Abstract
Background
One third of patients with Escherichia coli bacteraemia develop a dysregulated inflammatory response (sepsis/septic shock). Our objective was to investigate whether specific microbiological determinants of E. coli are associated to presentation with sepsis/shock.
Methods
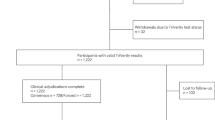
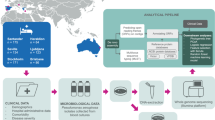
A matched case-control study was performed; 101 case-patients with E. coli bacteraemia presenting with sepsis (SEPSIS-3 criteria) and 101 control-patients with E. coli bacteraemia without sepsis were matched by service, sex, age, Charlson index, acquisition and source of the bacteraemia and empirical treatment. Whole genome sequencing of E. coli isolates was performed (Illumina MiSeq Inc.). Sequence type, serotype, fimH type, virulence factors, antibiotic resistance genes, plasmid replicons pathogenicity islands and prophages were determined. A multivariate model was built for presentation with sepsis/septic shock using conditional logistic regression. The predictive capacity on the observed data was measured with the area under the ROC curve (AUROC) with 95% confidence intervals (CI).
Results
Here we show that in the multivariate model (adjusted OR; 95% CI), the ST69 clone (7.53; 1.06-35.05) and pic gene (4.38; 1.53-12.54) are associated to presentation with sepsis/shock, while the genes papC (0.30; 0.12-0.74) and fdeC (0.18; 0.03-1.32) show a protective effect. The AUROC of this model is 0.81 (95% CI 0.74-0.87).
Conclusions
We identify E. coli bacterial factors associated with severe clinical presentation in patients with bacteraemia. Further studies would be needed to consider these factors as potential preventive or therapeutic targets.
Plain language summary
Escherichia coli is the most common cause of invasive infections, including bacteraemia that often progresses to severe conditions like sepsis or septic shock. While many host factors determine the severity of illness, this study looked at the bacterial factors that may contribute to sepsis severity. We directly compared E. coli-infected patients with similar traits but either with or without sepsis to control for patient factors Our analysis revealed that the ST69 clone and the presence of the pic gene were significantly associated with an increased risk of sepsis/septic shock, whereas the adhesion genes papC and fdeC were associated with a lower risk. These key findings underscore a role for specific E. coli genetic factors in determining clinical severity, thereby providing potential bacterial targets for the development of improved diagnostics and novel preventive or therapeutic interventions.
Similar content being viewed by others
Data availability
Source data for Figs. 1, 2 and 3 are accessible from Supplementary Data 1, 2 and 3, respectively. The complete genome sequence data are publicly available in the European Nucleotide Archive (ENA) under Bioproject PRJEB62601. Researchers may gain access to an anonymised and de-identified version of the dataset presented in this article. To obtain access, a proposed use must first be approved by an independent review committee and the requestor must sign a data access agreement with the senior authors’ institution. Please submit all inquiries and proposals to jesusrb@us.es
Code availability
The custom source code to generate the study’s main results is publicly available at https://data.mendeley.com/datasets/4j8tw9wnwb/1. This material is provided under the licence CC BY NC 3.0, meaning you are free to adapt, copy or redistribute the material, providing you attribute appropriately and do not use the material for commercial purposes. Quality analysis of genome assemblies, genome annotation and pan-genome analysis were performed in QUAST v5.2.0, Bakta v1.6.1 and Roary v3.13.0, respectively. Phylogenomic tree reconstruction based on the best-scoring maximum-likelihood (ML) inference tree for a DNA alignment was performed in RAxML v8.2.12 and best-scoring ML inference tree with branch lengths corrected to account for recombination events in ClonalFrameML v1.12. Other bioinformatics analyses, including Principal Coordinate Analysis (PCoA), clustering based on PCoA, Boruta algorithm and Random Forest Models were performed in R v4.3.1 using the following core packages: tidyverse (2.0.0), caret (6.0-94), Boruta (8.0.0), ranger (0.17.0), vegan (2.6-4) and pROC (1.18.5).
References
European Centre for Disease Prevention and Control. Antimicrobial Resistance in the EU/EEA (EARS-Net)-Annual Epidemiological Report 2022. (ECDC, 2023).
Bonten, M. et al. Epidemiology of Escherichia coli bacteremia: a systematic literature review. Clin. Infect. Dis.XXX72, 1211–1219 (2021).
Mora-Rillo, M. et al. Impact of virulence genes on sepsis severity and survival in Escherichia coli bacteremia. VirulenceXXX6, 93–100 (2015).
Denamur, E. et al. Genome wide association study of Escherichia coli bloodstream infection isolates identifies genetic determinants for the portal of entry but not fatal outcome. PLoS Genet.XXX18, e1010112 (2022).
Brumwell, A. et al. Escherichia coli ST131 associated with increased mortality in bloodstream infections from urinary tract source. J. Clin. Microbiol.XXX61, e0019923 (2023).
Lu, H. et al. Host genetic variants in sepsis risk: a field synopsis and meta-analysis. Crit. CareXXX23, 26 (2019).
Zeng, X. et al. Screening of key genes of sepsis and septic shock using bioinformatics analysis. J. Inflamm. Res.XXX14, 829–841 (2021).
Butler, J. M. et al. Pathogen-specific host response in critically ill patients with blood stream infections: a nested case-control study. EBioMedicineXXX117, 105799 (2025).
Pérez-Crespo, P. M. M. et al. Revisiting the epidemiology of bloodstream infections and healthcare-associated episodes: results from a multicentre prospective cohort in Spain (PRO-BAC Study). Int. J. Antimicrob. AgentsXXX58, 106352 (2021).
Singer, M. et al. The Third International Consensus Definitions for sepsis and septic shock (sepsis-3). JAMAXXX315, 801–810 (2016).
Maldonado, N. et al. Whole-genome characterisation of Escherichia coli isolates from patients with bacteraemia presenting with sepsis or septic shock in Spain: a multicentre cross-sectional study. Lancet MicrobeXXX5, e390–e399 (2024).
Schwengers, O. et al. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom.XXX7, 000685 (2021).
Page, A. J. et al. Roary: rapid large-scale prokaryote pan genome analysis. BioinformaticsXXX31, 3691–3693 (2015).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. BioinformaticsXXX30, 1312–1313 (2014).
Didelot, X. & Wilson, D. J. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput. Biol.XXX11, e1004041 (2015).
Letunic, I. & Bork, P. Interactive Tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res.XXX49, W293–W296 (2021).
Wang, H. E. et al. Chronic medical conditions and risk of sepsis. PLoS ONEXXX7, e48307 (2012).
D’Onofrio, V. et al. Virulence factor genes in invasive Escherichia coli are associated with clinical outcomes and disease severity in patients with sepsis: a prospective observational cohort study. MicroorganismsXXX11, 1827 (2023).
Jauréguy, F. et al. Host and bacterial determinants of initial severity and outcome of Escherichia coli sepsis. Clin. Microbiol. Infect.XXX13, 854–862 (2007).
Bone, R. C. et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM consensus conference committee. American College of Chest Physicians/Society of Critical Care Medicine. ChestXXX101, 1644–1655 (1992).
Rodríguez-Baño, J. et al. Virulence profiles of bacteremic extended-spectrum β-lactamase-producing Escherichia coli: association with epidemiological and clinical features. PLoS ONEXXX7, e44238 (2012).
Fröding, I. et al. Extended-spectrum-β-lactamase- and plasmid AmpC-producingEscherichia coli causing community-onset bloodstream infection: association of bacterial clones and virulence genes with septic shock, source of infection, and recurrence. Antimicrob. Agents Chemother.XXX64, e02351–19 (2020).
Kocsis, B., Gulyás, D. & Szabó, D. Emergence and dissemination of extraintestinal pathogenic high-risk international clones of Escherichia coli. LifeXXX12, 2077 (2022).
de Lastours, V. et al. Mortality in Escherichia coli bloodstream infections: antibiotic resistance still does not make it. J. Antimicrob. Chemother.XXX75, 2334–2343 (2020).
Lipworth, S. et al. Ten-year longitudinal molecular epidemiology study of Escherichia coli and Klebsiella species bloodstream infections in Oxfordshire, UK. Genome Med.XXX13, 144 (2021).
Jauneikaite, E. et al. Bacterial genotypic and patient risk factors for adverse outcomes in Escherichia coli bloodstream infections: a prospective molecular epidemiological study. J. Antimicrob. Chemother.XXX77, 1753–1761 (2022).
Kim, B., Kim, J. H. & Lee, Y. Virulence factors associated with Escherichia coli bacteremia and urinary tract infection. Ann. Lab. Med.XXX42, 203–212 (2022).
Vanstokstraeten, R. et al. Molecular characterization of extraintestinal and diarrheagenic Escherichia coli blood isolates. VirulenceXXX13, 2032–2041 (2022).
Zhou, M. et al. Long polar fimbriae contribute to pathogenic Escherichia coli infection to host cells. Appl. Microbiol. Biotechnol.XXX103, 7317–7324 (2019).
Fox, S. et al. A highly conserved complete accessory Escherichia coli type III secretion system 2 is widespread in bloodstream isolates of the ST69 lineage. Sci. Rep.XXX10, 4135 (2020).
Wang, X. et al. Genetic distribution, characterization, and function of Escherichia coli type III secretion system 2 (ETT2). iScienceXXX27, 109763 (2024).
Sheikh, J. et al. EilA, a HilA-like regulator in enteroaggregative Escherichia coli. Mol. Microbiol.XXX61, 338–350 (2006).
Maldonado, N. et al. Association of microbiological factors with mortality in Escherichia coli bacteraemia presenting with sepsis/septic shock: a prospective cohort study. Clin. Microbiol. Infect.XXX30, 1035–1041 (2024).
Henderson, I. R. et al. Characterization of Pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun.XXX67, 5587–5596 (1999).
Gutiérrez-Jiménez, J., Arciniega, I. & Navarro-García, F. The serine protease motif of Pic mediates a dose-dependent mucolytic activity after binding to sugar constituents of the mucin substrate. Microb. Pathog.XXX45, 115–123 (2008).
Navarro-Garcia, F. et al. Pic, an autotransporter protein secreted by different pathogens in the Enterobacteriaceae family, is a potent mucus secretagogue. Infect. Immun.XXX78, 4101–4109 (2010).
Harrington, S. M. et al. The Pic protease of enteroaggregative Escherichia coli promotes intestinal colonization and growth in the presence of mucin. Infect. Immun.XXX77, 2465–2473 (2009).
Abreu, A. G. et al. The serine protease Pic from enteroaggregative Escherichia coli mediates immune evasion by the direct cleavage of complement proteins. J. Infect. Dis.XXX212, 106–115 (2015).
Dutra, I. L. et al. Pic-producingEscherichia coli induces high production of proinflammatory mediators by the host leading to death by sepsis. Int. J. Mol. Sci.XXX21, 2068 (2020).
Abreu, A. G. et al. The serine protease Pic as a virulence factor of atypical enteropathogenic Escherichia coli. Gut MicrobesXXX7, 115–125 (2016).
Ruiz-Perez, F. et al. Serine protease autotransporters from Shigella flexneri and pathogenic Escherichia coli target a broad range of leukocyte glycoproteins. Proc. Natl. Acad. Sci. USAXXX108, 12881–12886 (2011).
Lara, F. B. et al. Virulence markers and phylogenetic analysis of Escherichia coli strains with hybrid EAEC/UPEC genotypes recovered from sporadic cases of extraintestinal infections. Front. Microbiol.XXX8, 146 (2017).
Boll, E. J. et al. Emergence of enteroaggregative Escherichia coli within the ST131 lineage as a cause of extraintestinal infections. mBioXXX11, e00353-20 (2020).
Del Carpio, A. M. G. et al. Genomic dissection of an enteroaggregative Escherichia coli strain isolated from bacteremia reveals insights into its hybrid pathogenic potential. Int. J. Mol. Sci.XXX25, 9238 (2024).
Luiz, B. M. et al. Enteroaggregative Escherichia coli (EAEC) isolates obtained from non-diarrheic children carry virulence factor-encoding genes from extraintestinal pathogenic E. coli (ExPEC). Braz. J. Microbiol.XXX55, 3551–3561 (2024).
Waksman, G. & Hultgren, S. J. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat. Rev. Microbiol.XXX7, 765–774 (2009).
Norgren, M., Båga, M., Tennent, J. M. & Normark, S. Nucleotide sequence, regulation and functional analysis of the papC gene required for cell surface localization of Pap pili of uropathogenic Escherichia coli. Mol. Microbiol.XXX1, 169–178 (1987).
Rodríguez-Baño, J. et al. Outcome of bacteraemia due to extended-spectrum β-lactamase-producing Escherichia coli: impact of microbiological determinants. J. Infect.XXX67, 27–34 (2013).
Nesta, B. et al. FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. mBioXXX3, e00010–e00012 (2012).
Moriel, D. G. et al. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc. Natl. Acad. Sci. USAXXX107, 9072–9077 (2010).
Xu, H. et al. Estimating the receiver operating characteristic curve in matched case control studies. Stat. Med.XXX38, 437–451 (2019).
Acknowledgements
This study was funded by the Instituto de Salud Carlos III through grant PI16/01432 and co-funded by the European Union (Development Regional Fund ‘A Way to Achieve Europe’). The funders had no role in the design, collection of data, analysis and writing of the manuscript or the decision to publish. We would like to thank all local clinical and microbiological researchers at participating hospitals, members of the PROBAC REIPI/GEIRAS-SEIMC/SAMICEI Group, who helped recruit patients and collect the data.
Author information
Authors and Affiliations
Consortia
Contributions
N.M., I.L.-H., J.R.-B. and A.P. were responsible for conceptualisation, formulating the overall research questions, methodology, formal analysis and writing of the original draft. N.M., I.L.-H., A.G.-M. and M.A.-R. were responsible for bioinformatics analysis. L.E.L.-C., P.M.M.-P.C., A.S.-D., A.J.-S., J.G., A.P.-N., L.B.-M., A.A., L.B.-P., A.d.-J., A.S.-A., J.M.S.-C., C.N.-K., J.M.R.-I., C.A.-C., F.G.-S., A.B., I.G.-L., C.C.-A., I.P.-C., T.M.-C., B.B.-C. participated by reviewing the design, recruiting patients and isolates and thoroughly reviewed the manuscript. J.K.-V. and P.N. participated reviewing the design, performing analysis and reviewed the manuscript. All authors have seen and approved the submitted version of this manuscript and accept responsibility for the decision to submit for publication. I.L.-H., J.R.-B. and A.P. were responsible for funding acquisition, project administration, supervision and coordinating the study.
Corresponding author
Ethics declarations
Competing interests
L.E.L.-C. reports consulting fees from Angelini Pharm and payments for presentations from Correvio Pharma Corp., Gilead Sciences, Inc. and ViiV Healthcare. L.B.-P. reports payments for presentations in educational events from Tillotts Pharma AG and Menarini Group and support for attending meetings and/or travel from Pfizer, Inc. All other authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Evdoxia Kyriazopoulou, Valentino D’Onofrio and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Maldonado, N., López-Hernández, I., Valik, J.K. et al. A matched case-control study on Escherichia coli factors contributing to sepsis and septic shock in bacteraemic patients. Commun Med (2026). https://doi.org/10.1038/s43856-025-01364-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-025-01364-x