Abstract
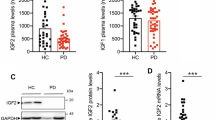
Beyond dyslipidemia, inflammation contributes to the development of atherosclerosis. However, intrinsic factors that counteract vascular inflammation and atherosclerosis remain scarce. Here we identify insulin-like growth factor binding protein 6 (IGFBP6) as a homeostasis-associated molecule that restrains endothelial inflammation and atherosclerosis. IGFBP6 levels are significantly reduced in human atherosclerotic arteries and patient serum. Reduction of IGFBP6 in human endothelial cells by siRNA increases inflammatory molecule expression and monocyte adhesion. Conversely, pro-inflammatory effects mediated by disturbed flow (DF) and tumor necrosis factor (TNF) are reversed by IGFBP6 overexpression. Mechanistic investigations further reveal that IGFBP6 executes anti-inflammatory effects directly through the major vault protein (MVP)–c-Jun N-terminal kinase (JNK)/nuclear factor kappa B (NF-κB) signaling axis. Finally, IGFBP6-deficient mice show aggravated diet- and DF-induced atherosclerosis, whereas endothelial-cell-specific IGFBP6-overexpressing mice protect against atherosclerosis. Based on these findings, we propose that reduction of endothelial IGFBP6 is a predisposing factor in vascular inflammation and atherosclerosis, which can be therapeutically targeted.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The original data files of RNA sequencing generated in this study were deposited on the Gene Expression Omnibus (GSE265787). The public datasets used in this study include GSE41571 (stable plaques versus ruptured plaques), GSE163154 (intraplaque hemorrhage (IPH) versus non-IPH), GSE20739 (UF versus DF), GSE87534 (UF versus static), GSE1176531 (atorvastatin versus DMSO) and STARNET database (600 patients with CAD versus 250 healthy individuals) (http://starnet.mssm.edu/). Proteomic resources were deposited on ProteomeXchange (PXD052204).
Code availability
No custom-made code was used in this study.
References
Libby, P. et al. Atherosclerosis. Nat. Rev. Dis. Primers 5, 56 (2019).
Hahn, C. & Schwartz, M. A. Mechanotransduction in vascular physiology and atherogenesis. Nat. Rev. Mol. Cell Biol. 10, 53–62 (2009).
Souilhol, C. et al. Endothelial responses to shear stress in atherosclerosis: a novel role for developmental genes. Nat. Rev. Cardiol. 17, 52–63 (2020).
Davies, P. F. Endothelial mechanisms of flow-mediated athero-protection and susceptibility. Circ. Res. 101, 10–12 (2007).
Kwak, B. R. et al. Biomechanical factors in atherosclerosis: mechanisms and clinical implications. Eur. Heart J. 35, 3013–3020 (2014).
Berk, B. C. Atheroprotective signaling mechanisms activated by steady laminar flow in endothelial cells. Circulation 117, 1082–1089 (2008).
Chiu, J.-J. & Chien, S. Effects of disturbed flow on vascular endothelium: pathophysiological basis and clinical perspectives. Physiol. Rev. 91, 327–387 (2011).
He, L., Zhang, C.-L., Chen, Q., Wang, L. & Huang, Y. Endothelial shear stress signal transduction and atherogenesis: from mechanisms to therapeutics. Pharmacol. Ther. 235, 108152 (2022).
Baeyens, N., Bandyopadhyay, C., Coon, B. G., Yun, S. & Schwartz, M. A. Endothelial fluid shear stress sensing in vascular health and disease. J. Clin. Invest. 126, 821–828 (2016).
Tamargo, I. A., Baek, K. I., Kim, Y., Park, C. & Jo, H. Flow-induced reprogramming of endothelial cells in atherosclerosis. Nat. Rev. Cardiol. 20, 738–753 (2023).
Gimbrone, M. A. & García-Cardeña, G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ. Res. 118, 620–636 (2016).
Bergheanu, S. C., Bodde, M. C. & Jukema, J. W. Pathophysiology and treatment of atherosclerosis: current view and future perspective on lipoprotein modification treatment. Neth. Heart J. 25, 231–242 (2017).
Ridker, P. M. The time to initiate anti-inflammatory therapy for patients with chronic coronary atherosclerosis has arrived. Circulation 148, 1071–1073 (2023).
Ridker, P. M. et al. Inflammation and cholesterol as predictors of cardiovascular events among patients receiving statin therapy: a collaborative analysis of three randomised trials. Lancet 401, 1293–1301 (2023).
Verma, S., Mazer, C. D. & Connelly, K. A. Inflammation and cholesterol at the crossroads of vascular risk. Cell Metab. 35, 1095–1098 (2023).
Nelson, K., Fuster, V. & Ridker, P. M. Low-dose colchicine for secondary prevention of coronary artery disease: JACC review topic of the week. J. Am. Coll. Cardiol. 82, 648–660 (2023).
Ridker, P. M. et al. IL-6 inhibition with ziltivekimab in patients at high atherosclerotic risk (RESCUE): a double-blind, randomised, placebo-controlled, phase 2 trial. Lancet 397, 2060–2069 (2021).
Ridker, P. M. et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N. Engl. J. Med. 377, 1119–1131 (2017).
Wang, Z. et al. IGFBP6 regulates vascular smooth muscle cell proliferation and morphology via cyclin E-CDK2. J. Cell. Physiol. 235, 9538–9556 (2020).
Zhang, C. et al. IGF binding protein-6 expression in vascular endothelial cells is induced by hypoxia and plays a negative role in tumor angiogenesis. Int. J. Cancer 130, 2003–2012 (2012).
Fu, P., Liang, G. J., Khot, S. S., Phan, R. & Bach, L. A. Cross-talk between MAP kinase pathways is involved in IGF-independent, IGFBP-6-induced Rh30 rhabdomyosarcoma cell migration. J. Cell. Physiol. 224, 636–643 (2010).
Liu, Y., Huan, W., Wu, J., Zou, S. & Qu, L. IGFBP6 is downregulated in unstable carotid atherosclerotic plaques according to an integrated bioinformatics analysis and experimental verification. J. Atheroscler. Thromb. 27, 1068–1085 (2020).
Shi, H. et al. CCN3 regulates macrophage foam cell formation and atherosclerosis. Am. J. Pathol. 187, 1230–1237 (2017).
Jönsson-Rylander, A.-C. et al. Role of ADAMTS-1 in atherosclerosis: remodeling of carotid artery, immunohistochemistry, and proteolysis of versican. Arter. Thromb. Vasc. Biol. 25, 180–185 (2005).
Björkegren, J. L. M., Kovacic, J. C., Dudley, J. T. & Schadt, E. E. Genome-wide significant loci: how important are they? Systems genetics to understand heritability of coronary artery disease and other common complex disorders. J. Am. Coll. Cardiol. 65, 830–845 (2015).
Hägg, S. et al. Multi-organ expression profiling uncovers a gene module in coronary artery disease involving transendothelial migration of leukocytes and LIM domain binding 2: the Stockholm Atherosclerosis Gene Expression (STAGE) study. PLoS Genet. 5, e1000754 (2009).
Koplev, S. et al. A mechanistic framework for cardiometabolic and coronary artery diseases. Nat. Cardiovasc. Res. 1, 85–100 (2022).
Xu, S. et al. Flow-dependent epigenetic regulation of IGFBP5 expression by H3K27me3 contributes to endothelial anti-inflammatory effects. Theranostics 8, 3007–3021 (2018).
Luo, J.-Y. et al. Endothelial UCP2 is a mechanosensitive suppressor of atherosclerosis. Circ. Res. 131, 424–441 (2022).
Sen-Banerjee, S. et al. Kruppel-like factor 2 as a novel mediator of statin effects in endothelial cells. Circulation 112, 720–726 (2005).
Niu, N. et al. Targeting mechanosensitive transcription factors in atherosclerosis. Trends Pharmacol. Sci. 40, 253–266 (2019).
Ben, J. et al. Major vault protein suppresses obesity and atherosclerosis through inhibiting IKK–NF-κB signaling mediated inflammation. Nat. Commun. 10, 1801 (2019).
Liu, Q. et al. Major vault protein prevents atherosclerotic plaque destabilization by suppressing macrophage ASK1-JNK signaling. Arter. Thromb. Vasc. Biol. 42, 580–596 (2022).
Marinou, D. et al. Major vault protein/lung resistance related protein: a novel biomarker for rheumatoid arthritis. Clin. Exp. Rheumatol. 39, 1033–1042 (2021).
Gracia-Sancho, J., Villarreal, G., Zhang, Y. & García-Cardeña, G. Activation of SIRT1 by resveratrol induces KLF2 expression conferring an endothelial vasoprotective phenotype. Cardiovasc. Res. 85, 514–519 (2010).
Sayed, N. et al. Clinical trial in a dish using iPSCs shows lovastatin improves endothelial dysfunction and cellular cross-talk in LMNA cardiomyopathy. Sci. Transl. Med. 12, eaax9276 (2020).
Monzavi, R. & Cohen, P. IGFs and IGFBPs: role in health and disease. Best Pract. Res. Clin. Endocrinol. Metab. 16, 433–447 (2002).
Rajwani, A. et al. Increasing circulating IGFBP1 levels improves insulin sensitivity, promotes nitric oxide production, lowers blood pressure, and protects against atherosclerosis. Diabetes 61, 915–924 (2012).
Wang, W. et al. The impact of circulating IGF-1 and IGFBP-2 on cardiovascular prognosis in patients with acute coronary syndrome. Front. Cardiovasc. Med. 10, 1126093 (2023).
Childs, B. G. et al. Senescent cells suppress innate smooth muscle cell repair functions in atherosclerosis. Nat. Aging 1, 698–714 (2021).
Harrington, S. C., Simari, R. D. & Conover, C. A. Genetic deletion of pregnancy-associated plasma protein-A is associated with resistance to atherosclerotic lesion development in apolipoprotein E-deficient mice challenged with a high-fat diet. Circ. Res. 100, 1696–1702 (2007).
Yagi, H. et al. Discovery of novel biomarkers for atherosclerotic aortic aneurysm through proteomics-based assessment of disease progression. Sci. Rep. 10, 6429 (2020).
Chen, C.-H. & Walterscheid, J. P. Plaque angiogenesis versus compensatory arteriogenesis in atherosclerosis. Circ. Res. 99, 787–789 (2006).
Wu, W. et al. Endothelial Gata6 deletion reduces monocyte recruitment and proinflammatory macrophage formation and attenuates atherosclerosis through Cmpk2-Nlrp3 pathways. Redox Biol. 64, 102775 (2023).
Bjørklund, M. M. et al. Induction of atherosclerosis in mice and hamsters without germline genetic engineering. Circ. Res. 114, 1684–1689 (2014).
Quan, M. et al. MST1 suppresses disturbed flow induced atherosclerosis. Circ. Res. 131, 748–764 (2022).
Tang, C. et al. Endothelial CCRL2 induced by disturbed flow promotes atherosclerosis via chemerin-dependent β2 integrin activation in monocytes. Cardiovasc. Res. 119, 1811–1824 (2023).
Liu, W. et al. In situ expansion and reprogramming of Kupffer cells elicit potent tumoricidal immunity against liver metastasis. J. Clin. Invest. 133, e157937 (2023).
Kumar, S., Kang, D.-W., Rezvan, A. & Jo, H. Accelerated atherosclerosis development in C57Bl6 mice by overexpressing AAV-mediated PCSK9 and partial carotid ligation. Lab. Invest. 97, 935–945 (2017).
Nam, D. et al. Partial carotid ligation is a model of acutely induced disturbed flow, leading to rapid endothelial dysfunction and atherosclerosis. Am. J. Physiol. Heart. Circ. Physiol. 297, H1535–H1543 (2009).
Yan, Y., Tao, H., He, J. & Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 15, 1829–1852 (2020).
Acknowledgements
This study was supported by grants from the National Key R&D Program of China (grant no. 2021YFC2500500), the National Natural Science Foundation of China (grant nos. 82370444, 82070464 and 12411530127) and the Strategic Priority Research Program of the Chinese Academy of Sciences (grant no. XDB38010100). This work was also supported by the Program for Innovative Research Team of The First Affiliated Hospital of University of Science and Technology of China (CXGG02) and the Anhui Provincial Natural Science Foundation (grant no. 2208085J08). This work was supported by the Hong Kong Research Grants Council (T12-101/23-N, SRFS2021-4S04, 11104823). P.C.E. was supported by the British Heart Foundation (RG/19/10/34506). S.X. is a recipient of a Humboldt Research Fellowship from the Alexander von Humboldt Foundation, Germany. The authors are grateful to Prof. Li Zhu (Soochow University, Suzhou, China) for expert suggestions in study design and insightful discussion on shear stress experiments.
Author information
Authors and Affiliations
Contributions
M.S., W.Z., S.X. and J.W. conceptualized and planned the experiments. M.S., W.Z., Hui Jiang, Y.Z., Zhaopeng Liao, Zhenghong Liu, M.X., S.J. and Lili Wu performed the experiments. Y.Y., Z.W., Z.Z., C.T. and Li Wang conducted experiments and software guidance. Hui Jiang supervised the clinical study and collected the clinical samples. Y.F., C.M., P.C.E., M.B., Hanjoong Jo, B.C.B., S.O., Y.H. and J.G. interpreted the data and revised the manuscript. M.S., S.X. and J.W. wrote and edited the final manuscript, with input and approval from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cardiovascular Research thanks the anonymous reviewer(s) for the contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 The gene expression values of IGFs and IGFBPs.
a, The mRNA expression counts of IGFs and IGFBPs in UF- and DF-treated HUVECs. (GEO accession No. GSE20739, n = 3). b, A heatmap showing the differentially expressed genes (DEGs) between the two groups. Upregulated genes are labelled in red and downregulated genes are shown in blue. c, The mRNA expression of IGFBP1-7 in HUVECs under static conditions (n = 3). Statistical analysis was performed by two-tailed Welch’s t test for a, by one-way ANOVA followed by the Tukey’s test for c.
Extended Data Fig. 2 Construction and genotype identification of IGFBP6 global knockout mice.
a, Identification of genotypes of Igfbp6-/- mice. b, The LDLR protein level in liver tissues of mice after PCSK9 injection as detected by Western blot. Liver tissues from male C57BL/6 J and Ldlr-/- mice were used as controls. c, Analysis of ALT, AST, TG, TC, HDL-C, and LDL-C levels in male WT and Igfbp6-/- mice (n = 13-15). Statistical analysis was performed by two-tailed Student’s t test, Mann‒Whitney U test and Welch’s t test for c.
Extended Data Fig. 3 Construction of EC-specific IGFBP6 knockout mice, genotype identification and serum biochemistry.
a, Schematic view of the generation of Igfbp6ECKO mice. b, Identification of genotypes of Igfbp6ECKO mice. c, The mRNA expression of Igfbps in the aortic intima of male Igfbp6ECKO mice and control mice (n = 5). d, Analysis of body weight, blood glucose, ALT, AST, TG, TC, HDL-C, and LDL-C levels (n = 7-11). Statistical analysis was performed by two-tailed Student’s t test and Mann‒Whitney U test for c, d, and by Welch’s t test for d (Male: LDL-C).
Extended Data Fig. 4 Validation and serum biochemistry of EC-specific IGFBP6 overexpression mice.
a, En face immunofluorescence staining IGFBP6 (red), VE-Cadherin (green), and DAPI (blue) in mouse aorta, showing increased IGFBP6 expression in the aortic endothelium of male AAV9-EC-Igfbp6 mice (n = 3). Scale bar: 50 μm. b, The mRNA expression of Igfbps in the aortic intima of male AAV9-EC-Igfbp6 mice and control mice (n = 5). c, Analysis of body weight, blood glucose, ALT, AST, TG, TC, HDL-C, and LDL-C levels in mouse serum (n = 10). Statistical analysis was performed by two-tailed Mann‒Whitney U test and by Student’s t test for b, c, and by Welch’s t test for c (ALT).
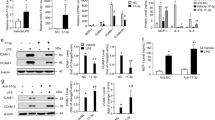
Extended Data Fig. 5 IGFBP6 suppresses inflammation in HAECs and RNA-sequencing of IGFBP6 overexpressed ECs indicates the anti-inflammatory effect of IGFBP6.
a and b, HAECs were transfected with control si-NC or si-IGFBP6 for 48 h and then treated with TNF-α for 6 h. The expression of IGFBP6, VCAM-1, ICAM-1 was determined by qRT-PCR (a, n = 3) and Western blot (b, n = 4). c and d, HAECs were transfected with Ad-NC or Ad-IGFBP6 for 24 h and then treated with TNF-α for 6 h. The expression of Flag-IGFBP6, VCAM-1, ICAM-1, SELE and CCL2 was determined by qRT-PCR (c) and Western blot (d) (n = 3). e and f, HAECs were treated as indicated and THP-1 monocyte adhesion assay was performed and the number of adherent monocytes were quantified (n = 3). Scale bar: 50 μm. g, HUVECs were transfected with Ad-NC or Ad-IGFBP6 for 24 h and then treated with TNF-α for 6 h. The treated cells were analyzed by transcriptome sequencing (n = 3). h, Bubble diagram of the KEGG pathway enrichment analysis of DEGs. i, Heatmap diagram showing the DEGs between the two groups. Upregulated genes are labelled in red and downregulated genes are shown in blue. Scale bar: 50 μm. Statistical analysis was performed by two-tailed paired t test for a (left panel), b, d, by two-way ANOVA (repeated measures) followed by the Bonferroni’s test for a (right panel), c, by Student’s t test for e, f.
Extended Data Fig. 6 KLF4 up-regulates the expression of IGFBP6 and MVP depletion promotes inflammation in HUVECs.
a and b, HUVECs were treated with Ad-NC or Ad-KLF4 for 24 h, and the expression of KLF4, NOS3 and IGFBP6 was determined by qRT-PCR (a, n = 5), and ELISA (b, n = 3). c, HUVECs were transfected with si-NC or si-MVP for 48 h and then treated with TNF-α for 6 h. The expression of MVP, ICAM-1and SELE was determined by qRT-PCR (n = 3). d, HUVECs were treated as indicated and THP-1 monocyte adhesion assay was performed and the number of adherent monocytes were quantified (n = 3). Scale bar: 50 μm. Statistical analysis was performed by two-tailed paired t test for a, by Student’s t test for b, by two-way ANOVA (repeated measures) followed by the Bonferroni’s test for c, and by two-way ANOVA followed by the Tukey’s test for d.
Extended Data Fig. 7 The anti-inflammatory effect of IGFBP6 depends on the binding site of IGFBP6 to MVP.
a, In HUVECs, Co-IP experiments were performed on MVP and ASK1 (n = 3). b, HUVECs were treated with adenovirus to overexpress IGFBP6. After 42 h of treatment, TNF-α was added for 6 h. After that, flag-tagged IGFBP6 was pulled down and ASK1 (n = 3) was detected. c, Representative Co-IP and Western blot assays revealed dimerization of ASK1 in HEK293T cells transfected with indicated plasmid vectors (n = 3). d, Molecular docking was performed using Alphafold3 to predict the binding sites of IGFBP6 and MVP. e, Representative Co-IP and Western blot assays showing the binding domains of IGFBP6 to MVP in HEK293T cells (n = 3). f, HUVECs were treated with IGFBP6 and mutated IGFBP6 adenovirus for 42 h, followed by TNF-α for 6 h. The expression levels of Flag, VCAM-1, ICAM-1 were detected by Western blot (n = 3).
Extended Data Fig. 8 Secreted IGFBP6 and MVP do not affect inflammation in HUVECs.
a and b, HUVECs were treated with adenoviruses (Ad-MVP and Ad-IGFBP6) for 7 days, during which the supernatant was continuously collected. Ultrafiltration tubes were used to concentrate the protein in the supernatant, and flag (a, n = 3) or HA (b, n = 3) antibodies were used for Co-IP, respectively, and finally verified by Western blot. c, HUVECs were treated with different concentrations of IGFBP6 recombinant protein for 42 h, and then treated with TNF-α for 6 h, and indicated proteins were detected by Western blot (n = 3). d, HUVECs were treated with different concentrations of IGFBP6 recombinant protein for 3 h, and then treated with TNF-α for 10 mins, and indicated proteins were detected by Western blot (n = 3). e, 80% of confluent HUVECs were treated with Ad-NC or Ad-IGFBP6 in the presence of TNF. After that, whole cell lysate was collected for Western blot (left 4 lanes, control). In parallel experiments, the treated cells were washed with PBS and new complete medium was supplemented. Then, conditioned media were collected to treat HUVECs before lysate was collected for Western blot (right 4 lanes) (n = 3). f, HUVECs were treated with different concentrations of MVP recombinant protein for 3 h, and then treated with TNF-α for 10 mins, and indicated proteins were detected by Western blot (n = 3).
Extended Data Fig. 9 Graphical Abstract.
IGFBP6 serves as a novel endothelial homeostasis-associated molecule which is mechanoresponsive and confers anti-inflammation and atheroprotection via binding with intracellular MVP and suppresses JNK and p65 phosphorylation.
Extended Data Fig. 10 Immunostaining of murine aortic roots and human coronary arteries with isotype control IgG.
a and b, Representative images of immunofluorescence staining with rat IgG, rabbit IgG or mouse IgG for mouse and human plaque tissues (n = 3). Scale bar = 50 μm.
Supplementary information
Supplementary information
Supplementary Tables 1–5.
Source data
Source Data Figs. 1–7
Statistical source data.
Source Data Extended Data Figs. 1–10
Statistical source data.
Source Data Figs. 1–7
Unprocessed western blots and/or gels.
Source Data Extended Data Figs. 1–10
Unprocessed western blots and/or gels.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Su, M., Zhao, W., Jiang, H. et al. Endothelial IGFBP6 suppresses vascular inflammation and atherosclerosis. Nat Cardiovasc Res 4, 145–162 (2025). https://doi.org/10.1038/s44161-024-00591-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44161-024-00591-0
This article is cited by
-
Persistent immune, coagulation and cardiac dysregulation are correlated with later post-discharge mortality in children with severe malnutrition
BMC Medicine (2026)
-
CASC15 participated in the damage of vascular endothelial cells in atherosclerosis through interaction with miR-940
BMC Medical Genomics (2025)
-
Revisiting the role of GDF15 in atherosclerosis in mouse and human
Acta Pharmacologica Sinica (2025)
-
Endothelial major vault protein alleviates vascular remodeling via promoting Parkin-mediated mitophagy
Nature Communications (2025)
-
IGFBP6 contributes to vascular resilience
Nature Cardiovascular Research (2025)