools, leading to more effective interventions and improved health outcomes. While barriers to the adoption of data science tools in the African context, such as infrastructural constraints and the need for capacity strengthening in computational and data science skills, should not be neglected, overcoming these challenges will require concerted efforts from funding agencies, academic institutions and policymakers to invest in infrastructure development, data governance frameworks and educational programmes tailored to the needs of the African community. Importantly, risks and challenges such as bias, safety, model explainability and governance should not be overlooked.
Data science tools have the potential to transform infectious disease drug discovery in Africa; indeed, they are already doing so. In this review, we have highlighted a few representative examples of drug discovery initiatives primarily driven by African researchers, which, when compounded and complemented by valuable global efforts such as those of the Global Health Innovation Technology Fund and the Tokyo International Conference on African Development, set the stage for a strong and data-rich drug discovery community in the region. By harnessing the power of data science tools, the efficiency and effectiveness of drug discovery efforts can be augmented to improve public health outcomes across the continent. It needs to be emphasised that the provision of access to affordable and reliable power, digital infrastructure and data—complemented by skilled human resources—are the basic ingredients needed for data science tools to help leapfrog progress and avoid widening the inequalities between Africa and the rest of the world.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support the findings of this review were derived from resources available in the public domain.
References
WHO. Global health estimates. https://www.who.int/data/gho/data/themes/mortality-and-global-health-estimates/global-health-estimates-leading-causes-of-dalys (2020).
UN. World Population Prospects 2022: Summary of Results (United Nations Fund for Population Activities, 2023).
Mora, C. et al. Over half of known human pathogenic diseases can be aggravated by climate change. Nat. Clim. Change 12, 869–875 (2022).
Malhi, Y., Adu-Bredu, S., Asare, R. A., Lewis, S. L. & Mayaux, P. African rainforests: past, present and future. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120312 (2013).
Antimicrobial Resistance Collaborators The burden of bacterial antimicrobial resistance in the WHO African region in 2019: a cross-country systematic analysis. Lancet Glob. Health 12, e201–e216 (2024).
Popejoy, A. B. & Fullerton, S. M. Genomics is failing on diversity. Nature 538, 161–164 (2016).
Turon, G., Njoroge, M., Mulubwa, M., Duran-Frigola, M. & Chibale, K. AI can help to tailor drugs for Africa—but Africans should lead the way. Nature 265, 267 (2024).
Veale, C. G. L., Edkins, A. L., Winks, S., Njoroge, M. & Chibale, K. Including African data in drug discovery and development. Nat. Rev. Drug Discov. 22, 521–522 (2023).
Venkatesan, P. The 2023 WHO World Malaria Report. Lancet Microbe. https://doi.org/10.1016/S2666-5247(24)00016-8 (2024).
Kariuki, S., Kering, K., Wairimu, C., Onsare, R. & Mbae, C. Antimicrobial resistance rates and surveillance in sub-Saharan Africa: Where are we now?. Infect. Drug Resist. 15, 3589–3609 (2022).
Mohs, R. C. & Greig, N. H. Drug discovery and development: role of basic biological research. Alzheimers Dement. 3, 651–657 (2017).
Wouters, O. J., McKee, M. & Luyten, J. Estimated research and development investment needed to bring a new medicine to market, 2009-2018. JAMA 323, 844–853 (2020).
De Rycker, M., Baragaña, B., Duce, S. L. & Gilbert, I. H. Challenges and recent progress in drug discovery for tropical diseases. Nature 559, 498–506 (2018).
Hikaambo, C. N., Shakela, N., Woodland, J. G., Wicht, K. J. & Chibale, K. Drug discovery in Africa tackles zoonotic and related infections. Sci. Transl. Med. 15, eadj0035 (2023).
Adepoju, P. African coronavirus surveillance network provides early warning for world. Nat. Biotechnol. 40, 147–148 (2022).
Adebisi, Y. A., Rabe, A. & Lucero-Prisno Iii, D. E. COVID-19 surveillance systems in African countries. Health Promot. Perspect. 11, 382–392 (2021).
Mathebula, L., Runeyi, S., Wiysonge, C. & Ndwandwe, D. Clinical trial registration during COVID-19 and beyond in the African context: What have we learned?. Trials 23, 460 (2022).
Olatunji, G. et al. Enhancing clinical and translational research in Africa: a comprehensive exploration of challenges and opportunities for advancement. J. Clin. Transl. Res. https://doi.org/10.18053/jctres.09.202305.23-00079 (2023).
Mukhwana, A., Shorinola, O., Ndlovu, D. F. & Osaso, J. Slow, difficult and expensive: How the lab supplies market is crippling African science. Nature https://www.nature.com/natureindex/news/slow-difficult-expensive-how-lab-supplies-market-crippling-african-science (2024).
Atickem, A. et al. Build science in Africa. Nature 570, 297–300 (2019).
Dike, V. N. et al. Obstacles facing Africa’s young climate scientists. Nat. Clim. Change 8, 447–449 (2018).
Capuano, S. & Marfouk, A. African brain drain and its impact on source countries: What do we know and what do we need to know?. J. Comp. Pol. Anal. Res. Pract. 15, 297–314 (2013).
African Union Commission & OECD. Africa’s Development Dynamics 2024 (OECD, 2024).
Marincola, E. & Kariuki, T. Quality research in Africa and why it is important. ACS Omega 5, 24155–24157 (2020).
UNESCO. UNESCO Science Report: The Race against Time for Smarter Development (UNESCO Publishing, 2021).
Nyasse, B. Overview of current drug discovery activities in Africa and their links to international efforts to combat tropical infectious diseases. In Drug Discovery in Africa (eds Chibale, K., Davies-Coleman, M. & Masimirembwa, C.) 1–28 (Springer Berlin Heidelberg, 2012).
Ekins, S. et al. Exploiting machine learning for end-to-end drug discovery and development. Nat. Mater. 18, 435–441 (2019).
Vamathevan, J. et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 18, 463–477 (2019).
Pun, F. W., Ozerov, I. V. & Zhavoronkov, A. AI-powered therapeutic target discovery. Trends Pharmacol. Sci. 44, 561–572 (2023).
Wong, F., de la Fuente-Nunez, C. & Collins, J. J. Leveraging artificial intelligence in the fight against infectious diseases. Science 381, 164–170 (2023).
Paquet, T. et al. Antimalarial efficacy of MMV390048, an inhibitor of Plasmodium phosphatidylinositol 4-kinase. Sci. Transl. Med. 9, eaad9735 (2017).
Sinxadi, P. et al. Safety, tolerability, pharmacokinetics, and antimalarial activity of the novel plasmodium phosphatidylinositol 4-kinase inhibitor MMV390048 in healthy volunteers. Antimicrob. Agents Chemother. 64, e01896-19 (2020).
Winks, S., Woodland, J. G., Pillai, G. ’colin’ & Chibale, K. Fostering drug discovery and development in Africa. Nat. Med. 28, 1523–1526 (2022).
Grand Challenges. Global Grand Challenges. https://bit.ly/499KJnP.
LifeArc. LifeArc makes multi-million pound investment to support drug discovery in sub-Saharan Africa. LifeArc. https://www.globenewswire.com/news-release/2024/01/31/2820723/0/en/LifeArc-makes-multi-million-pound-investment-to-support-drug-discovery-in-sub-Saharan-Africa.html (2024).
Dao, F. et al. Malian field isolates provide insight into Plasmodium malariae intra-erythrocytic development and invasion. PLoS Negl. Trop. Dis. 19, e0012790 (2025).
Maiga, M. et al. Towards clinically relevant dose ratios for cabamiquine and pyronaridine combination using P. falciparum field isolate data. Nat. Commun. 15, 7659 (2024).
Kanji, C. R., Mbavha, B. T., Masimirembwa, C. & Thelingwani, R. S. Analytical validation of GenoPharm a clinical genotyping open array panel of 46 pharmacogenes inclusive of variants unique to people of African ancestry. PLoS One 18, e0292131 (2023).
Mwangi, K. et al. Draft genome sequences of two strains of Staphylococcus aureus isolated from mastitis-infected camel in Kajiado County, Kenya. Microbiol. Resour. Announc. 12, e0025423 (2023).
Amewu, R. K. et al. Drug discovery research in Ghana, challenges, current efforts, and the way forward. PLoS Negl. Trop. Dis. 16, e0010645 (2022).
Namba-Nzanguim, C. T. et al. Artificial intelligence for antiviral drug discovery in low resourced settings: a perspective. Front. Drug Discov. 2, 1013285 (2022).
WellcomeTrust. Unlocking the Potential of AI in Drug Discovery Wellcome Trust Reports. (WellcomeTrust, 2023).
Stokes, J. M. et al. A deep learning approach to antibiotic discovery. Cell 180, 688–702.e13 (2020).
Liu, G. et al. Deep learning-guided discovery of an antibiotic targeting Acinetobacter baumannii. Nat. Chem. Biol. 19, 1342–1350 (2023).
Das, P. et al. Accelerated antimicrobial discovery via deep generative models and molecular dynamics simulations. Nat. Biomed. Eng. 5, 613–623 (2021).
Ma, J. et al. Few-shot learning creates predictive models of drug response that translate from high-throughput screens to individual patients. Nat. Cancer 2, 233–244 (2021).
Aldridge, B. B. et al. The Tuberculosis Drug Accelerator at year 10: What have we learned? Nat. Med. 27, 1333–1337 (2021).
Yang, T. et al. MalDA, accelerating malaria drug discovery. Trends Parasitol. 37, 493–507 (2021).
Blaskovich, M. A. T., Zuegg, J., Elliott, A. G. & Cooper, M. A. Helping chemists discover new antibiotics. ACS Infect. Dis. 1, 285–287 (2015).
Kondo, T. S. & Diwani, S. A. Artificial intelligence in Africa: a bibliometric analysis from 2013 to 2022. Discov. Artif. Intell. 3, 34 (2023).
Turon, G. et al. First fully-automated AI/ML virtual screening cascade implemented at a drug discovery centre in Africa. Nat. Commun. 14, 5736 (2023).
van Heerden, A., van Wyk, R. & Birkholtz, L.-M. Machine learning uses chemo-transcriptomic profiles to stratify antimalarial compounds with similar mode of action. Front. Cell. Infect. Microbiol. 11, 688256 (2021).
van Heerden, A., Turon, G., Duran-Frigola, M., Pillay, N. & Birkholtz, L.-M. Machine learning approaches identify chemical features for stage-specific antimalarial compounds. ACS Omega. 8, 43813–43826 (2023).
Djaout, K. et al. Predictive modeling targets thymidylate synthase ThyX in Mycobacterium tuberculosis. Sci. Rep. 6, 27792 (2016).
Adams, J., Agyenkwa-Mawuli, K., Agyapong, O., Wilson, M. D. & Kwofie, S. K. EBOLApred: A machine learning-based web application for predicting cell entry inhibitors of the Ebola virus. Comput. Biol. Chem. 101, 107766 (2022).
Mendez, D. et al. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 47, D930–D940 (2018).
Kim, S. et al. PubChem protein, gene, pathway, and taxonomy data collections: bridging biology and chemistry through target-centric views of PubChem data. J. Mol. Biol. 434, 167514 (2022).
Wishart, D. S. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2017).
Liu, T., Lin, Y., Wen, X., Jorissen, R. N. & Gilson, M. K. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res. 35, D198–D201 (2006).
Cai, C. et al. Transfer learning for drug discovery. J. Med. Chem. 63, 8683–8694 (2020).
Altae-Tran, H., Ramsundar, B., Pappu, A. S. & Pande, V. Low data drug discovery with one-shot learning. ACS Cent. Sci. 3, 283–293 (2017).
Huang, K. et al. A foundation model for clinician-centered drug repurposing. Nat Med 30, 3601–3613 (2024).
Zeng, Z., Yao, Y., Liu, Z. & Sun, M. A deep-learning system bridging molecule structure and biomedical text with comprehension comparable to human professionals. Nat. Commun. 13, 1–11 (2022).
Seidl, P., Vall, A., Hochreiter, S. & Klambauer, G. Enhancing activity prediction models in drug discovery with the ability to understand human language. ICML'23: Proceedings of the 40th International Conference on Machine Learning, article No. 1263, 30458–30490 (2023).
Wilke, G. Development of an In Vitro Culture System for Cryptosporidium Parvum (Washington University, 2020).
Simwela, N. V. & Waters, A. P. Current status of experimental models for the study of malaria. Parasitology 149, 729–750 (2022).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Baek, M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021).
Qiu, D. et al. A G358S mutation in the Plasmodium falciparum Na+ pump PfATP4 confers clinically-relevant resistance to cipargamin. Nat. Commun. 13, 1–18 (2022).
Ros-Lucas, A., Martinez-Peinado, N., Bastida, J., Gascón, J. & Alonso-Padilla, J. The use of AlphaFold for in silico exploration of drug targets in the parasite Trypanosoma cruzi. Front. Cell. Infect. Microbiol. 12, 944748 (2022).
Gutnik, D., Evseev, P., Miroshnikov, K. & Shneider, M. Using AlphaFold predictions in viral research. Curr. Issues Mol. Biol. 45, 3705–3732 (2023).
Scardino, V., Di Filippo, J. I. & Cavasotto, C. N. How good are AlphaFold models for docking-based virtual screening?. iScience 26, 105920 (2023).
Google DeepMind AlphaFold Team, Isomorphic Labs Team. Performance and structural coverage of the latest, in-development AlphaFold model. https://storage.googleapis.com/deepmind-media/DeepMind.com/Blog/a-glimpse-of-the-next-generation-of-alphafold/alphafold_latest_oct2023.pdf (2023).
Baptista, D. et al. AlphaFold models of host-pathogen interactions elucidate the prevalence and structural modes of molecular mimicry. Preprint at bioRxivorg https://doi.org/10.1101/2025.06.04.657796 (2025).
Kim, D.-K. et al. A proteome-scale map of the SARS-CoV-2–human contactome. Nat. Biotechnol. 41, 140–149 (2022).
Homma, F., Huang, J. & van der Hoorn, R. A. L. AlphaFold-Multimer predicts cross-kingdom interactions at the plant-pathogen interface. Nat. Commun. 14, 6040 (2023).
Penn, B. H. et al. An Mtb-Human protein-protein interaction map identifies a switch between host antiviral and antibacterial responses. Mol. Cell 71, 637–648.e5 (2018).
Cole, M. A., Elliott, R. J. R., Occhiali, G. & Strobl, E. Power outages and firm performance in Sub-Saharan Africa. J. Dev. Econ. 134, 150–159 (2018).
Vuruma, S. K. R., Margetts, A., Su, J., Ahmed, F. & Srivastava, B. From cloud to edge: rethinking generative AI for low-resource design challenges. Preprint at arXiv https://doi.org/10.48550/arXiv.2402.12702 (2024).
Pandey, M. et al. The transformational role of GPU computing and deep learning in drug discovery. Nat. Mach. Intell. 4, 211–221 (2022).
NIH. Harnessing data science for health discovery and innovation in Africa (DS-I Africa). https://commonfund.nih.gov/AfricaData (2022).
Yamey, G., Batson, A., Kilmarx, P. H. & Yotebieng, M. Funding innovation in neglected diseases. Br. Med. J. 360, k1182 (2018).
Williamson, A. E. et al. Open source drug discovery: highly potent antimalarial compounds derived from the Tres Cantos arylpyrroles. ACS Cent. Sci. 2, 687–701 (2016).
Tse, E. G. et al. An open drug discovery competition: experimental validation of predictive models in a series of novel antimalarials. J. Med. Chem. 64, 16450–16463 (2021).
Zhang, H., Guo, J., Li, H. & Guan, Y. Machine learning for artemisinin resistance in malaria treatment across in vivo-in vitro platforms. iScience 25, 103910 (2022).
Turon, G., Arora, D. & Duran-Frigola, M. The Ersilia Model Hub: a repository of AI/ML for neglected tropical diseases. https://doi.org/10.5281/zenodo.7274646 (2024).
Boby, M. L. et al. Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors. Science 382, eabo7201 (2023).
Guleid, F. H. et al. A bibliometric analysis of COVID-19 research in Africa. BMJ Glob. Health 6, e005690 (2021).
Ban, F. et al. Best practices of computer-aided drug discovery: lessons learned from the development of a preclinical candidate for prostate cancer with a new mechanism of action. J. Chem. Inf. Model. 57, 1018–1028 (2017).
Neves, B. J. et al. Deep Learning-driven research for drug discovery: tackling Malaria. PLoS Comput. Biol. 16, e1007025 (2020).
Atanasov, A. G., Zotchev, S. B., Dirsch, V. M. & Supuran, C. T. Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discov. 20, 200–216 (2021).
Thomford, N. E. et al. Natural products for drug discovery in the 21st century: innovations for novel drug discovery. Int. J. Mol. Sci. 19, 1578 (2018).
Diallo, B. N., Glenister, M., Musyoka, T. M., Lobb, K. & Tastan Bishop, Ö SANCDB: an update on South African natural compounds and their readily available analogs. J. Cheminform. 13, 37 (2021).
Ntie-Kang, F. et al. NANPDB: A resource for natural products from northern African sources. J. Nat. Prod. 80, 2067–2076 (2017).
Grisoni, F. et al. Scaffold hopping from natural products to synthetic mimetics by holistic molecular similarity. Commun. Chem. 1, 1–9 (2018).
Mshelia Halilu, E. Cultivation and conservation of African medicinal plants for pharmaceutical research and socio-economic development. In Medicinal Plants (ed. Kumar, S.) (IntechOpen, 2022).
Richard, K., Andrae-Marobela, K. & Tietjen, I. An ethnopharmacological survey of medicinal plants traditionally used by the BaKalanga people of the Tutume subdistrict in Central Botswana to manage HIV/AIDS, HIV-associated conditions, and other health conditions. J. Ethnopharmacol. 316, 116759 (2023).
Mulder, N. et al. H3Africa: current perspectives. Pharmacogenomics Pers. Med. 11, 59–66 (2018).
Ndong Sima, C. A. A., Othman, H. & Möller, M. & Project Africa GRADIENT Consortium. Advancing pharmacogenetics research in Africa: the ‘Project Africa GRADIENT’ initiative. Drug Discov. Today 29, 103939 (2024).
Taylor-Robinson, S. D., Spearman, C. W. & Suliman, A. A. A. Why is there a paucity of clinical trials in Africa? QJM 114, 357–358 (2021).
Chattu, V. K. et al. Advancing African medicines agency through global health diplomacy for an equitable pan-African universal health coverage: a scoping review. Int. J. Environ. Res. Public Health 18, 11758 (2021).
Harrer, S., Shah, P., Antony, B. & Hu, J. Artificial intelligence for clinical trial design. Trends Pharmacol. Sci. 40, 577–591 (2019).
Acknowledgements
This manuscript is part of a broader writing project on the State of Data Science for Health in Africa (https://bit.ly/StateDataSciAfrica). The project is led by three scientific co-chairs: Catherine Kyobutungi of the African Population and Health Research Centre (APHRC, Kenya), Emile R. Chimusa of Northumbria University Newcastle (United Kingdom) and A. Kofi Amegah of the University of Cape Coast (Ghana). The project is coordinated and supported by the Centre for Global Health Studies at the Fogarty International Centre, U.S. National Institutes of Health (NIH), the African Population and Health Research Centre (APHRC), Wellcome through Grant No. 228261/Z/23/Z, and the Gates Foundation through Grant No. INV-058418, in collaboration with other partner organisations. K.C. is the Neville Isdell Chair in African-centric Drug Discovery and Development and thanks Neville Isdell for generously funding the Chair.
Author information
Authors and Affiliations
Contributions
G.T. and J.W. outlined, wrote and revised the manuscript. J.H., M.D.F. and K.C. wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Alan Talevi, Fabrice Boyom and Yash Gupta for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Turon, G., Hlozek, J., Duran-Frigola, M. et al. Addressing infectious diseases in Africa by accelerating drug discovery through data science. Commun Med 5, 498 (2025). https://doi.org/10.1038/s43856-025-01211-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s43856-025-01211-z