Abstract
Background
Escherichia coli clonal complex 38 (CC38) is a genetically diverse lineage increasingly linked to antimicrobial resistance and extraintestinal infections in humans. Despite its clinical and epidemiological relevance, its population structure, zoonotic potential, and ecological associations remain poorly understood.
Methods
We analyzed 242 human E. coli CC38 bloodstream isolates collected through Danish national surveillance, 83 isolates from food and production animals, and 2313 international genomes to investigate host associations and transmission dynamics. Phylogenetic reconstruction, Bayesian host prediction based on mobile genetic elements, and statistical testing of plasmid–host associations were used to delineate population structure and identify potential host-associated markers.
Results
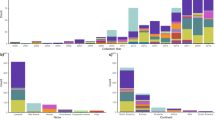
Here we show that Danish CC38 isolates belong to multiple sub-lineages, with no evidence of foodborne outbreaks and limited hospital transmission. Bayesian host prediction supports a poultry origin for several distinct human sub-lineages. Global analyses of 2638 genomes reveal two major clusters: a poultry-associated Cluster I and a predominantly human-associated Cluster II, which subdivides into eight sub-lineages with distinct host, resistance, and virulence profiles. Two small plasmids, ColRNAI and Col(MG828), are strongly enriched in poultry and livestock isolates but largely absent from human-associated sub-clusters, indicating their value as host-associated genetic markers.
Conclusions
Our findings refine the phylogenetic structure of E. coli CC38 and identify plasmid markers that may enhance genomic surveillance of zoonotic transmission. These results highlight the importance of a One Health approach to monitor antimicrobial resistance across human, food, and animal reservoirs. Together, these insights support data-driven One Health surveillance and intervention strategies.
Plain language summary
Antibiotic-resistant bacteria are a growing concern for both human and animal health. This study examined a specific group of E. coli bacteria, which can cause serious infections. By comparing the genomes of bacteria, isolated from people, food, and animals in Denmark and around the world, the researchers mapped how different lineages are related and where they are most likely to come from. The results show that some strains found in humans may have originated in poultry. The study also identified two small DNA elements strongly linked to bacteria from poultry and livestock. These elements could help trace how resistant bacteria move between animals and humans, supporting more effective disease surveillance and antibiotic resistance prevention.
Similar content being viewed by others
Data availability
All E. coli CC38 sequence data generated as part of the Danish National Surveillance that have not previously been published are deposited in the European Nucleotide Archive (ENA) as part of the BioProjects National Surveillance of ESBL-producing Escherichia coli (BioProject PRJEB75816) and National Surveillance of Carbapenemase-Producing Organisms (BioProject PRJEB75178). Accession numbers for the raw sequencing read data included in this study are provided in Supplementary Data 1. In addition, global Escherichia coli CC38 genome sequences analyzed in this study were obtained from public repositories, including the ENA Sequence Read Archive (SRA), NCBI SRA, and EnteroBase. Accession numbers and assembly barcodes for all publicly available genomes are listed in Supplementary Data 1. The EnteroBase E. coli collection is accessible at https://enterobase.warwick.ac.uk/species/index/ecoli. The source data for Fig. 1 are provided in Supplementary Data 5, and the corresponding phylogenetic tree is proved in Supplementary Data 6. The source data for Fig. 2 are provided in Supplementary Data 7. The phylogenetic tree for Fig. 3 is provided in Supplementary Data 8, and source file for Fig. 3 and Table 1 are provided in Supplementary Data 1 and 4.
References
Murray, C. J. et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
Roy Chowdhury, P. et al. Phylogenomic analysis of a global collection of Escherichia coli ST38: evidence of interspecies and environmental transmission? mSystems 8, e0123622 (2023).
Ahlstrom, C. A., Woksepp, H., Sandegren, L., Ramey, A. M. & Bonnedahl, J. Exchange of carbapenem-resistant Escherichia coli Sequence Type 38 intercontinentally and among wild bird, human, and environmental niches. Appl Environ. Microbiol. 89, 1–8 (2023).
Clark, J. R. & Maresso, A. M. Comparative pathogenomics of Escherichia coli: Polyvalent vaccine target identification through virulome analysis. Monack D., editor. Infect Immun [Internet]. 2021 Jul 15;89. Available from: https://journals.asm.org/doi/10.1128/IAI.00115-21
Moez, N. M., Mashouf, R. Y., Sedighi, I., Shokoohizadeh, L. & Taheri, M. Phylogroup classification and investigation the relationships between phylogroups and antibiotic resistance patterns of uropathogenic E. coli isolated from pediatric urinary tract infection. Gene Rep. 20, 100758 (2020).
Mo, S. S. et al. Escherichia coli multilocus sequence type 38 from humans and broiler production represent distinct monophyletic groups. Front Microbiol. 14, 1173287 (2023).
Massella, E. et al. Antimicrobial resistance rrofile and EXPEC virulence potential in commensal Escherichia coli of multiple sources. Antibiotics 10, 351 (2021).
Chattaway, M. A. et al. Evidence of evolving extraintestinal enteroaggregative Escherichia coli ST38 clone. Emerg. Infect. Dis. 20, 1935–1937 (2014).
Mostafa, H. H. et al. Genomic surveillance of ceftriaxone-resistant Escherichia coli in Western New York suggests the extended-spectrum β-lactamase blaCTX-M-27 is emerging on distinct plasmids in ST38. Front Microbiol. 11, 1747 (2020).
Lindemann, P. C. et al. Intraregional hospital outbreak of OXA-244-producing Escherichia coli ST38 in Norway, 2020. Eurosurveillance 28, 1–8 (2023).
Hammerum, A. M. et al. Surveillance of OXA-244-producing Escherichia coli and epidemiologic investigation of cases, Denmark, January 2016 to August 2019. Eurosurveillance. 25, 1–9 (2020).
Falgenhauer, L. et al. Cross-border emergence of clonal lineages of ST38 Escherichia coli producing the OXA-48-like carbapenemase OXA-244 in Germany and Switzerland. Int J. Antimicrob. Agents 56, 106157 (2020).
DANMAP 2022 - Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. [Internet]. 2023. Available from: www.danmap.org
Alghoribi, M. F. et al. Antibiotic-resistant ST38, ST131 and ST405 strains are the leading uropathogenic Escherichia coli clones in Riyadh, Saudi Arabia. J. Antimicrob. Chemother. 70, 2757–2762 (2015).
Guenther, S. et al. Chromosomally encoded ESBL genes in Escherichia coli of ST38 from Mongolian wild birds. J. Antimicrob. Chemother. 72, 1310–1313 (2017).
Vogt, D. et al. Occurrence and genetic characteristics of third-generation cephalosporin-resistant Escherichia coli in Swiss retail meat. Micro Drug Resist 20, 485–494 (2014).
Fonseca, E. L., Morgado, S. M., Caldart, R. V. & Vicente, A. C. Global Genomic epidemiology of Escherichia coli (EXPEC) ST38 lineage revealed a virulome associated with human infections. Microorganisms 10, 2482 (2022).
Xu, C. et al. Characterization of an ST38 carbapenem-resistant and highly virulent Escherichia coli carrying conjugatively transferable ColV virulence–resistance and blaNDM-5-positive resistance plasmids. J. Antimicrob. Chemother. 79, 447–452 (2024).
Grevskott, D. H., Ghavidel, F. Z., Svanevik, C. S. & Marathe, N. P. Resistance profiles and diversity of β-lactamases in Escherichia coli strains isolated from city-scale sewage surveillance in Bergen, Norway mimic clinical prevalence. Ecotoxicol. Environ. Saf. [Internet] 226, 112788 (2021).
Belotindos, L. et al. Prevalence and characterization of quinolone-resistance determinants in Escherichia coli isolated from food-producing animals and animal-derived food in the Philippines. Antibiotics 10, 413 (2021).
Souvorov, A., Agarwala, R. & Lipman, D. J. SKESA: Strategic k-mer extension for scrupulous assemblies. Genome Biol. 19, 1–13 (2018).
Wirth, T. et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol [Internet] 60, 1136–1151 (2006).
Clausen, P. T. L. C., Aarestrup, F. M. & Lund, O. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinforma. 19, 1–8 (2018).
Waters, N. R., Abram, F., Brennan, F., Holmes, A. & Pritchard, L. Easy phylotyping of Escherichia coli via the ezclermont web app and command-line tool. Access Microbiol. 2, Available from https://doi.org/10.1099/acmi.0.000143 (2022).
Malberg Tetzschner, A. M., Johnson, J. R., Johnston, B. D., Lund, O. & Scheutz, F. In silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J. Clin. Microbiol. 58, 1–13 (2020).
Sherry, N. L. et al. An ISO-certified genomics workflow for identification and surveillance of antimicrobial resistance. Nat. Commun. 14, 60 (2023).
Carattoli, A. et al. In Silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903 (2014).
Liu C. M. et al. Escherichia coli ST131-H22 as a foodborne uropathogen. Mobley H. L. T., Ravel J., editors. Mbio. 28;9(4) Available from: https://doi.org/10.1128/mbio.00470-18 (2018).
Sahl, J. W. et al. NASP: an accurate, rapid method for the identification of snps in WGS datasets that supports flexible input and output formats. Micro Genomics 2, 1–12 (2016).
Delcher, A. L. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30, 2478–2483 (2002).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinforma 25, 1754–1760 (2009).
Mckenna, A. et al. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Croucher, N. J. et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43, e15 (2015).
Price, M. N., Dehal, P. S. & Arkin, A. P. Fasttree 2-approximately maximum-likelihood trees for large alignments. Plos One 5, e9490 (2010).
Letunic, I. & Bork, P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245 (2016).
Roer, L. et al. WGS-based surveillance of third-generation cephalosporin-resistant Escherichia coli from bloodstream infections in Denmark. J. Antimicrob. Chemother. 72, 1922–1929 (2017).
Roer, L. et al. Escherichia coli sequence type 410 is causing new international high-risk clones. Castanheira M., editor. Msphere [Internet]. 2018 Jul 18;3(4):e00337-18. Avaiable fromle fr http://msphere.asm.org/lookup/doi/10.1128/msphere.00337-18
Schmidt, M. et al. The Danish National patient registry: A review of content, data quality, and research potential. Clin. Epidemiol. 7, 449–490 (2015).
Liu, C. M. et al. Using source-associated mobile genetic elements to identify zoonotic extraintestinal E. coli infections. One Heal 16, 100518 (2023).
R. Core Team. R: A language and environment for statistical computing [Internet]. Vienna, Austria: R Foundation for Statistical Computing; 2022. Available from: https://www.r-project.org/
European Centre for Disease Prevention and Control. Increase in OXA-244-producing Escherichia coli in the European Union / European Economic Area and the UK since 2013, first update - 20 July 2021. Stockholm; (2021).
Kipper, D. et al. Genomic characterization of Salmonella Minnesota clonal lineages associated with poultry production in Brazil. Anim 10, 2043 (2020).
Wang, W. et al. Genomic characterization of Salmonella isolated from retail chicken and humans with diarrhea in Qingdao, China. Front Microbiol 14, 1–13 (2023).
Flatgard, B. M. et al. Tracking antimicrobial resistance transmission in urban and rural communities in Bangladesh: a One Health study of genomic diversity of ESBL-producing and carbapenem-resistant Escherichia coli. Babiker A., editor. Microbiol Spectr [Internet]. 2024 Jun 4;12(6). https://doi.org/10.1128/spectrum.03956-23
Sugita, K. et al. Molecular analysis of blaKPC-2-Harboring plasmids: Tn4401a interplasmid transposition and Tn4401a-carrying ColRNAI plasmid mobilization from Klebsiella pneumoniae to Citrobacter europaeus and Morganella morganii in a single patient. Msphere 6, e0085021 (2021).
Roer, L., Hansen, F., Hasman, H., Hammerum, A. M. & Cavaco, L. M. Characterisation of extended-spectrum β-lactamase/plasmid ampc-β-lactamase-producing Escherichia coli isolates from long-term recurrent bloodstream infections. Int J. Antimicrob. Agents 56, 10–14 (2020).
Emery, A., Hocquet, D., Bonnet, R. & Bertrand, X. Genotypic characteristics and antimicrobial resistance of Escherichia coli ST141 clonal group. Antibiotics 12, 1–10 (2023).
Roer, L. et al. Development of a web tool for Escherichia coli subtyping based on fimh alleles. Diekema D. J., editor. J Clin Microbiol [Internet]. 2017 Aug; 55(8):2538–43.vailable from: http://jcm.asm.org/lookup/doi/10.1128/JCM.00737-17
Bunduki, G. K. et al. Virulence factors and antimicrobial resistance of uropathogenic Escherichia coli (UPEC) isolated from urinary tract infections: a systematic review and meta-analysis. BMC Infect. Dis. 21, 1–13 (2021).
Koga, V. L. et al. Characterization of CMY-2-type beta-lactamase-producing Escherichia coli isolated from chicken carcasses and human infection in a city of South Brazil. BMC Microbiol. 19, 174 (2019).
Pietsch, M. et al. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genomics19, 601 (2018).
Díaz-Jiménez, D. et al. Genomic characterization of Escherichia coli isolates belonging to a new hybrid aEPEC/EXPEC pathotype O153:H10-A-ST10 eae-beta1 occurred in meat, poultry, wildlife and human diarrheagenic samples. Antibiotics 9, 192 (2020).
Manageiro, V., Jones-Dias, D., Ferreira, E. & Caniça, M. Plasmid-mediated colistin resistance (mcr-1) in Escherichia coli from non-imported fresh vegetables for human consumption in Portugal. Microorg 8, 429 (2020).
Fu Y., Nawrocki E. M., m’ikanatha N. M., Dudley E. G. Host species shapes genotype, antimicrobial resistance, and virulence profiles of enterotoxigenic Escherichia coli (ETEC) from livestock in the United States. Dozois C. M., editor. Appl Environ Microbiol [Internet]. 2024 Aug 21;90. Available from: https://doi.org/10.1128/aem.00749-24
De Lagarde, M., Vanier, G., Arsenault, J. & Fairbrother, J. M. High risk clone: A proposal of criteria adapted to the One Health context with application to enterotoxigenic Escherichia coli in the pig population. Antibiotics 10, 244 (2021).
Hayashi, W. et al. Acquisition of mcr-1 and cocarriage of virulence genes in avian pathogenic Escherichia coli isolates from Municipal wastewater influents in Japan. Elkins CA, editor. Appl Environ. Microbiol [Internet] 85, 1–11 (2019).
Mandomando, I. et al. Escherichia coli ST131 clones harbouring Aggr and AAF/V fimbriae causing bacteremia in Mozambican children: Emergence of new variant of fimH27 subclone. Blanco J., editor. Plos Negl Trop Dis [Interne]. 2020 May 1;14(5):e0008274. Available from: https://dx.plos.org/10.1371/journal.pntd.0008274
Kidsley, A. K. et al. Companion animals are spillover hosts of the multidrug-resistant human extraintestinal Escherichia coli pandemic clones ST131 and ST1193. Front Microbiol [Internet] 11, 1–10 (2020).
Mukerji, S. et al. Resistance to critically important antimicrobials in Australian silver gulls (Chroicocephalus novaehollandiae) and evidence of anthropogenic origins. J. Antimicrob. Chemother. [Internet] 74, 2566–2574 (2019).
Acknowledgements
Pia Thurø Hansen, Line Toft Madsen and Søren Iversen at Statens Serum Institut are thanked for their excellent laboratory assistance. We also thank Jette Sejer Kjeldgaard, Pimlapas Shinny Leekitcharoenphon, and Natasia Rebekka Thornval at the National Food Institute, Technical University of Denmark, for assistance with sequencing data. Part of this work was supported by the Danish Ministry of Health.
Author information
Authors and Affiliations
Contributions
L.R. and M.S. contributed conceptualization, data curation, formal analysis, investigation, methodology, project administration, software, validation, visualization, writing—original draft, and writing—review and editing. A.M.H. and H.H. contributed conceptualization, original draft, and writing—review and editing. A.R., L.E.B.C., R.S., D.E.P., and M.A. contributed bioinformatic analyses, figure preparation, and writing—review and editing. F.H. contributed laboratory analyses, sequencing, and registration of Danish surveillance isolates, as well as writing—review and editing. F.S. contributed virulence profiling and writing—review, and editing. S.A. and L.B.P. contributed resources, supervision, and writing—review and editing, with additional input on conceptualization and interpretation. R.S.H., B.D.J., J.R.J., B.J.H., L.S., K.S., D.B.H., U.S.J., C.Ø., T.S.S., M.T.K.N., M.W., and H.L.N. contributed resources (isolate collection), data curation, and writing—review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Reuben Maghembe and Ivan Sserwadda for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Roer, L., Rasmussen, A., Hansen, F. et al. Genomic characterization and sub-clustering of Escherichia coli clonal complex 38 reveal host associated genetic markers. Commun Med (2026). https://doi.org/10.1038/s43856-026-01402-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-026-01402-2