Abstract
Background
Accurate detection of tuberculosis (TB) treatment failure and recurrence can improve disease control, but current sputum-based monitoring tools pose significant limitations. This study aimed to identify sputum-independent biomarkers for detecting and predicting TB treatment failure and recurrence.
Methods
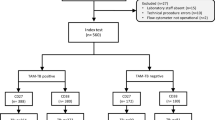
Within the Pan-African TB Sequel study, we conducted a matched case-control study with 40 participants who had recurrent TB or treatment failure and 37 successfully treated controls matched by sex, age, and HIV status. Cases were classified as (a) non-converters with persistently positive sputum Mycobacterium tuberculosis (MTB) results during treatment, (b) reverters at the end of treatment (EOT), or (c) recurrence after EOT. Peripheral blood was collected at baseline, months 2, 4, 6, 9, and 12, and at suspected recurrence. MTB-specific T-cell activation markers (CD38, CD27, HLA-DR, Ki67) and transcriptomic signatures (Sweeney3, Risk6, MAMS6) were assessed and compared to the reference standard MTB culture and smear results.
Results
Here, we show that both MTB-specific T-cell activation and transcriptomic signatures detected non-conversion and TB recurrence at month 9 or 12 after treatment initiation. CD38 expression demonstrates 100% sensitive (95% CI: 56.6–100%) and 78% specific (95% CI: 56.5–99.4%) for detecting TB recurrence, with an AUC of 0.98 (95% CI: 91–100%). Among transcriptomic signatures, MAMS6, RISK6, and Sweeney3 achieve 75% sensitivity (95% CI: 50–100%) and 87–93% specificity (95% CI: MAMS6 0–100%, RISK6 0–93%, Sweeney3 0–100%), with comparable AUCs (0.78–0.83). Neither marker detected TB reversion at EOT.
Conclusion
These sputum-independent biomarkers effectively identify TB disease, non-conversion and recurrence TB after EOT, whereas their utility in detecting TB reversion during treatment remains limited.
Plain Language Summary
Tuberculosis (TB) is a serious infectious disease that can be fatal if untreated. While most patients recover with treatment, some do not respond well or develop TB again after completing therapy. Monitoring how well patients respond to TB treatment currently relies on tests using sputum samples, which can be slow and may be less reliable during treatment. This study aimed to identify alternative host-based markers in blood that could help detect patients with poor treatment response or TB recurrence. We found that specific blood markers can reliably identify patients with TB and detect poor treatment response during therapy, as well as TB recurrence after treatment completion. These findings may help improve early detection, guide treatment decisions, and reduce TB transmission.
Similar content being viewed by others
Data availability
Source data underlying the main figures, including the TAM-TB cohort and the relevant clinical datasets, are provided as a single Excel workbook in the Supplementary Data (Supplementary Data 1). The workbook contains multiple sheets corresponding to the individual figures. RNA sequencing data will be deposited in the ENA repository (Accession number PRJEB101203).
Code availability
The code for the analysis of the RNA signatures is provided at https://github.com/TropI-LMU/BauerEtAl2025.
References
Global Tuberculosis Report 2024. 1st ed. [cited 1 Mar 2025]. Available from: https://ebookcentral.proquest.com/lib/kxp/detail.action?docID=31850656 (World Health Organization, 2024).
Companion handbook to the WHO guidelines for the programmatic management of drug-resistant tuberculosis. Available from: https://permalink.obvsg.at/ (World Health Organization, 2014).
Vega, V., Rodríguez, S., van der Stuyft, P., Seas, C. & Otero, L. Recurrent TB: a systematic review and meta-analysis of the incidence rates and the proportions of relapses and reinfections. Thorax 76, 494–502 (2021).
van der Werf, M. J., Langendam, M. W., Huitric, E. & Manissero, D. Multidrug resistance after inappropriate tuberculosis treatment: a meta-analysis. Eur. Respir. J. 39, 1511–1519 (2012).
Espinal, M. A. et al. Determinants of drug-resistant tuberculosis: analysis of 11 countries. Int J. Tuberc. Lung Dis. 5, 887–893 (2001).
Romanowski, K. et al. Predicting tuberculosis relapse in patients treated with the standard 6-month regimen: an individual patient data meta-analysis. Thorax 74, 291–297 (2019).
Chaves Torres, N. M., Quijano Rodríguez, J. J., Porras Andrade, P. S., Arriaga, M. B. & Netto, E. M. Factors predictive of the success of tuberculosis treatment: a systematic review with meta-analysis. PLoS One 14, e0226507 (2019).
Chang, K. C., Leung, C. C., Yew, W. W., Ho, S. C. & Tam, C. M. A nested case-control study on treatment-related risk factors for early relapse of tuberculosis. Am. J. Respir. Crit. Care Med. 170, 1124–1130 (2004).
Vega, V. et al. Risk factors for pulmonary tuberculosis recurrence, relapse and reinfection: a systematic review and meta-analysis. BMJ Open Respir. Res. 11 (2024).
Imperial, M. Z. et al. A patient-level pooled analysis of treatment-shortening regimens for drug-susceptible pulmonary tuberculosis. Nat. Med. 24, 1708–1715 (2018).
Heyckendorf, J. et al. Tuberculosis treatment monitoring and outcome measures: new interest and new strategies. Clin. Microbiol Rev. 35, e0022721 (2022).
Adekambi, T. et al. Biomarkers on patient T cells diagnose active tuberculosis and monitor treatment response. J. Clin. Investig. 125, 1827–1838 (2015).
Portevin, D. et al. Assessment of the novel T-cell activation marker-tuberculosis assay for diagnosis of active tuberculosis in children: a prospective proof-of-concept study. Lancet Infect. Dis. 14, 931–938 (2014).
Zak, D. E. et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet 387, 2312–2322 (2016).
Riou, C. et al. Disease extent and anti-tubercular treatment response correlates with Mycobacterium tuberculosis-specific CD4 T-cell phenotype regardless of HIV-1 status. Clin. Transl. Immunol. 9, e1176 (2020).
Penn-Nicholson, A. et al. RISK6, a 6-gene transcriptomic signature of TB disease risk, diagnosis and treatment response. Sci. Rep. 10, 8629 (2020).
Heyckendorf, J. et al. Prediction of anti-tuberculosis treatment duration based on a 22-gene transcriptomic model. Eur. Respir. J. 58 (2021).
Sivakumaran, D. et al. Combining host-derived biomarkers with patient characteristics improves signature performance in predicting tuberculosis treatment outcomes. Commun. Biol. 3, 359 (2020).
Ahmed, M. I. M. et al. Phenotypic changes on mycobacterium tuberculosis-specific CD4 T cells as surrogate markers for tuberculosis treatment efficacy. Front. Immunol. 9, 2247 (2018).
Riou, C., Berkowitz, N., Goliath, R., Burgers, W. A. & Wilkinson, R. J. Analysis of the phenotype of mycobacterium tuberculosis-specific CD4+ T cells to discriminate latent from active tuberculosis in HIV-uninfected and HIV-infected individuals. Front. Immunol. 8, 968 (2017).
Kroidl, I. et al. Assessment of tuberculosis disease activity in people infected with Mycobacterium tuberculosis and living with HIV: a longitudinal cohort study. EClinicalMedicine 49, 101470 (2022).
Roe, J. et al. Blood transcriptomic stratification of short-term risk in contacts of tuberculosis. Clin. Infect. Dis. 70, 731–737 (2020).
Singhania, A., Wilkinson, R. J., Rodrigue, M., Haldar, P. & O’Garra, A. The value of transcriptomics in advancing knowledge of the immune response and diagnosis in tuberculosis. Nat. Immunol. 19, 1159–1168 (2018).
Sweeney, T. E., Braviak, L., Tato, C. M. & Khatri, P. Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. Lancet Respir. Med 4, 213–224 (2016).
Kaforou, M. et al. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med 10, e1001538 (2013).
Verhagen, L. M. et al. A predictive signature gene set for discriminating active from latent tuberculosis in Warao Amerindian children. BMC Genom. 14, 74 (2013).
Berry, M. P. R. et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466, 973–977 (2010).
Bloom, C. I. et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One 8, e70630 (2013).
Da Laux Costa, L. et al. A real-time PCR signature to discriminate between tuberculosis and other pulmonary diseases. Tuberculosis 95, 421–425 (2015).
Darboe, F. et al. Diagnostic performance of an optimized transcriptomic signature of risk of tuberculosis in cryopreserved peripheral blood mononuclear cells. Tuberculosis 108, 124–126 (2018).
Suliman, S. et al. Four-Gene Pan-African Blood Signature Predicts Progression to Tuberculosis. Am. J. Respir. Crit. Care Med. 197, 1198–1208 (2018).
Jenum, S. et al. BLR1 and FCGR1A transcripts in peripheral blood associate with the extent of intrathoracic tuberculosis in children and predict treatment outcome. Sci. Rep. 6, 38841 (2016).
Thompson, E. G. et al. Host blood RNA signatures predict the outcome of tuberculosis treatment. Tuberculosis 107, 48–58 (2017).
van Doorn, C. L. R. et al. Transcriptional profiles predict treatment outcome in patients with tuberculosis and diabetes at diagnosis and at two weeks after initiation of anti-tuberculosis treatment. EBioMedicine 82, 104173 (2022).
Cliff, J. M. et al. Excessive cytolytic responses predict tuberculosis relapse after apparently successful treatment. J. Infect. Dis. 213, 485–495 (2016).
Reimann, M. et al. The TB27 transcriptomic model for predicting Mycobacterium tuberculosis culture conversion. Pathog. Immun. 10, 120–139 (2024).
Rachow, A. et al. TB sequel: incidence, pathogenesis and risk factors of long-term medical and social sequelae of pulmonary TB—a study protocol. BMC Pulm. Med. 19, 4 (2019).
Linh NN, et al. World Health Organization treatment outcome definitions for tuberculosis: 2021 update. Eur. Respir. J. 58, 2100804(2021).
Behr, M. A., Edelstein, P. H. & Ramakrishnan, L. Revisiting the timetable of tuberculosis. BMJ 362, k2738 (2018).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Robin, X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinforma. 12, 77 (2011).
R Core Team. R: A Language and Environment for Statistical Computing. R version 4.1.3. Vienna, Austria; 2022. Available from: https://www.R-project.org/.
Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Second edition. (Use R!). Available from https://ebookcentral.proquest.com/lib/kxp/detail.action?docID=4546676 (Springer International Publishing, 2016).
Ralph, A. P. et al. A simple, valid, numerical score for grading chest X-ray severity in adult smear-positive pulmonary tuberculosis. Thorax 65, 863–869 (2010).
Malherbe, S. T. et al. Persisting positron emission tomography lesion activity and Mycobacterium tuberculosis mRNA after tuberculosis cure. Nat. Med. 22, 1094–1100 (2016).
Bryant, J. M. et al. Whole-genome sequencing to establish relapse or re-infection with Mycobacterium tuberculosis: a retrospective observational study. Lancet Respir. Med. 1, 786–792 (2013).
Burman, W. J. et al. The incidence of false-positive cultures for Mycobacterium tuberculosis. Am. J. Respir. Crit. Care Med. 155, 321–326 (1997).
Ruddy, M. et al. Estimation of the rate of unrecognized cross-contamination with Mycobacterium tuberculosis in London microbiology laboratories. J. Clin. Microbiol. 40, 4100–4104 (2002).
Pai, M. et al. Tuberculosis. Nat. Rev. Dis. Prim. 2, 16076 (2016).
Lindestam Arlehamn, C. S. et al. A quantitative analysis of complexity of human pathogen-specific CD4 T cell responses in healthy M. tuberculosis-infected South Africans. PLoS Pathog. 12, e1005760 (2016).
Warsinske, H. C. et al. Assessment of the validity of a blood-based 3-gene signature score for progression and diagnosis of tuberculosis, disease severity, and treatment response. JAMA Netw. Open 1, e183779 (2018).
Ronacher, K. et al. Distinct serum biosignatures are associated with different tuberculosis treatment outcomes. Tuberculosis 118, 101859 (2019).
Olbrich, L. et al. Rapid and accurate diagnosis of pediatric tuberculosis disease (RaPaed-TB): a diagnostic accuracy study for pediatric tuberculosis. Pediatr. Infect. Dis. J. 42, 353–360 (2023).
Acknowledgements
The authors would like to thank all colleagues and partners involved in the TB Sequel project for their dedication and invaluable contributions. Special thanks are extended to all study participants in South Africa, Gambia, Mozambique, and Tanzania, whose involvement made this project possible. The TB Sequel project is funded by the German Ministry for Education and Research (BMBF, 01KA1613) and is part of the Research Networks for Health Innovations in Sub-Saharan Africa. The experimental and data analytical work presented here was supported by the German Ministry for Education and Research through funding from the Deutsches Zentrum für Infektionsforschung (DZIF, TTU-TB personalized medicine TTU 02_813). The funders did not influence the study design, data collection, analysis, or interpretation; the writing of the manuscript; or the decision to submit the paper for publication. All authors confirm they had complete access to the study data and take full responsibility for the decision to submit the manuscript for publication.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Consortia
Contributions
C.G., A.R., and K.H. were responsible for study development, funding acquisition, supervision of study conduct, and data analysis strategy. The setup and conduct of the clinical study, including data and sample collection, were carried out by M.R., S.C., J.S., A.K., M.C., N.N., and C.K. Laboratory data collection and analysis were performed by B.B., O.B., M.A., and A.B. B.B. and C.G. developed the paper and wrote the first draft. Critical review of the paper was performed by C.G., K.H., J.S. and A.R. All co-authors read, commented on, and approved the final version of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors (M.A., O.B., M.H., K.H., C.G.) have submitted a European patent application related to the MAMS_6 transcriptomic signature presented in the manuscript. The application is currently unpublished and pending. Otherwise, the authors have no competing interests.
Peer review
Peer review information
Communications Medicine thanks Simon Mendelsohn and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bauer, B., Ahmed, M.I.M., Baranov, O. et al. Host response biomarkers of tuberculosis recurrence and treatment failure. Commun Med (2026). https://doi.org/10.1038/s43856-026-01424-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s43856-026-01424-w