Abstract
Background
Relatively few studies have investigated HIV-1 persistence in tissues, especially in healthy people-living-with-HIV-1 (PLWH) on a successful antiretroviral regimen containing second generation integrase inhibitors.
Methods
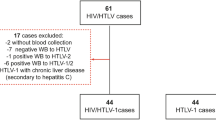
In the ANRS EP64 DOLUVOIR, we explore HIV-1 persistence in five accessible anatomical sites in 20 PLWH on an efficient first-line ART regimen containing dolutegravir with virological load <50 copies/mL: PBMCs, rectum, adipose tissue, lymph node and sperm. We quantify total HIV-DNA and cell-associated HIV-1 RNA in different compartments. We sequence HIV-1 DNA for searching drug resistance mutations (DRM) (in RT and INT) and for studying HIV diversity within tissues (ENV). Intact proviral DNA is estimated in PBMCs with an adapted IPDA assay.
Results
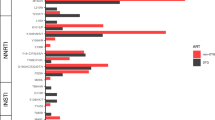
Broad ranges of total HIV-DNA and transcripts levels are detected in lymph nodes, PBMCs, adipose tissue and rectum with the highest levels being found in lymph nodes (2.77 log copies HIV-1-DNA/106 cells and 1.50 log copies of HIV-1 cell-associated-RNA/µg RNA). HIV-1 DNA is undetected in all sperm samples (n = 19) except for one (1.52 log copies HIV-1-DNA/106 cells). No difference is noted between the diversity in the four compartments. DRMs to the current regimen are found archived in compartments of six participants. Only two major DRMs to dolutegravir (G118R and R263K) are found archived in two participants. They are the results of APOBEC hypermutations.
Conclusions
Despite ongoing transcriptional activity, persistence of HIV-1 in deep tissues is not associated with the selection of DRMs to dolutegravir on intact proviruses. Our results suggest that the detectable transcriptional activity stems predominantly from defective proviral DNA.
Plain language summary
The main obstacle for the eradication of Human immunodeficiency Virus (HIV-1) is that the virus persists deep in the human body. In this present study, we explore this persistence by measuring the level of infection and expression of viral genes in five parts of the body: blood, rectum, lymph nodes, sperm and fat. We look in 20 People-living-with-HIV-1 on successful treatment with a combination of medicines including one called Dolutegravir. We demonstrate that the levels of infection are highest in lymph nodes. By testing HIV-1 DNA for drug resistance, we show that this persistence in the body does not lead to major resistance to Dolutegravir.
Similar content being viewed by others
Data availability
The quantification data that support the findings of this study and the source data presented in figures can be found in Supplementary Data 1. Sequenced fastq files are accessible on the European Nucleotide Archive with the following accession number: PRJEB104864 via https://www.ebi.ac.uk/ena/browser/view/PRJEB104864?show=reads.
References
Kalada, W. & Cory, T. J. The importance of tissue sanctuaries and cellular reservoirs of HIV-1. Curr. HIV Res. 20, 102–110 (2022).
Cohn, L. B., Chomont, N. & Deeks, S. G. The biology of the HIV-1 latent reservoir and implications for cure strategies. Cell Host Microbe 27, 519–530 (2020).
Chomont, N. et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 15, 893–900 (2009).
Lorenzo-Redondo, R. et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 530, 51–56 (2016).
Fletcher, C. V. et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc. Natl. Acad. Sci. USA 111, 2307–2312 (2014).
Joseph, S. B. et al. Human immunodeficiency virus type 1 RNA detected in the central nervous system (CNS) after years of suppressive antiretroviral therapy can originate from a replicating CNS reservoir or clonally expanded cells. Clin. Infect. Dis. 69, 1345–1352 (2019).
Martinez-Picado, J. & Deeks, S. G. Persistent HIV-1 replication during antiretroviral therapy. Curr. Opin. HIV AIDS 11, 417 (2016).
Miller, R. L. et al. HIV diversity and genetic compartmentalization in blood and testes during suppressive antiretroviral therapy. J. Virol. 93, e00755-19 (2019).
Kariuki, S. M. et al. Compartmentalization and clonal amplification of HIV-1 in the male genital tract characterized using next-generation sequencing. J. Virol. 94, https://doi.org/10.1128/jvi.00229-20 (2020).
Gianella, S. et al. Compartmentalized HIV rebound in the central nervous system after interruption of antiretroviral therapy. Virus Evol. 2, vew020 (2016).
Dufour, C. et al. Near full-length HIV sequencing in multiple tissues collected postmortem reveals shared clonal expansions across distinct reservoirs during ART. Cell Rep. 42, 113053 (2023).
Chaillon, A. et al. HIV persists throughout deep tissues with repopulation from multiple anatomical sources. J. Clin. Invest. 130, 1699–1712 (2020).
Cochrane, C. R. et al. Intact HIV proviruses persist in the brain despite viral suppression with ART. Ann. Neurol. 92, 532–544 (2022).
Bozzi, G. et al. No evidence of ongoing HIV replication or compartmentalization in tissues during combination antiretroviral therapy: implications for HIV eradication. Sci. Adv. 5, eaav2045 (2019).
Pardons, M. et al. Blood and tissue HIV-1 reservoirs display plasticity and lack of compartmentalization in virally suppressed people. Nat. Commun. 16, 2173 (2025).
Fletcher, C. V. et al. Persistent HIV transcription and variable antiretroviral drug penetration in lymph nodes during plasma viral suppression. AIDS 36, 985–990 (2022).
Vellas, C. et al. Intact proviruses are enriched in the colon and associated with PD-1+TIGIT- mucosal CD4+ T cells of people with HIV-1 on antiretroviral therapy. EBioMedicine 100, 104954 (2024).
Avettand-Fenoel, V. et al. HIV-DNA in rectal cells is well correlated with HIV-DNA in blood in different groups of patients, including long-term non-progressors. AIDS 22, 1880–1882 (2008).
Damouche, A. et al. Adipose tissue is a neglected viral reservoir and an inflammatory site during chronic HIV and SIV infection. PLoS Pathog. 11, e1005153 (2015).
Dupin, N. et al. HIV and antiretroviral drug distribution in plasma and fat tissue of HIV-infected patients with lipodystrophy. AIDS 16, 2419–2424 (2002).
Koethe, J. R. et al. Adipose tissue is enriched for activated and late-differentiated CD8+ T cells and shows distinct CD8+ receptor usage, compared with blood in HIV-infected persons. J. Acquir. Immune Defic. Syndr. 77, e14–e21 (2018).
Couturier, J. et al. Human adipose tissue as a reservoir for memory CD4+ T cells and HIV. AIDS 29, 667–674 (2015).
De Scheerder, M. A. et al. HIV rebound is predominantly fueled by genetically identical viral expansions from diverse reservoirs. Cell Host Microbe 26, 347–358.e7 (2019).
Moron-Lopez, S. et al. Tissue-specific differences in HIV DNA levels and mechanisms that govern HIV transcription in blood, gut, genital tract and liver in ART-treated women. J. Int. AIDS Soc. 24, e25738 (2021).
Imaz, A. et al. Dynamics of the decay of human immunodeficiency virus (HIV) RNA and distribution of bictegravir in the genital tract and rectum in antiretroviral-naive adults living with HIV-1 treated with Bictegravir/Emtricitabine/Tenofovir alafenamide (Spanish HIV/AIDS Research Network, PreEC/RIS 58). Clin. Infect. Dis. 73, e1991–e1999 (2021).
Ferrara, M. et al. Antiretroviral concentrations in post-mortem tissues: preliminary results from the Last Gift Program. https://www.croiconference.org/abstract/antiretroviral-concentrations-in-post-mortem-tissues-preliminary-results-from-the-last-gift-program/ (2024).
Labarthe, L. et al. Pharmacokinetics and tissue distribution of tenofovir, emtricitabine and dolutegravir in mice. J. Antimicrob. Chemother. 77, 1094–1101 (2022).
Avettand-Fènoël, V. et al. LTR real-time PCR for HIV-1 DNA quantitation in blood cells for early diagnosis in infants born to seropositive mothers treated in HAART area (ANRS CO 01). J. Med. Virol. 81, 217–223 (2009).
Bruner, K. M. et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566, 120–125 (2019).
Rose, P. P. & Korber, B. T. Detecting hypermutations in viral sequences with an emphasis on G --> A hypermutation. Bioinformatics 16, 400–401 (2000).
Knyazev, S. et al. Accurate assembly of minority viral haplotypes from next-generation sequencing through efficient noise reduction. Nucleic Acids Res. 49, e102 (2021).
Deeks, S. G. et al. Research priorities for an HIV cure: International AIDS Society Global Scientific Strategy 2021. Nat. Med. 27, 2085–2098 (2021).
Banga, R. & Perreau, M. The multifaceted nature of HIV tissue reservoirs. Curr. Opin. HIV AIDS 19, 116–123 (2024).
Busman-Sahay, K., Starke, C. E., Nekorchuk, M. D. & Estes, J. D. Eliminating HIV reservoirs for a cure: the issue is in the tissue. Curr. Opin. HIV AIDS 16, 200–208 (2021).
Abdel-Mohsen, M. et al. Recommendations for measuring HIV reservoir size in cure-directed clinical trials. Nat. Med. 26, 1339–1350 (2020).
Avettand-Fènoël, V. et al. Total HIV-1 DNA, a marker of viral reservoir dynamics with clinical implications. Clin. Microbiol. Rev. 29, 859–880 (2016).
Estes, J. D. et al. Defining total-body AIDS-virus burden with implications for curative strategies. Nat. Med. 23, 1271–1276 (2017).
Mchantaf, G. et al. The build-up of stock of stable integrated proviruses overtime explains the difficulty in reducing HIV-1 DNA levels when treatment is initiated at the chronic stage of the infection. J. Virus Erad. 9, 100357 (2023).
Hocqueloux, L. et al. Long-term antiretroviral therapy initiated during primary HIV-1 infection is key to achieving both low HIV reservoirs and normal T cell counts. J. Antimicrob. Chemother. 68, 1169–1178 (2013).
Massanella, M. et al. Long-term effects of early antiretroviral initiation on HIV reservoir markers: a longitudinal analysis of the MERLIN clinical study. Lancet Microbe 2, e198–e209 (2021).
Collora, J. A. & Ho, Y. C. The loud minority: transcriptionally active HIV-1-infected cells survive, proliferate, and persist. Cell 185, 227–229 (2022).
Fombellida-Lopez, C., Berkhout, B., Darcis, G. & Pasternak, A. O. Persistent HIV-1 transcription during ART: time to reassess its significance?. Curr. Opin. HIV AIDS 19, 124–132 (2024).
Scrimieri, F. et al. Transcriptionally active defective HIV-1 proviruses and their association with immunological nonresponse to antiretroviral therapy. J. Infect. Dis. 229, 1786–1790 (2024).
Singh, K. et al. Long-term persistence of transcriptionally active “defective” HIV-1 proviruses: implications for persistent immune activation during antiretroviral therapy. AIDS 37, 2119–2130 (2023).
Martin, H. A. et al. New assay reveals vast excess of defective over intact HIV-1 transcripts in antiretroviral therapy-suppressed individuals. J. Virol. 96, e0160522 (2022).
Dorman, A. et al. Nuclear retention of unspliced HIV-1 RNA as a reversible post-transcriptional block in latency. Nat. Commun. 16, 2078 (2025).
George, A. F. et al. Anatomical, subset, and HIV-dependent expression of viral sensors and restriction factors. Cell Rep. 44, 115202 (2025).
Mangeat, B. et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424, 99–103 (2003).
Jary, A. et al. M184V/I does not impact the efficacy of abacavir/lamivudine/dolutegravir use as switch therapy in virologically suppressed patients. J. Antimicrob. Chemother. 75, 1290–1293 (2020).
Santoro, M. M. et al. Virological efficacy of switch to DTG plus 3TC in a retrospective observational cohort of suppressed HIV-1 patients with or without past M184V: the LAMRES study. J. Glob. Antimicrob. Resist. 31, 52–62 (2022).
Mchantaf, G. et al. Learning from full characterization of HIV proviruses in people receiving long-acting cabotegravir/rilpivirine with a history of replication on the antiretroviral classes. Open Forum Infect. Dis. 12, ofae748 (2025).
Acknowledgements
The study was funded and promoted by the ANRS-MIE. ViiV Healthcare acted as a cofounder. The funders had no role in the study design, data collection, data analysis, data interpretation or the writing of the manuscript.
Author information
Authors and Affiliations
Contributions
V.A.F., A.C., L.M. and A.B.T. conceptualized and designed the protocol. A.C. obtained funding. V.A.F. supervised virological analyses. G.M., A.C. and V.A.F. designed experiments. G.M., A.M. and E.G. carried out experiments. L.A. participated to data generation. V.A.F., G.M., A.M., K.D.S., A.Cha. and F.L. analyzed data. L.M. supervised data monitoring. S.O. and F.C. participated to the project administrative management. B.L., J.G., O.R. and J.P.V. included participants. G.M. and V.A.F. interpreted and analyzed results and wrote the original manuscript. All authors critically reviewed the manuscript and contributed to the final version.
Corresponding author
Ethics declarations
Competing interests
V.A.F. has received institutional grants from ViiV Healthcare and honoraria and travel grants from ViiV Healthcare and Gilead Sciences for participation in educational meetings and conferences. A.C. has received institutional grants from ViiV Healthcare and travel grants from Gilead Sciences. O.R. has received consulting fees and payment/honoraria for lectures from Gilead, MSD, Pfizer, and ViiV Healthcare. J.G. reports honoraria for consulting from Gilead Sciences, ViiV Healthcare, Bavarian Nordic and GSK. All other authors declare no competing interests.
Peer review
Peer review information
Communications Medicine thanks Maria C. Puertas, Stefano Rusconi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mchantaf, G., Melard, A., Da Silva, K. et al. HIV persistence in tissues on dolutegravir-based therapy is not associated with resistance mutations to dolutegravir. Commun Med (2026). https://doi.org/10.1038/s43856-026-01405-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-026-01405-z