Abstract
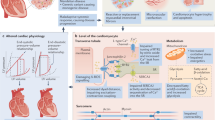
Heart failure (HF) has limited therapeutic options. In this study, we differentiated the pathophysiological underpinnings of the HF subtypes—HF with reduced ejection fraction (HFrEF) and HF with preserved ejection fraction (HFpEF)—and uncovered subtype-specific therapeutic strategies. We investigated the causal roles of the human proteome and transcriptome using Mendelian randomization on more than 420,000 participants from the Million Veteran Program (27,799 HFrEF and 27,579 HFpEF cases). We created therapeutic target profiles covering efficacy, safety, novelty, druggability and mechanism of action. We replicated findings on more than 175,000 participants of diverse ancestries. We identified 70 HFrEF and 10 HFpEF targets, of which 58 were not previously reported; notably, the HFrEF and HFpEF targets are non-overlapping, suggesting the need for subtype-specific therapies. We classified 14 previously unclassified HF loci as HFrEF. We substantiated the role of ubiquitin–proteasome system, small ubiquitin-related modifier pathway, inflammation and mitochondrial metabolism in HFrEF. Among druggable genes, IL6R, ADM and EDNRA emerged as potential HFrEF targets, and LPA emerged as a potential target for both subtypes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The MVP GWAS summary statistics used in this study will be available through the database of Genotypes and Phenotypes (dbGAP) (study accession number phs001672.v11.p1). The only restriction is that use of the data is limited to health/medical/biomedical purposes and does not include the study of population origins or ancestry. Use of the data does include methods development research (for example, development and testing of software or algorithms), and requesters must agree to make the results of studies using the data available to the larger scientific community. Full summary statistics of the risk factor and safety traits that used data from the Million Veteran Program are publicly available and can be downloaded at the VA’s Centralized Interactive Phenomics Resource (CIPHER) web portal. All supporting annotation files are also available for download at CIPHER. Detailed visualizations pertaining to the Mendelian randomization and co-localization results and orthogonal sources of support are provided in the supplementary figures.
Code availability
We used publicly available software for the analyses, and all software used is listed and described in the Methods section of our paper. Statistical analyses were conducted in R version 3.6.3. Mendelian randomization analyses were conducted using the TwoSampleMR package in R (version 0.5.6, release date 25 March 2021); genetic co-localization analyses were conducted using the coloc (version 5.2.3) package in R; and standardization of GWAS summary statistics was conducted using MungeSumstats (release 3.20). Imputation was conducted using Minimac4 (version 1.0.0). The GWAS for HFrEF and HFpEF was conducted using Plink 2.0. The GWAS findings were functionally annotated using FUMA (version 1.3.8). Meta-analysis of GWAS summary statistics was prepared using METAL (version released 6 October 2020).
References
Heidenreich, P. A. et al. 2022 AHA/ACC/HFSA Guideline for the Management of Heart Failure: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 145, e895–e1032 (2022).
Jhund, P. S. et al. Dapagliflozin across the range of ejection fraction in patients with heart failure: a patient-level, pooled meta-analysis of DAPA-HF and DELIVER. Nat. Med. 28, 1956–1964 (2022).
Shah, S. J. et al. Research priorities for heart failure with preserved ejection fraction: National Heart, Lung, and Blood Institute Working Group Summary. Circulation 141, 1001–1026 (2020).
Rucker, D. & Joseph, J. Defining the phenotypes for heart failure with preserved ejection fraction. Curr. Heart Fail. Rep. 19, 445–457 (2022).
Trajanoska, K. et al. From target discovery to clinical drug development with human genetics. Nature 620, 737–745 (2023).
Rasooly, D. et al. Genome-wide association analysis and Mendelian randomization proteomics identify drug targets for heart failure. Nat. Commun. 14, 3826 (2023).
Shah, S. et al. Genome-wide association and Mendelian randomisation analysis provide insights into the pathogenesis of heart failure. Nat. Commun. 11, 163 (2020).
Levin, M. G. et al. Genome-wide association and multi-trait analyses characterize the common genetic architecture of heart failure. Nat. Commun. 13, 6914 (2022).
Joseph, J. et al. Genetic architecture of heart failure with preserved versus reduced ejection fraction. Nat. Commun. 13, 7753 (2022).
Ferkingstad, E. et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 53, 1712–1721 (2021).
Pietzner, M. et al. Mapping the proteo-genomic convergence of human diseases. Science 374, eabj1541 (2021).
Zhang, J. et al. Plasma proteome analyses in individuals of European and African ancestry identify cis-pQTLs and models for proteome-wide association studies. Nat. Genet. 54, 593–602 (2022).
GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Võsa, U. et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 53, 1300–1310 (2021).
Roden, D. M. et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin. Pharmacol. Ther. 84, 362–369 (2008).
Sun, B. B. et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature 622, 329–338 (2023).
Zhao, H. et al. Proteome-wide Mendelian randomization in global biobank meta-analysis reveals multi-ancestry drug targets for common diseases. Cell Genom. 2, 100195 (2022).
Wu, K.-H. H. et al. Polygenic risk score from a multi-ancestry GWAS uncovers susceptibility of heart failure. Preprint at bioRxiv https://doi.org/10.1101/2021.12.06.21267389 (2021).
Henry, A. et al. Therapeutic targets for heart failure identified using proteomics and Mendelian randomization. Circulation 145, 1205–1217 (2022).
Zhou, W. et al. Global Biobank Meta-analysis Initiative: powering genetic discovery across human disease. Cell Genom. 2, 100192 (2022).
Schmidt, A. F. et al. Druggable proteins influencing cardiac structure and function: implications for heart failure therapies and cancer cardiotoxicity. Sci. Adv. 9, eadd4984 (2023).
Guo, H. et al. Integration of disease association and eQTL data using a Bayesian colocalisation approach highlights six candidate causal genes in immune-mediated diseases. Hum. Mol. Genet. 24, 3305–3313 (2015).
Aung, N. et al. Genome-wide analysis of left ventricular image-derived phenotypes identifies fourteen loci associated with cardiac morphogenesis and heart failure development. Circulation 140, 1318–1330 (2019).
MacNamara, A. et al. Network and pathway expansion of genetic disease associations identifies successful drug targets. Sci. Rep. 10, 20970 (2020).
Pitt, B. et al. The effect of spironolactone on morbidity and mortality in patients with severe heart failure. Randomized Aldactone Evaluation Study Investigators. N. Engl. J. Med. 341, 709–717 (1999).
Li, X., Peng, W., Wu, J., Yeung, S.-C. J. & Yang, R. Advances in immune checkpoint inhibitors induced-cardiotoxicity. Front. Immunol. 14, 1130438 (2023).
Ochoa, D. et al. Open Targets Platform: supporting systematic drug–target identification and prioritisation. Nucleic Acids Res. 49, D1302–D1310 (2021).
Tsimikas, S., Moriarty, P. M. & Stroes, E. S. Emerging RNA therapeutics to lower blood levels of Lp(a): JACC Focus Seminar 2/4. J. Am. Coll. Cardiol. 77, 1576–1589 (2021).
Hoekstra, M. et al. Genome-wide association study highlights APOH as a novel locus for lipoprotein(a) levels—brief report. Arterioscler. Thromb. Vasc. Biol. 41, 458–464 (2021).
O’Donoghue, M. L. et al. Small interfering RNA to reduce lipoprotein(a) in cardiovascular disease. N. Engl. J. Med. 387, 1855–1864 (2022).
Morley, M. P. et al. Cardioprotective effects of MTSS1 enhancer variants. Circulation 139, 2073–2076 (2019).
Buyandelger, B. et al. ZBTB17 (MIZ1) is important for the cardiac stress response and a novel candidate gene for cardiomyopathy and heart failure. Circ. Cardiovasc. Genet. 8, 643–652 (2015).
de Bruin, R. G., Rabelink, T. J., van Zonneveld, A. J. & van der Veer, E. P. Emerging roles for RNA-binding proteins as effectors and regulators of cardiovascular disease. Eur. Heart J. 38, 1380–1388 (2017).
Cheng, H. et al. Loss of enigma homolog protein results in dilated cardiomyopathy. Circ. Res. 107, 348–356 (2010).
Huang, Z.-P. et al. Cardiomyocyte-enriched protein CIP protects against pathophysiological stresses and regulates cardiac homeostasis. J. Clin. Invest. 125, 4122–4134 (2015).
Pagan, J., Seto, T., Pagano, M. & Cittadini, A. Role of the ubiquitin proteasome system in the heart. Circ. Res. 112, 1046–1058 (2013).
Mason, B. & Laman, H. The FBXL family of F-box proteins: variations on a theme. Open Biol. 10, 200319 (2020).
Spielmann, N. et al. Extensive identification of genes involved in congenital and structural heart disorders and cardiomyopathy. Nat. Cardiovasc. Res. 1, 157–173 (2022).
Georgiopoulos, G. et al. Cardiovascular toxicity of proteasome inhibitors: underlying mechanisms and management strategies: JACC: CardioOncology state-of-the-art review. JACC CardioOncol. 5, 1–21 (2023).
Bedford, L., Lowe, J., Dick, L. R., Mayer, R. J. & Brownell, J. E. Ubiquitin-like protein conjugation and the ubiquitin-proteasome system as drug targets. Nat. Rev. Drug Discov. 10, 29–46 (2011).
Mendler, L., Braun, T. & Müller, S. The ubiquitin-like SUMO system and heart function: from development to disease. Circ. Res. 118, 132–144 (2016).
Dib, M.-J. et al. Proteomic associations of adverse outcomes in human heart failure. J. Am. Heart Assoc. 13, e031154 (2024).
Adamo, L., Rocha-Resende, C., Prabhu, S. D. & Mann, D. L. Reappraising the role of inflammation in heart failure. Nat. Rev. Cardiol. 17, 269–285 (2020).
Wang, M. et al. MCC950, a selective NLRP3 inhibitor, attenuates adverse cardiac remodeling following heart failure through improving the cardiometabolic dysfunction in obese mice. Front. Cardiovasc. Med. 9, 727474 (2022).
Olsen, M. B. et al. Targeting the inflammasome in cardiovascular disease. JACC Basic Transl. Sci. 7, 84–98 (2022).
Felker, G. M. et al. Underlying causes and long-term survival in patients with initially unexplained cardiomyopathy. N. Engl. J. Med. 342, 1077–1084 (2000).
Brown, D. A. et al. Mitochondrial function as a therapeutic target in heart failure. Nat. Rev. Cardiol. 14, 238–250 (2017).
Guo, K., Diemer, E. W., Labrecque, J. A. & Swanson, S. A. Falsification of the instrumental variable conditions in Mendelian randomization studies in the UK Biobank. Eur. J. Epidemiol. 38, 921–927 (2023).
Bielinski, S. J. et al. A robust e-epidemiology tool in phenotyping heart failure with differentiation for preserved and reduced ejection fraction: the Electronic Medical Records and Genomics (eMERGE) Network. J. Cardiovasc. Transl. Res. 8, 475–483 (2015).
Rasooly, D. & Patel, C. J. Conducting a reproducible Mendelian randomization analysis using the R analytic statistical environment. Curr. Protoc. Hum. Genet. 101, e82 (2019).
Rasooly, D., Peloso, G. M. & Giambartolomei, C. Bayesian genetic colocalization test of two traits using coloc. Curr. Protoc. 2, e627 (2022).
Wallace, C. A more accurate method for colocalisation analysis allowing for multiple causal variants. PLoS Genet. 17, e1009440 (2021).
Avsec, Ž. et al. Effective gene expression prediction from sequence by integrating long-range interactions. Nat. Methods 18, 1196–1203 (2021).
Aragam, K. G. et al. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nat. Genet. 54, 1803–1815 (2022).
Nauffal, V. et al. Genetics of myocardial interstitial fibrosis in the human heart and association with disease. Nat. Genet. 55, 777–786 (2023).
Thanaj, M. et al. Genetic and environmental determinants of diastolic heart function. Nat. Cardiovasc. Res. 1, 361–371 (2022).
Karczewski, K. J. et al. Systematic single-variant and gene-based association testing of thousands of phenotypes in 394,841 UK Biobank exomes. Cell Genom. 2, 100168 (2022).
Sun, B. B. et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018).
Sakaue, S. et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 53, 1415–1424 (2021).
Young, W. J. et al. Genetic analyses of the electrocardiographic QT interval and its components identify additional loci and pathways. Nat. Commun. 13, 5144 (2022).
Acknowledgements
We are grateful to all the MVP investigators. A list of MVP investigators can be found in the Supplementary Information.
This research is based on data from the Million Veteran Program, Office of Research and Development, Veterans Health Administration, and was supported by awards MVP037 (BLR&D Merit Award BX005831), Veterans Affairs Grant I01CX001922 (principal investigator (PI): J.J.) and MVP001 (I01-BX004821). This publication does not represent the views of the Department of Veterans Affairs or the United States Government. We also acknowledge the VA Merit Grant I01-CX001025 (PIs: P.W.F.W. and K.C.).
The British Heart Foundation funded the manual analysis to create a cardiovascular MRI reference standard for the UK Biobank imaging resource in 5,000 CMR scans (https://www.bhf.org.uk/; PG/14/89/31194). This work acknowledges the support of the National Institute for Health and Care Research Barts Biomedical Research Centre (NIHR203330), a delivery partnership of Barts Health NHS Trust, Queen Mary University of London, St. George’s University Hospitals NHS Foundation Trust and St. George’s University of London. This article is supported by the London Medical Imaging and Artificial Intelligence Centre for Value Based Healthcare (AI4VBH), which is funded by the Data to Early Diagnosis and Precision Medicine strand of the government’s Industrial Strategy Challenge Fund, managed and delivered by Innovate UK on behalf of UK Research and Innovation (UKRI). Views expressed are those of the authors and not necessarily those of the AI4VBH Consortium members, the NHS, Innovate UK or UKRI. This work was funded by UKRI Programme Grant MC_UU_00002/18. S.E.P. acknowledges support from the ‘SmartHeart’ EPSRC Programme Grant (https://www.nihr.ac.uk/; EP/P001009/1). N.A. acknowledges support from the Medical Research Council for his Clinician Scientist Fellowship (MR/X020924/1). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Consortia
Contributions
D.R., J.P.C. and J.J. conceptualized the project. D.R., C.G., G.M.P., H.D., B.R.F., D.G., G.B., G.P., G.G.T. and N.A. contributed to formal analysis. D.R., C.G., G.M.P., B.R.F., D.G., G.B., G.P., G.G.T., N.A., A.R.V.R.H., M.P., E.H.F.-E. and Q.S.W. provided data curation. D.R., C.G., G.M.P., H.D., B.R.F., N.M.K., P.W.F.W., L.S.P., P.B.M., S.E.P., K.C., J.M.G., A.R.L., J.W., C.L., N.A., Y.V.S., A.C.P., J.P.C. and J.J. contributed to investigation. D.R., C.G., J.P.C. and J.J. wrote the original draft, and all authors contributed to the review and editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.E.P. provides consultancy to Circle Cardiovascular Imaging, Inc. J.W. holds membership of scientific advisory boards/consultancy for Relation Therapeutics and Silence Therapeutics and ownership of GlaxoSmithKline shares. J.P.C. is employed full-time by the Novartis Institute of Biomedical Interest (his major contributions to this project were while employed at the VA Boston Healthcare System). The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Cardiovascular Research thanks Ruth Frikke-Schmidt, Scott Ritchie and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Venn diagram illustrating the overlap of genes among pQTLs and eQTLs.
Figure A shows the overlap of genes among ARIC (1,342 genes), Fenland (1,383 genes), and deCODE (1,493 genes). Figure B shows the overlap of genes among eQTLGen (8,054 genes) and GTEx v8 (11,985 genes).
Extended Data Fig. 2 MR effect on atrial fibrillation against the MR effect on HFpEF and HFrEF for genes that pass the Bonferroni threshold only.
The error bars indicate the 95% confidence interval of the MR beta coefficient. The best-fit line derived through ordinary least squares is shown in black. The sample size used to derive statistics for the MR effects on HFpEF is 27,579 participants with HFpEF, 27,799 participants with HFrEF, and 367,267 control individuals. The sample size used to derive statistics for the MR effects on atrial fibrillation is 91,703 cases and 776,176 controls. All results are shown for MR on atrial fibrillation analyses that have passed p-value \(< 5\times {10}^{-5}\). The predicted model for HFpEF is y = -0.0016 + 0.98x (left plot) and for HFrEF is y = -0.003 + 0.65x (right plot). The correlation between the MR betas for atrial fibrillation and HFpEF is 0.99 and between the MR betas for atrial fibrillation and HFrEF is 0.94. Data are presented as mean values +/- 1.96*standard error. All tests were two-sided without adjustment for multiple testing.
Extended Data Fig. 3 MR effect on atrial fibrillation against the MR effect on HFpEF and HFrEF for genes that pass the Bonferroni threshold and including HFpEF suggestive genes.
The plot is shown for HFpEF findings that pass the Bonferroni-adjusted threshold of p-value \(< 2.06\times {10}^{-6}\) (in red) and for findings that pass the false discovery rate (FDR) of 5% (p-value \(< 6.80\times {10}^{-4}\)) (in blue). The sample size used to derive statistics for the MR effects on HFpEF is 27,579 participants with HFpEF, 27,799 participants with HFrEF, and 367,267 control individuals. The sample size used to derive statistics for the MR effects on atrial fibrillation is 91,703 cases and 776,176 controls. The predicted model is y = 0.0079 + 0.83x. The correlation between the MR betas for atrial fibrillation and HFpEF is 0.93. The errors bars represent the 95% confidence interval, defined as 1.96*standard error. All tests were two-sided without adjustment for multiple testing.
Extended Data Fig. 4 The number of unique and shared genes among the present study categorized as HFpEF/HFrEF MR, and prior studies on HF GWAS, HFpEF/HFrEF GWAS, HF MR, and cardiomyopathy presented as a Venn diagram and as an upset plot sorted by cardinality.
In Extended Data Fig. 3A, the percentage presented in parentheses represents the proportion of the total data set that falls within each section of the Venn diagram. Sets with no genes are not annotated.
Extended Data Fig. 5 Schematic for two-step Mendelian randomization study design for assessing mediation.
The causal effect of Lp(a) levels on the mediator, coronary artery disease (CAD), \({\beta }_{1},\) was determined via two-sample MR. The causal effect of the mediator, CAD, on HFpEF and HFrEF, \({\beta }_{2},\) was determined via two-sample MR. The instrumental variables utilized in the second step are independent of those utilized in the first step. The total effect, \({\beta }_{0}\), represents the effect of Lp(a) levels on HFrEF or HFpEF. The indirect effect of the mediator, CAD, was calculated as \({\beta }_{1}* {\beta }_{2}\) and the proportion mediated was calculated as the indirect effect divided by the total effect,\({(\beta }_{1}* {\beta }_{2})/{\beta }_{0}\).
Supplementary information
Supplementary Information
Supplementary Figs. 1–80
Supplementary Table
Supplementary Tables 1–28
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rasooly, D., Giambartolomei, C., Peloso, G.M. et al. Large-scale multi-omics identifies drug targets for heart failure with reduced and preserved ejection fraction. Nat Cardiovasc Res 4, 293–311 (2025). https://doi.org/10.1038/s44161-025-00609-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s44161-025-00609-1
This article is cited by
-
Integrating multi-omics and machine learning strategies to explore the “gene-protein-metabolite” network in ischemic heart failure with Qi deficiency and blood stasis syndrome
Chinese Medicine (2025)
-
Discovery of drug targets for heart failure with preserved and reduced ejection fraction
Nature Cardiovascular Research (2025)