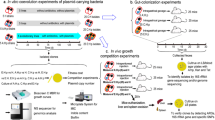
Abstract
The rapid emergence of multidrug-resistant virulent bacteria poses a global health concern. It is enabled through the lateral transfer of plasmid-carrying antibiotic resistance and virulence genes between bacteria. The guts of animals, particularly chickens, are recognized as reservoirs for this plasmid transfer, yet the regulation of this process by host factors remains poorly understood. Here, we examined the role of chicken intestinal tissue in the lateral transfer of the antimicrobial resistance plasmid pAPEC-O2-211A-ColV from its natural bacterial host, E. coli APEC-O2-211, to the plasmid-free E. coli HS-4 using an explant model, and how chicken host miRNAs contribute to this process. Ceca tissues collected from commercial and heritage birds were exposed to bacterial conjugation mixtures. Host small RNAs were extracted from chicken ceca tissue and evaluated for their effect on conjugation in vitro. We observed the expected bactericidal effect of chicken host tissues on both donor and recipient populations, alongside a variable response of bacterial conjugation influenced by different host genetic backgrounds. Additionally, we observed a correlation between small RNAs, mimic RNAs and bacterial plasmid conjugation in vitro. This study shed light on the role of small RNAs derived from host tissue in the differential regulation of bacterial plasmid conjugation.
Similar content being viewed by others
Introduction
Bacterial plasmid conjugation (BPC) is a driving force behind the emergence and rapid spread of novel multi-drug-resistance (MDR) and virulent bacteria in the clinical, agricultural, and consumer environments1,2,3. Additionally, the gut of animals serves as a potent reservoir for BPC between microorganisms, allowing for expansion of novel pathogenic and commensal bacteria with the potential to cause severe infections in humans and other animals4,5,6,7,8,9,10. As a result, considerable attention has been given to identifying novel compounds that can regulate BPC11,12,13,14,15,16. However, there have been few experimental studies conducted in animals to assess the effectiveness of conjugation inhibitors (COIN) or conjugation stimulators (COST) in the gut or gut-associated environments16,17. To tackle this issue, in vitro models simulating the animal gut have been proposed, utilized, and previously reviewed18,19,20.
Food producing animals, such as cows, pigs, and chickens, are increasingly being studied for their contributions to this MDR emergence21,22,23,24. The chicken, in particular, is of interest because it serves as a natural reservoir for asymptomatic colonization with Salmonella spp. Additionally, chickens can be hosts for pathogenic Escherichia (E.) coli infections, including extraintestinal infections linked to Avian Pathogenic E. coli (APEC)25,26,27,28,29. These pathogens are often associated with the presence of large antimicrobial resistance (AR) plasmids that carry and disseminate antibiotic resistance, virulence and metabolic genes25,30. The transfer of these gene vectors to resident microbes has the potential to create novel, potentially pathogenic organisms17,29,31,32. Studies on the host-associated factors that influence the occurrence of BPC in the gut have been limited10,33. However, recent advancements in animal gut models have enabled the examination of specific host factors, such as genetics, diet, and inflammation, on the molecular process involved in bacterial conjugation within the gut environment and this topic has been reviewed extensively9,19,20,34,35.
Recent discoveries in RNA biology suggest that RNA secreted by host organisms plays a regulatory role in bacterial gene expression and physiology36,37. Liu et al. demonstrated that there were significant alterations in gene expression in E. coli (yegH) and Fusobacterium nucleatum (16S rRNA), which resulted in increased growth observed in vitro37. However, in mice with a Dicer gene knockout in secretory epithelial cells (Dicer1ΔIEC), which leads to a lack of micro-RNA (miRNA) secretion into the lumen, the authors observed microbial dysbiosis and chronic colitis37. We previously demonstrated a correlation between the concentration of chicken small RNAs in the ceca and the incidence of plasmids in cecal bacterial isolates38. Furthermore, we showed a positive correlation between the concentrations of cecal small RNA extracts and plasmid conjugation in in vitro assays38. However, more detailed studies are needed to explore the role of host-secreted RNA species on BPC to determine whether secretory RNAs operate through gene-specific mechanisms and regulate gene and protein expression related to the BPC process. In addition, we have recently shown the effectiveness of using a chicken explant model to investigate BPC in host tissue-associated environments, where dietary short-chain fatty acids are identified as another host factor affecting BPC17. Chicken tissue co-culture conjugation assays provide a more comprehensive presentation of host factors through in vitro models, potentially bridging the gap between host-independent in vitro models and in vivo animal experiments17,39.
In this study, we utilized the chicken ceca explant co-culture conjugation assay with tissues derived from genetically diverse chickens to evaluate the incidence of BPC in response to host tissue and miRNA. Using in silico hybridization methods, we further assessed the role of a known gastrointestinally expressed miRNA species, as well as the entire mature Gallus gallus miRNA library, to identify potential interactions with bacterial chromosome and plasmid sequences; we aimed to determine how these interactions may regulate bacterial conjugation. Subsequently, we conducted in vitro broth conjugation assays to examine the role of these miRNAs and analyzed the correlation between their expression and the observations made during explant conjugations. Through our experimentation and investigation, we identified an miRNA species, gga-miR-183, expressed in the gastrointestinal tract, which is capable of regulating BPC in vitro. Finally, we investigated the impact of miRNA mimic supplementation on the expression of genes necessary for efficient plasmid transfer. Overall, our studies reveal a novel regulatory role of host-secreted miRNA in the conjugal transfer of AR and virulence plasmids among bacteria. Additionally, we provide evidence supporting the further exploration of miRNA mimics as effective interventions with characteristics of COIN and COST.
Results
Commercial chicken ceca tissues demonstrated an antimicrobial response independent of conjugation frequency
To determine whether the genetic background of commercially available chickens influences the incidence of BPC differentially, we conducted in vitro conjugation assays using ceca tissue from various chicken breeds: Dekalb White (DW), Hy-Line North America (W80x W36) (R-38), or Lohmann Select Layer (LSL). The ceca tissue was treated with antibiotics and rinsed before being added to conjugation assays involving E. coli strains APEC-O2-211 and HS-4 (Fig. 1).
Log10 donor E. coli APEC O2-211 (A), recipient E. coli HS-4 (B) transconjugant E. coli HS-4(pAPEC-O2-211A-ColV) (C), and conjugation efficiency (D) of conjugation reactions supplemented without (white) or with 1 cm2 chicken ceca tissue biopsies from 10-week-old Dekalb White (pink), 14-week-old Hy-line North America (R-38) (green), and 6-week-old Lohman Select Leghorn (purple) birds. Bars represent the mean and error lines represent the standard deviation around the mean (A–C) or median and standard error around the median (D), and symbols represent individual replicates. *p value < 0.05; ***p value < 0.0005; ****p value < 0.00005.
The results revealed that conjugation mixtures containing R-38 and LSL tissues exhibited significantly reduced populations of donor bacteria compared to the tissue-free control (p < 0.0005). In contrast, the DW-supplemented conjugations showed a significant increase in detected donor populations (p < 0.05) (Fig. 1A). Among the tissue-supplemented treatment groups, the DW-supplemented conjugations had significantly greater donor populations compared to the R-38- and LSL-supplemented treatment groups (p < 0.0005) (Fig. 1A).
Regarding recipient populations, LSL-supplemented conjugations demonstrated a significant reduction (p < 0.05), while DW- and R-38-supplemented conjugations either showed significantly greater recipients (p < 0.00005) or no change in recipient populations (p > 0.05), respectively (Fig. 1B). Significantly fewer recipients were detected in R-38- and LSL-supplemented groups compared to the DW-supplemented treatment groups (p < 0.0005) (Fig. 1B).
Additionally, the DW-supplemented groups exhibited bactericidal activity at 16 weeks of age, demonstrating significant bactericidal effects against donor strains (p < 0.00005) and transconjugant strains (p < 0.05). In contrast, the 2-week- and 10-week-old birds showed growth-promoting effects on recipient populations (p < 0.0005) (Fig. S2A–C).
There was no significant difference (p > 0.05) in transconjugant populations between R-38- or LSL-supplemented conjugations and the tissue-free control. However, a significant increase (p < 0.05) in transconjugant populations was observed in the DW-supplemented conjugations compared to both tissue-free and R-38-supplemented conjugations (Fig. 1C).
To evaluate whether changes in transconjugant populations were masked by the bactericidal effect of tissue supplementation, we calculated the Log10 conjugation efficiency of conjugations according to the following equation:
Where T is the total transconjugant population, D is the total donor population, and R is the total recipient population40.
In the DW-supplemented reactions, the only age group that showed a significant difference in conjugation efficiency compared to the control was 10-week group (p < 0.01), although this finding was not consistent when factoring in multiple genetic lines for comparison (Figs. S2D and 1D). Significant increases in conjugation efficiency were observed in the R-38 (p < 0.05) and LSL (p < 0.0005) supplemented conjugations when compared to the tissue-free conjugation (Fig. 1D). Conversely, a numerical but non-significant reduction (p > 0.05), in conjugation efficiency was noted for the DW-supplemented conjugation reactions (Fig. 1D). Within the tissue treatment groups, both R-38- and LSL-supplements resulted in significantly greater (p < 0.05 and p < 0.0005 respectively) median Log10 conjugation efficiency compared to the DW-supplemented reactions (Fig. 1D).
Heritage chicken ceca tissue demonstrated a robust antimicrobial response with decreased conjugation
Ceca tissues were collected from three historically inbred heritage chicken lines: GHS-6, Line-8, and M-15.2 at 73 days of age. These tissues were further evaluated in explant tissue conjugations as with commercial tissues (Fig. 2). The addition of ceca tissues from any of the three genetic lines led to significant reductions (p < 0.0005) in donor and recipient populations when compared to the no-tissue treatments controls. There was a less significant reduction (p < 0.005) in recipient populations in Line-8-supplemented conjugations (Fig. 2A, B). No significant differences were observed when comparing donors or recipients across the different tissue-supplemented treatment groups (Fig. 2A, B).
Log10 donor E. coli APEC O2-211 (A), recipient E. coli HS-4 (B) transconjugant E. coli HS-4(pAPEC-O2-211A-ColV) (C), and conjugation efficiency (D) of conjugation reactions supplemented without (white) or with 1 cm2 chicken ceca tissue biopsies from GHS-6 (green), Line-8 (brown), and M-15.2 (purple) birds. Bars represent the mean and standard deviation (A–C) or median and standard error around the median (D), and symbols represent individual replicates. **p value < 0.005; ***p value < 0.0005; ****p value < 0.00005.
Regarding the transconjugant populations, the reactions supplemented with Line-8- and M-15.2 ceca tissues showed significantly fewer transconjugants compared to the no-tissue control (p < 0.00005 and p < 0.0005 respectively) (Fig. 2C). In comparisons between tissue-supplemented conjugations, the GHS-6-supplemented conjugation resulted in a significantly greater number of transconjugants than either the Line-8- (p < 0.00005) or the M-15.2-supplemented conjugations (p < 0.0005) (Fig. 2C). Similarly to commercial chicken conjugations, the efficiency of plasmid transfer was calculated using Eq. (1) (Fig. 2D). Line-8-supplemented conjugations showed a significant reduction in Log10 conjugation efficiency compared to the tissue-free control conjugations (p < 0.05) and the M-15.2- (p < 0.05) and GHS-6- (p < 0.00005) supplemented conjugations (Fig. 2D). No significant differences were observed between the tissue-free, and the M-15.2- and GHS-6-supplemented conjugations (Fig. 2D).
Small RNAs extracted from heritage ceca tissues demonstrated varied inhibitory effects on bacterial plasmid conjugation in vitro
To evaluate the role of host cecal small RNAs in regulating BPC observed in tissue explant experiments, total small RNAs were extracted from content-free cecal tissue of both conventional and heritage chickens. These small RNAs were then supplemented into in vitro conjugation assays (Figs. 3 and 4). In conventional cecal small RNA-supplemented conjugations, no significant effects were observed on donor, recipient, or transconjugant populations, nor on conjugation efficiency (Fig. 3).
Log10 donor E. coli APEC O2-211 (A), recipient E. coli HS-4 (B) transconjugant E. coli HS-4(pAPEC-O2-211A-ColV) (C), and conjugation efficiency (D) of conjugation reactions supplemented without (white) or with 5 μM final concentration of total small RNA extracts isolated from Dekalb White (pink), Hy-line North America (R-38) (green), and Lohman Select Leghorn (purple) birds. Bars represent the mean and error lines represent the standard deviation around the mean (A–C) or median and standard error around the median (D), and symbols represent individual replicates. *p value < 0.05; **p value < 0.005; ***p value < 0.0005; ****p value < 0.00005.
Log10 donor E. coli APEC O2-211 (A), recipient E. coli HS-4 (B) transconjugant E. coli HS-4(pAPEC-O2-211A-ColV) (C), and conjugation efficiency (D) of conjugation reactions supplemented without (white) or with 5 μM final concentration of total small RNA extracts isolated from GHS-6 (green), Line-8 (brown), and M-15.2 (purple) chicken ceca tissue. Bars represent the mean and error lines represent the standard deviation around the mean (A–C) or median and standard error around the median (D), and symbols represent individual replicates. *p value < 0.05; **p value < 0.005; ***p value < 0.0005; ****p value < 0.00005.
Although small and insignificant variations in donor and recipient populations (p > 0.05) were noted, conjugations containing Line-8 and GHS-6 small RNAs at a concentration of 5 μM resulted in significantly fewer transconjugants compared to those observed in nuclease-free double-distilled water (NF-ddH2O) supplemented control conjugations (p < 0.00005, Line-8 and M-15.2; p < 0.0005 GHS-6) (Fig. 4C). Additionally, in RNA-supplemented conjugations, significantly greater populations of transconjugants were observed in the M-15.2 and GHS-6 treatments compared to the conjugation reactions treated with the Line-8 cecal small RNA extract (p < 0.05) (Fig. 4C).
Again, to determine the rate of plasmid transfer among viable donor and recipient cells, we calculated the Log10 conjugation efficiency using Eq. (1). We observed significant reductions (p < 0.005) in Log10 conjugation efficiency for Line-8 and M-15.2 small RNA supplemented conjugations. In contrast, there was a numerical but not statistically significant decrease (p > 0.05) in conjugation efficiency of the GHS-6 small RNA supplemented reactions compared to the control (Fig. 4D). Although no significant differences were found between the small RNA-supplemented treatment groups, a similar numerical trend to that seen in the heritage ceca tissue explant conjugations was noted (Figs. 2C and 4C).
In silico analyses detected potential miRNAs involved in BPC
Based on specific binding criteria outlined in material and methods, in silico analysis identified several miRNAs that may play a role in regulating BPC. These miRNAs include those that target (1) start codons and/or coding DNA sequence (CDS) within the tra operon (gga-miR-12207-3p, gga-miR-12233-5p, gga-miR-12235-5p, gga-miR-12263-5p, gga-miR-12279-5p, gga-miR-12282-5p), (2) promoter 1304 (tra) (gga-miR-12289-3p, gga-miR-183), (3) National Center for Biotechnology Information (NCBI) blast alignment with pAPEC-O2-211A-ColV (gga-miR-6642-3p, gga-miR-1306-3p), or (4) bindings to various other promoter sequences in addition to descriptions of activity in literature (gga-miR-130b-5p, gga-miR-1794, gga-miR-7447-5p) (Tables S1 and S2). Among these significant findings, seven miRNAs identified through promoter binding, NCBI blast alignments, and prior literature were chosen for further in vitro testing. Notably, gga-miR-183 was found to be the only mimic that significantly influenced conjugation efficiency (Fig. S1D).
Potential hybridizations were specifically detected between the mature gga-miR-183 sequence and both the whole genome of the E. coli APEC O2-211 and plasmid pAPEC-O2-211A-ColV (Table 1). Several hybridizations involving chromosomally encoded protein coding sequences were identified with hypothetical P-values below the threshold set in this study (p < 0.05). The top five chromosomal hybridizations, ranked by minimum free energy of hybridization (MFE) consisting of the targets; Non-oxidative hydroxyarylic acid decarboxylases subunit D (MFE: −33.5, WP_000562976.1), class II fructose-bisphosphatase (MFE: −33.3, WP_000987308.1), class 1 fructose-bisphosphatase (MFE: −33.2, WP_000853753.1), L-threonylcarbamoyladenylate synthase type 1 TsaC (MFE: −33.1, WP_001297709.1), and the 4-hydroxy-2-oxovalerate aldolase (MFE: −33, WP_001013518.1) (Table 1).
Similarly, RNAhybrid analysis identified three hybridizations with the large AMR and virulence plasmid pAPEC-O2-211A-ColV (NZ_CP030791.1). These hybridizations included the coding sequences (CDS) for the major facilitator superfamily (MFS) transporter (MFE: −32.5, WP_000095528.1), the conjugal transfer complement resistance protein TraT (MFE: −32, WP_000850416.1), and an IS91 family transposase (MFE: −25.8, APECO2_RS26905) (Table 1). No significant hybridizations were detected between the mature gga-miR-183 sequence and the two accessory plasmids of E. coli APEC O2-211 pAPEC-O2-211B, and pAPEC-2-211C. Although the specific role of miR-183 as a regulator of BPC is not currently known, the vast bioactivity and tentative in silico predictions suggest that it may have a significant regulatory role, which was evaluated in this study.
The miRNA species gga-miR-183 conferred stimulatory plasmid conjugation in vitro and was overexpressed in the GHS-6 ceca tissues
To identify if the predicted miRNA binding identified in silico correlates with changes in conjugations in vitro, we evaluated the mature gga-miR-183 species, obtained as a miRVana miRNA mimic, through in vitro conjugation assays for its effect on plasmid conjugation (Fig. 5). Although minor numerical differences were observed in donor and recipient populations supplemented with the gga-miR-183 miRNA mimic, no significant differences were found (p > 0.05). However, we detected a significantly greater number of transconjugants in gga-miR-183-supplemented reactions compared to the NF-ddH2O-supplemented control conjugations. To confirm the impact of this alteration in the transconjugants population relative to the donor and recipient populations, we calculated the Log10 conjugations efficiency was calculated as in Eq. (1). The results showed significantly greater median conjugation efficiency (p < 0.05) in gga-miR-183 supplemented conjugations compared to the NF-ddH2O supplemented conjugation (Fig. 5A–D). Other potential miRNA mimics were assessed for their regulatory effects on the donor, recipient, transconjugant populations, and conjugation efficiency, however, they did not demonstrate any significant differences from the control (Fig. S1). To determine whether the differential expression of the miRNA species in the cecal small RNA populations correlates to the observed changes in conjugation frequency associated with ceca explants from the same birds, we evaluated the relative expression of gga-miR-183 using miRNA specific reverse-transcription (RT) qPCR (Fig. 5E). No significant differences (p > 0.05) in the relative fold change of gga-miR-183 gene expression were detected between the Line-8 M-15.2, and conventional cecal small RNA populations. However, a significant increase in gga-miR-183-ranging from two- to four-fold was observed in GHS-6 cecal small RNA extracts compared to the normalized Line-8 (p < 0.005), LSL (p < 0.005), and M-15.2 (p < 0.0005) cecal small RNA populations (Fig. 5E). Additionally, gastrointestinally expressed gga-miR-183 showed potentially high-affinity binding to donor chromosomal and plasmid sequences (Table S2).
In vitro miRNA mimic conjugations (A–D). Log10 donor E. coli APEC O2-211 (A), recipient E. coli HS-4 (B) transconjugant E. coli HS-4(pAPEC-O2-211A-ColV) (C), and conjugation efficiency (D) of conjugation reactions supplemented with NF-ddH2O (control, white), and gga-miR-183 mimic RNA (blue). Relative 2−ΔΔCT fold change in gene expression of the gga-miR-183 RNA in small RNA extracts from GHS-6 (green), Line-8 (brown), and M-15.2 (purple), Dekalb White (pink), Hy-line North America (R-38) (green), and Lohman Select Leghorn (purple) ceca tissues (E). Hypothetical in silico hybridizations of gga-miR-183 towards the CDS of pAPEC-O2-211A-ColV (F). Bars represent the mean and lines represent the standard deviation around the mean (A–C) or median and standard error around the median (D, E). Symbols represent individual replicates. *p value < 0.05; **p value < 0.005; ***p value < 0.0005.
To identify potential interactions between the gga-miR-183 small RNA species expressed in the ceca and the total chromosome and plasmid genes of the E. coli APEC O2-211A donor (chromosome, NZ_CP006834.2; pAPEC-O2-211A-ColV, NZ_CP030791.1; pAPEC-O2-211B, NZ_CP030792.1; and pAPEC-O2-211C, NZ_CP030793.1) an in silico analysis was conducted using RNAhybrid along with the mature Gallus gallus miRNA database. This analysis generated hypothetical hybridizations between the mature gga-miR-183 miRNA and the total CDSs from both the whole genome and the plasmid encoded tra genes (Table 1). Overall, 51 unique hybridizations were identified with tentative p values below 0.05. Of these, 48 targeted various chromosomal encoded genes, while 3 targeted plasmid encoded genes (Fig. 5F and Table 1). The plasmid-targeted genes included the MFS transporter (WP_000095528.1), the conjugal transfer complement resistance protein TraT (WP_000850416.1), and the IS91 family transposase (APECO2_RS26905). The IS91 family transposase is annotated as; frameshifted, incomplete, partial in the middle of a contig, or missing an N-terminus according to the NCBI Prokaryotic Genome Annotation Pipeline (PGAP).
Plasmid conjugations supplemented with the miRNA mimic gga-miR-183 showed a significant change in the expression of the tra gene
To investigate the mechanism by which gga-miR-183 enhances plasmid conjugation, we analyzed the expression of genes associated with plasmid replication (rep) and transfer (tra) using RT-qPCR (Fig. 6). We found no significant differences in the expression of rep genes between conjugations supplemented with gga-miR-183 and NF-ddH20-treated controls (Fig. 6A). However, the expression of the tra relaxosome formation genes, traY and traI, was significantly increased in the gga-miR-183 supplemented conjugations (p < 0.01) (Fig. 6B). Additionally, the expression of topoisomerase formation gene traE also showed significant increase in gga-miR-183 supplemented conjugations (p < 0.05) (Fig. 6B). The only gene that showed a significant decrease in expression in the conjugations treated with gga-miR-183 was the surface exclusion protein-encoding gene traS (p < 0.05) (Fig. 6B).
Log2 fold changes in expression of replication (rep, A) and transfer (tra, B) genes in in vitro plasmid conjugation reactions supplemented with NF-ddH2O carrier control (white) or 5 nM miRNA mimic gga-miR-183 (red). Genes are presented in the order observed in the plasmid sequence and all genes are in the same orientation. The locations of known promoters are represented by vertical dashed lines. Bars represent triplicate reactions evaluated in duplicate. Horizontal lines in the center of each bar represent the arithmetic mean. Log fold change in gene expression was calculated via the 2−ΔΔCt method. p values below or equal to 0.05 were considered significant. *p value < 0.05; **p value < 0.005.
Discussion
The animal gut serves as a significant reservoir for the lateral exchange of bacterial plasmid DNA4,8,10,18,41. While much is known about the bacterial-host interactions that contribute to pathology and disease, there is limited knowledge about the bacterial-host interactions that regulate the process of BPC in the gut18. In this study, we utilized an avian in vitro ceca explant model to investigate the role of chicken host cecal tissues in BPC across commercial and heritage genetic lines. Our objective also included identifying the potential role of miRNAs’ as components of the complex regulation of this process in vitro.
Previous studies have shown that variations in chicken host responses to bacteria are influenced by genetics, involving both major histocompatibility complex (MHC) and non-MHC loci42,43,44. For instance, links have been established between single nucleotide polymorphisms (SNP) and differential responses of genetic lines to Salmonella Enteritidis infections, as well as the relationship between MHC structure and resistance to APEC based on divergent chicken host genetics42,44,45.
In our experiments, conjugations of the donor E. coli APEC-O2 and the recipient E. coli HS-4 in the presence of ceca tissues of either DW, R-38, or LSL chickens yielded an expected antimicrobial response towards both donor and recipient bacteria in commercial (R-38 and LSL) and heritage (GHS-6, Line-8, M-15.2) chicken tissues. Surprisingly, there was a significant increase in the number of donor and recipient bacteria in conjugations supplemented with commercial DW chicken tissues.
These findings align with previous research indicating that GHS-6 chickens exhibit a more generalist and lower immune response compared to Line-8 and Fayoumi chickens. In contrast, conjugations with Line-8 and Fayoumi tissues showed an antimicrobial or bacteriostatic effect, while GHS-6 tissues displayed a growth-promoting effect46. Although both Line-8 and GHS-6 birds are Leghorn in background, they exhibit substantial genetic differences due to their divergent origins and subsequent inbreeding. Line-8 is the oldest inbred line globally, having been inbred since 192546, whereas the GHS-6 and Fayoumi lines have been maintained and inbred since 195446.
In a previous study using a murine model for gut mediated BPC, we demonstrated that host genetics play a significant role in the variability observed in conjugation within the gut of mice, while keeping all other factors constant19. We speculated that this variability might stem from known differences in the MHC genes of the two murine genetic backgrounds we utilized, 129S6/SvEv and C3H/HeN mice19.
In this study, we tested conjugations between a chicken pathogenic and commensal E. coli often associated with the human gut, using ceca tissues from both commercial and heritage chicken genetic lines. We found that GHS-6 was unique in presenting transconjugant populations that were not significantly lower than that those observed in tissue-free controls47. To rule out the possibility that the reduction in transconjugants was not specifically related in the antimicrobial propriety of the explant tissues, we normalized transconjugant levels to account for changes in donor and recipient populations.
Our data indicated high conjugation efficiency in two out of the three commercial chicken lines, specifically in R-38 and LSL. In contrast, we observed either a decrease in conjugation efficiency in the heritage chicken line Line-8 or no changes in lines M-15.2 and GHS-6. These findings suggest that certain genetic lines used in commercial farms may contribute to the emergence of AR bacteria, while heritage lines may possess genetics that lead to a lower occurrence of AR bacteria. Our team has previously identified the widespread of colistin-resistant E. coli, along with multidrug resistant Acinetobacter and extended-spectrum β-lactamase-producing Enterobacteriaceae in cage-free and conventionally caged birds at various stages of maturity29.
We then wondered whether the selection of commercial ceca tissue for increased BPC efficiency was a result of selective breeding for layer traits or if it is a universal characteristic associated with avian ceca environments (Fig. 1). The historically inbred heritage birds, such as GHS-6, Line-8, and M-15.2 chickens, selected as representative hosts of interest for the chicken gut, have historically conserved genetic backgrounds. Unlike commercial lines, these heritage lines have not undergone years of selective breeding for economic traits, which may have led to health complications48. Variations between these inbred lines have been previously characterized, including viral pathogen resistance, reproductive capacity, and response to environmental stressors49,50. Of particular interest to this study, distinct immune capabilities are also identified between inbred lines, including MHC class I expression variation identifying Line-8 and Fayoumi lines as probable specialist antigen binders, whereas GHS lines GHS-6 and GHS-13 demonstrated more generalist capabilities and lower immune response46.
Although multiple host factors may be associated with BPC response, this study focused on the miRNA, a class of small RNAs recently suggested as potential host-secreted factors that can influence bacterial gene expression and metabolism in the gut environment37. Previously, we showed a correlation between the concentration of cecal small RNAs in the cecum and both the incidence of plasmids in gut isolates as well as on the induction of BPC in vitro38.
Here, supplementation of cecal small RNAs of the commercial birds to conjugation reactions did not affect BPC, which is in contrast with the pro-conjugation effects observed when commercial tissue was supplemented. In contrast, the supplementation of ceca small RNAs from heritage birds resulted in broad inhibition of BPC across lines, except GHS-6. This exception may be due to the presence of conflating pro-conjugation factors in the GHS-6 tissues that are absent in the small RNA populations.
While it has been previously described that small RNAs can positively regulate BPC, it is important to note that these small RNAs were contained within the conjugative E. coli itself50. Our results support our previous study that indicated there may be a relationship between host-secreted small RNAs, such as miRNA, and the regulation of BPC. However, that study did not identify the specific miRNAs involved in this regulatory process38.
The evolutionarily conserved miR-183 miRNA species is expressed in the gastrointestinal tract of humans, and its homologs have been identified in the genomes of various animal species such as goats, humans, rats, mice, and chickens51. Current research evidence shows that miR-183 is present in several cell types, such as colonic, retinal, and kidney cells, and is secreted into both the bloodstream and the gut lumen in exosomes37,52.
Moreover, miR-183 plays a role in the regulation of various cancers, including colorectal, lymphatic, gastric, liver, and prostate cancer, and is being evaluated as a biomarker for early detection of cancer-related diseases52,53,54,55. Additionally, miR-183 is considered essential for the development and function of sensory neurons55,56. These preliminary studies suggest that miR-183 is evolutionarily conserved to perform multiple functions in both homeostatic (healthy) and heterotactic (pathological) host function56.
Among the three hypothetical pAPEC-O2-211A-ColV targets identified through in silico analysis, traT stands out as the most promising target with potential regulatory effects on BPC. The TraT protein, which is encoded by the traT gene, plays a crucial role in surface exclusion and the disaggregation of microbial mating pair complexes following the transfer of DNA. Additionally, it interferes with the TraN–OmpA interaction, which is essential for stabilizing subsequent conjugations57,58.
The impact of gga-miR-183 hybridization with traT on the functional expression of TraT remains unclear. One possibility is that this binding could stabilize traT mRNA, leading to an increased translation of traT mRNA into protein59. Conversely, hybridization could also result in an increased cellular turnover of traT mRNA, ultimately causing downregulation of traT translation37,59,60. Recently, we developed a qPCR to evaluate tra operon gene expression40.
It is important to acknowledge that RNAhybrid analysis of bacterial gene targets has inherent limitations due to constraints in prokaryotic gene annotation. In eukaryotes, miRNA hybridization predictions typically focus on the 3’-untranslated region (3’-UTR) of mRNA, where it is generally believed that miRNA regulation occurs61. Unfortunately, this type of information is not currently annotated in prokaryotic genomes, primarily because prokaryotic genes lack the clear translation start and stop signals that are present in eukaryotic genes62. As such, in this study, a more a more limited CDS approach was utilized for analysis. However, if full transcript sequences of the plasmid expressed genes were available, it would enhance both the resolution and predictive capabilities of the analysis.
We have not yet identified any RNA-seq studies or entries in public databases, such as the NCBI sequence read archive (SRA), that have been completed under conditions conductive to conjugal protein expression. Our ongoing studies are focused on bridging this gap in experimental coverage and knowledge using approaches like 5’ and term-seq methods. Furthermore, small RNAs, including miRNA, may act post translationally on proteins directly. Currently, we are not aware of any tools available for analyzing miRNA binding to protein structures, which could represent a potential mechanism for miRNA-mediated interactions in bacteria. However, recent developments in the field of RNA aptamers have highlighted the potential role of RNA as a post-translational regulator both in biological and in synthetic applications. This topic is also the focus of our current studies63,64.
As observed through qPCR, a significantly higher expression of gga-miR-183 in the GHS-6 genetic heritage line is seen compared to both the Line-8 and M-15.2 genetic lines. This suggests that gga-miR-183 may play a stimulating role in regulating of, or preventing the general inhibitory response of heritage chicken tissues on BPC. Additionally, the higher abundance of gga-miR-183 correlates with a reduced inhibitory effect observed in plasmid conjugations supplemented with the small RNA ceca extract populations from this line. Furthermore, the significantly elevated expression of gga-miR-183 in the GHS-6 birds compared to LSL birds, as well as to the numerically higher levels compared to DW and R-38 birds, could indicate the involvement of other unexamined sRNA that may neutralize the effect on BPC in conventional birds.
Increased transcription of the tra genes, traY and traI, observed in gga-mir-183 supplemented conjugations sheds light on the mechanism involved. Both of these genes are associated with enhanced plasmid transfer through the formation of the relaxosome65. Additionally, elevated transcription of the tra gene, which is responsible for the activity of plasmid-associated topoisomerase, was also noted66. The only gene that showed a significant decrease in expression was traS, which is required for surface exclusion and the prevention of multiplicity transfer of plasmid DNA. This reduction in the expression of this surface exclusion protein may facilitate greater DNA transfer between bacterial mating pairs, through it poses a risk of potential zygotic stress and cell death67,68. While the tra genes identified through qPCR do not overlap with those identified in silico, these findings still provide valuable insights into the mechanism behind increased BPC that reasonably supports the observed results. It must be noted that both in silico and in vitro methods carry their own limitations on predictive capability and observational insight, and one result does not necessarily negate the other. Although these observations do not directly clarify why we see a decrease in transconjugant populations and conjugation frequency in conjugations supplemented with Line-8 and M-15.2 cecal small RNA populations, they may help explain the reduced inhibition observed in the GHS-6 tissue-supplemented conjugations compared to the either Line-8 or M-15.2 small RNA treated conjugations.
Overall, this study describes the potential role of gga-miR-183 as a stimulator of BPC in host-associated explant models, particularly in the gut environment. However, the mechanism behind the inhibition of conjugation efficiency and the formation of transconjugant populations is more complex. More research is needed to address these gaps by developing new target sequence resources, such as full-length mRNA sequences for plasmid genes of interest. There is a need for bioinformatic tools that can predict in silico hybridization between host secreted miRNA species and bacterial genes. The interactions between miRNAs produced by the host animal and the host microbiome are extensive, and the observed effects on BPC are likely influenced by multiple miRNA species, which may act either positively or negatively. Although this study has shown gga-miR-183 as a positive regulator of BPC, the effects seen in sRNA-supplemented conjugations may be driven by miRNAs that have not yet been screened through computational methods. Additionally, it is possible that miRNAs work together to influence BPC in a host environment, indicating a need for further investigations among multiple host miRNA and their effects on BPC. This study identifies a novel potential regulator for BPC that is expressed in the host and correlates with increased levels of BPC in ceca explant. Further evaluation is required to determine the specific nature of these interactions within a living host and to explore what other miRNA species might contribute to the variable responses observed in conjugation reactions supplemented with host cecal small RNA.
Methods
Bacterial strains, plasmids
The E. coli strain APEC O2-211 was used as the plasmid donor in both in vitro and ex vivo conjugation experiments. APEC O2-211 harbors one large antimicrobial resistance plasmid, pAPECO2-211A-ColV, and two small plasmids pAPEC-O2-211B and pAPEC-O2-211C26. The pAPEC-O2-211A-ColV plasmid is a hybrid IncFIB/IncFIC plasmid that confers resistance to macrolides, nitroimidazole, aminocoumarin, fluoroquinolone, tetracycline, carbapenem, and aminoglycoside antibiotics26. Here, tetracycline resistance was used as the selective marker for conjugation assays described in the following assays26. The plasmidless E. coli strain HS-4 was used as a recipient in all conjugations and was originally isolated from a healthy human gut and has been modified to maintain a spontaneous chromosomal resistance to rifampicin69. Prior to each experiment, fresh bacterial cultures were streaked onto MacConkey agar supplemented with antibiotics targeting each resistance marker (APEC-O2-211, 15 ug/mL tetracycline; HS-4, 100 μg/mL rifampicin).
Explant conjugation media
The bacterial conjugation broth used in explants comprised a complete CO2-independent growth medium (RPMI 1641, 10% heat inactivated fetal bovine serum, 4 mM L-Glutamine). A final OD600nm of ~1.0 of each donor and recipient cells were added to this medium and immediately used in conjugation assays17.
Chicken strains and tissue collection
Commercial male and female DW, R-38, or LSL chickens of various ages (DW at 2, 10, and 16 weeks, R-38 at 14 weeks, and LSL at 6 weeks) used in in vitro explant assays were obtained from a commercial farm located in southern Iowa. Heritage bird tissues were collected from Line-8, M-15.2, and GHS-6 strains raised at the Iowa State University Robert T. Hamilton Poultry Teaching and Research Farm. The highly inbred chicken lines GHS-6 (Leghorn), Line-8 (Leghorn), and M-15.2 (Fayoumi) were selected as hosts of interest and evaluated for the effect of cecal tissue exposure on BPC. GHS-6, Line-8, and M-15.2 lines have been maintained under inbreeding conditions for 64–96 years, are diverse from each other, and represent genetic characteristics of chicken lines prior to intense selective breeding for layer and broiler characteristics70. Twelve 73-day-old birds were used per experimental group with equal representation of male and female birds. Chickens were euthanized, then immediately surface decontaminated with 70% ethanol and opened aseptically in a biosafety cabinet (Cat. #302489100 Labconco, Kansas City, MO). Both ceca lobes were harvested, and one ceca lobe was placed in RNALater while the other lobe was placed in tissue collection medium (RPMI 1641, 50 ug/mL Gentamycin, 1% Penicillin/Streptomycin) and transferred to the lab for further processing, as necessary.
Explant co-culture and conjugation
Ceca tissues placed in tissue collection medium were removed, opened longitudinally, and rinsed twice in double volumes of sterile phosphate-buffered saline (PBS) and shaken at 225 RPM for 15 min to remove trace antibiotics and fecal material. Rinsed tissues were then segmented with sterile razor blades into 1 cm2 sections and placed into individual 1.5 mL microcentrifuge tubes containing 100 μL volumes of 1.0 mm sterile silica glass beads. Bacterial strains were prepared from overnight cultures as before17. To these vessels, 1 mL of the donor and recipient strains resuspended in complete growth medium was added. Tubes containing silica glass beads without tissues with 1 mL of bacterial conjugation matrix were used as a no-tissue control group for comparisons. Completed tubes were incubated for 6 h at 40 °C, vortex homogenized for 1 min, and serial dilutions prepared and plated onto selective MacConkey agar as described previously17.
The Log10 conjugation efficiency was used to normalize transconjugant populations to changes in the donor and recipient populations associated with bactericidal effects.
Tissue homogenization and large and small RNA extraction
Ceca tissues stored in RNALater were segmented into 1 cm2 portions, placed into new tubes containing 1 mL of fresh RNALater, and stored at −80 °C for further processing. Prior to RNA extraction, tissues were cryogenically homogenized using a liquid nitrogen microcentrifuge mortar and pestle (Cat #H37260-0100, Bel-Art, Wayne NJ, USA) following the manufacturer’s instructions. Briefly, tissues were removed from −80 °C, the total volume of RNALater was removed and the tissues were flash-frozen in liquid nitrogen. The samples were then physically homogenized via the liquid nitrogen-cooled micro mortar and pestle. The mortar was cleaned between samples using a progression of (1) 10% bleach, (2) diethyl-pyrocarbonate (DEPC)-treated double distilled (dd) H2O, (3) RNAse Zap, and (4) a final rinse with DEPC-treated ddH2O. The sample and pestle were then both acclimated to liquid nitrogen temperature prior to each use. Homogenized tissues were transferred back to −80 °C until RNA extraction. To extract both Large and Small RNAs from tissues, the RNAzol RT extraction reagent was used according to the manufacturer’s instructions (Molecular Research Center, Inc; Cincinnati OH; USA) with few modifications. Briefly, tissues were resuspended in RNAzol RT and vortexed for 1 min each. Suspensions were incubated at room temperature for 15 min, and then the large particulates, aqueous phase, carbohydrates, and fats were separated by centrifugation at 8000 × g for 10 min. Following centrifugation, the aqueous phase containing the protein, DNA, and RNA was removed and placed into new sterile tubes. The standard manufacturer’s instructions were completed following this point. Large and small RNA pellets were resuspended in 50 μL DEPC-treated ddH2O, enumerated via a NanoDrop Lite spectrophotometer (Thermo Fisher, Waltham, MA), and stored at -20°C for further analysis.
In vitro cecal RNA conjugation
To determine the role of cecal small RNAs on bacterial conjugation, in vitro broth conjugations with or without cecal small RNAs supplementations were conducted. Briefly, aliquots of small RNAs (50 ng) or NF-ddH2O (vehicle) were added to broth conjugation assays in 1:10 dilutions for total reaction volumes of 100 μL. When necessary, NF-ddH2O was used to complete reaction volumes to 100 μL. Bacterial cultures were prepared as before and resuspended in Luria Bertani (LB, Miller Formulation) to a final OD600nm of ~ 0.117. Cultures were mixed 1:1 and transferred to RNAse-free microcentrifuge tubes in 90 μL aliquots and supplemented with either cecal small RNA or NF-ddH2O and briefly vortexed to homogenize suspensions. The conjugation mixtures were incubated at the bacterial optimal growth temperature of 37 °C, standing, for 1 h, then serially diluted and plated on selective MacConkey agar plates as before. One-hour incubation was selected in order to prevent the contribution of transconjugant replication on the calculation of conjugation efficiency and to limit bacterial outgrowth of the finite amount of supplemented small RNA40. All work with small RNA extracts was performed with stringent care taken to prevent RNAse contamination, including UV sterilizing tools, decontamination of surfaces using RNAse-ZAP, and preparation of DEPC-treated chemicals and reagents.
In silico RNA hybridization
To identify hypothetical interactions between chicken host miRNAs and the plasmid-encoded tra operon genes of pAPEC-O2-211A-ColV, an in silico analysis was performed. The complete mature miRNA database for chickens (Gallus gallus), were obtained from miRBase (https://www.mirbase.org/summary.shtml?org=gga). The complete CDS record for the whole genome (GCF_001021635.2) and each tra gene from plasmid pAPEC-O2-211A-ColV (NZ_CP030791.1) were obtained from NCBI. The entire mature Gallus gallus miRNA database was used as the input query and either the whole genome or the pAPEC-O2-211A-ColV plasmid tra CDS were used as the target sequence set. The RNAHybrid RNA target hybridization software was used (options: -m 100000 -c -p 0.05), to generate theoretical hybridizations between target sequences and all Gallus gallus mature miRNA sequences71. RNAHybrid outputs the MFE of each theoretical binding and bindings with a significance value (p) less than 0.05 were considered significant theoretical bindings for further investigations. A select subset of miRNA queries with potential tra gene hybridizations to either the region directly ahead of a CDS, within a CDS, or to the mRNA gene transcript were obtained as mirVana® miRNA mimics (Cat. #MH10415, Thermo Fisher, Waltham, MA, USA) (Tables 2 and S2). Additional miRNA candidates were identified for testing via primary literature search for miRNAs that are shown to have effects in the gut and those targeting tra operon genes (Table S2).
In vitro miRNA mimic conjugations
Bacterial cultures were prepared as previously described and resuspended in LB (Miller formulation) to a final OD600nm of ~0.117. Cultures were mixed 1:1 and transferred to individual RNAse-free 1.5 mL microcentrifuge tubes in 180 μL aliquots. To each conjugation aliquot, either NF-ddH2O or NF-ddH2O containing 50 nM miRNA mimics were added (5 nM final mimic) (Table 2). Conjugation mixtures were incubated at the bacterial optimal growth conditions of 37 °C for 1 h, then vortex homogenized and serially diluted and plated on selective MacConkey agar plates as described previously. All work with RNA mimics was performed with stringent care taken to prevent RNAse contamination, including UV sterilizing tools, decontamination of surfaces using RNAse-ZAP, and preparation of DEPC-treated chemicals and reagents.
Relative miRNA specific reverse transcription qPCR
The relative abundance of the miRNA mimic gga-miR-183 between heritage genetic cecal small RNA extracts was determined using a miRNA-specific reverse transcription and qPCR approach40. Briefly, the extracted small RNAs were reverse transcribed using the TaqMan™ MicroRNA Reverse Transcription Kit (Cat. #4366596, Applied Biosystems, Waltham, MA, USA) following the manufacturer’s instructions. The resulting miRNA cDNAs were used as the template for qPCR on a QuantStudio 3 using the TaqMan™ Universal Master Mix II, with UNG (Cat. # 4440042, Waltham, MA, USA) according to the manufacturer’s instructions. The two-step “fast” qPCR cycle was used, consisting of an initial denaturation phase (20 s, 95 °C), 40 cycles of amplification phase (1 s, 95 °C; 20 s, 60 °C), and a continuous melt phase (1 s, 95 °C; 20 s, 60 °C; 1 s, 95 °C with a ramp speed of 0.1 °C/s). Fluorescence was measured during the combined annealing and extension phase for amplification and the temperature ramp in the melt curve phase.
Transcribed cDNA was evaluated for gga-miR-183 expression using a miR-183 specific TaqMan miRNA Assay (Cat. #4427975, Applied Biosystems, Waltham, MA, USA) according to the manufacturer’s instructions. The miR-451a housekeeping miRNA was used as an internal reference control for normalization between miRNA libraries, and the miR-451a-specific TaqMan miRNA Assay (Cat. # A25576, Applied Biosystems, Waltham, MA, USA) was used according to the manufacturer’s instructions. The geometric mean of the gga-miR-451 was used as the reference index for each sample. The 2−ΔΔCT method for relative gene expression was used, and statistics were conducted on the log10-transformed 2−ΔΔCT values to normalize sample distribution and prevent skewness72.
Quantitative RT-qPCR assays for detecting changes in plasmid replication and transfer genes expression
Primers for rep and tra genes expression analysis were prepared previously40. For the RT-qPCR reactions, cell pellets of in vitro conjugation with and without gga-miR-183 supplementation were harvested by centrifugation at 1 h of incubation and stored at −80 °C until processing. Conjugation reactions used in RNA extraction and RT-qPCR were conducted in triplicate, and each triplicate was measured in duplicate. Pellets were thawed by the addition of room-temperature RNAzol RT reagent, and total RNA was extracted following the manufacturer’s instructions (Cat. RN190, Molecular Research Center, Cincinnati, OH, USA). Genomic DNA was removed from RNA by RNase-free DNase I treatment following the manufacturer’s recommendations (Cat #: EN0521, Thermo Fisher, Waltham, MA, USA). RNA purity and concentration were evaluated using a NanoDrop™ Lite spectrophotometer (Thermo Fisher, Waltham, MA, USA), according to the manufacturer’s instructions. DNA-free RNA was then converted to cDNA via a High-Capacity cDNA Reverse Transcription kit (Cat #: 4368813, Thermo Fisher, Waltham, MA) according to the manufacturer instructions.
The resulting cDNA was used as the template for qPCR on a QuantStudio 3 using the PowerTrack™ SYBR Green qPCR Master Mix according to the manufacturer’s instructions (Cat #: A46109, Applied Biosystems, Waltham, MA). The two-step “fast” qPCR cycle was used, consisting of an initial denaturation phase (20 s, 95 °C), 40 cycles of amplification phase (1 s, 95 °C; 20 s, 60 °C), and a continuous melt phase (1 s, 95 °C; 20 s, 60 °C; 1 s, 95 °C with a ramp speed of 0.1 °C/s). Fluorescence was measured during the combined annealing and extension phases for amplification and the temperature ramp in the melt curve phase. The qPCR primers used in this study are described in Table 3 and were designed previously40. The geometric mean of the 16S rRNA reference gene for each sample was used as the reference index. The 2−ΔΔCT method for relative gene expression was used, and statistics were conducted on the Log10-transformed 2−ΔΔCT values to normalize sample distribution and prevent skewness72.
Statistics and reproducibility
All statistical analysis and graphing were performed using the GraphPad Prism software suite (version 6.0.0). Comparisons between treated and untreated conditions were performed using one-way ANOVA with post-hoc Bonferroni correction for multiple comparisons with 3 degrees of freedom, p values at or below 0.05 were considered significant during experimental analysis. For miRNA mimic conjugations, all miRNA were compared to their NF-ddH2O controls. For relative qPCR, the Log10-transformed 2−ΔΔCT was used to prevent skewness associated with log fold change expression.
Ethics statement
Animal studies were conducted using protocols approved by Iowa State University Institutional Animal Care and Use Committee, Log numbers IACUC-24-009 (MM) and IACUC-19-287 (SL).
Data availability
Data are provided within the manuscript or Supplementary Material. Raw data generated over the course of the study are available upon request from the corresponding author.
References
Dolejska, M. & Papagiannitsis, C. C. Plasmid-mediated resistance is going wild. Plasmid 99, 99–111 (2018).
Robicsek, A., Jacoby, G. A. & Hooper, D. C. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6, 629–640 (2006).
Nordmann, P. & Poirel, L. Plasmid-mediated colistin resistance: an additional antibiotic resistance menace. Clin. Microbiol. Infect. 22, 398–400 (2016).
Penders, J., Stobberingh, E. E., Savelkoul, P. H. M. & Wolffs, P. F. G. The human microbiome as a reservoir of antimicrobial resistance. Front. Microbiol. 4, 87 (2013).
Sommer, M. O. A., Dantas, G. & Church, G. M. Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325, 1128–1131 (2009).
Rolain, J.-M. Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front. Microbiol. 4, 173 (2013).
Liu, L. et al. The human microbiome: a hot spot of microbial horizontal gene transfer. Genomics 100, 265–270 (2012).
Salyers, A., Gupta, A. & Wang, Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12, 412–416 (2004).
Zeng, X. & Lin, J. Factors influencing horizontal gene transfer in the intestine. Anim. Health Res. Rev. 18, 153–159 (2017).
Shterzer, N. & Mizrahi, I. The animal gut as a melting pot for horizontal gene transfer. Can. J. Microbiol. 61, 603–605 (2015).
Jia, Y. et al. A broad-spectrum horizontal transfer inhibitor prevents transmission of plasmids carrying multiple antibiotic resistance genes. Transbound. Emerg. Dis. 2024, 7063673 (2024).
Fernandez-Lopez, R. et al. Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology 151, 3517–3526 (2005).
Palencia-G ndara, C. et al. Conjugation inhibitors effectively prevent plasmid transmission in natural environments. mBio 12, 1–5 (2021).
Getino, M. et al. Tanzawaic acids, a chemically novel set of bacterial conjugation inhibitors. PLoS ONE 11, e0148098 (2016).
Oyedemi, B. O. M. et al. Novel R-plasmid conjugal transfer inhibitory and antibacterial activities of phenolic compounds from Mallotus philippensis (Lam.) Mull. Arg. J. Glob. Antimicrob. Resist. 5, 15–21 (2016).
Cabezn, E., de la Cruz, F. & Arechaga, I. Conjugation inhibitors and their potential use to prevent dissemination of antibiotic resistance genes in bacteria. Front. Microbiol. 8, 1–7 (2017).
Ott, L. C. & Mellata, M. Short chain fatty acids inhibit bacterial plasmid transfer through conjugation in vitro and in ex vivo chicken tissue explants. Front. Microbiol. 15, 1414401 (2024).
Ott, L. C. & Mellata, M. Models for gut-mediated horizontal gene transfer by bacterial plasmid conjugation. Front. Microbiol. 13, 2371 (2022).
Ott, L. C., Stromberg, Z. R., Redweik, G. A. J., Wannemuehler, M. J. & Mellata, M. Mouse genetic background affects transfer of an antibiotic resistance plasmid in the gastrointestinal tract. mSphere 5, 1–13 (2020).
Ott, L. C., Engelken, M., Scott, S. M., McNeill, E. M. & Mellata, M. Drosophila model for gut-mediated horizontal transfer of narrow- and broad-host-range plasmids. mSphere 6, 1-11 (2021).
Mobasseri, G., JuTeh, C. S., Ooi, P. T. & Thong, K. L. The emergence of colistin-resistant Klebsiella pneumoniae strains from swine in Malaysia. J. Glob. Antimicrob. Resist. 7, 227–232 (2019).
Moawad, A. A. et al. Antimicrobial resistance in Enterobacteriaceae from healthy broilers in Egypt: emergence of colistin-resistant and extended-spectrum β-lactamase-producing Escherichia coli. Gut Pathog. 10, 1–12 (2018).
Liu, Y.-Y. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168 (2016).
Oppegaard, H., Steinum, T. M. & Wasteson, Y. Horizontal transfer of a multi-drug resistance plasmid between coliform bacteria of human and bovine origin in a farm environment. Appl. Environ. Microbiol. 67, 3732–3734 (2001).
Casadess, J. The virulence plasmids of Salmonella. Int. Microbiol. 2, 177–184 (1999).
Nielsen, D. W. et al. Complete genome sequence of avian pathogenic Escherichia coli strain APEC O2-211. Microbiol. Resour. Announc 7, 1–2 (2018).
Smith, H. W. The transfer of antibiotic resistance between strains of enterobacteria in chicken, calves and pigs. J. Med. Microbiol. 3, 165–180 (1970).
Tonu, N. S. et al. Pathological study on colibacillosis in chickens and detection of Escherichia coli By PCR. Bangladesh J. Vet. Med. 9, 17–25 (2012).
Jochum, J. M., Redweik, G. A. J., Ott, L. C. & Mellata, M. Bacteria broadly-resistant to last resort antibiotics detected in commercial chicken farms. Microorganisms 9, 1–16 (2021).
Mellata, M., Touchman, J. W. & Curtiss, R. Full sequence and comparative analysis of the plasmid pAPEC-1 of avian pathogenic E. coli x7122 (O78:K80:H9). PLoS ONE 4, e4232 (2009).
Mellata, M. Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog. Dis. 10, 916–932 (2013).
Mellata, M., Johnson, J. R. & Curtiss, R. Escherichia coli isolates from commercial chicken meat and eggs cause sepsis, meningitis and urinary tract infection in rodent models of human infections. Zoonoses Public Health 65, 103–113 (2018).
Lerner, A., Matthias, T. & Aminov, R. Potential effects of horizontal gene exchange in the human gut. Front. Immunol. 8, 27 (2017).
Stecher, B. et al. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. USA 109, 1269–1274 (2012).
Liu, W. et al. Factors and mechanisms influencing conjugation in vivo in the gastrointestinal tract environment: a review. Int. J. Mol. Sci. 24, 5919 (2023).
Zhou, X., Li, X. & Wu, M. miRNAs reshape immunity and inflammatory responses in bacterial infection. Signal Transduct. Target. Ther. 3, 1–13 (2018).
Liu, S. et al. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 19, 32–43 (2016).
Redweik, G. A. J., Horak, M. K., Hoven, R., Ott, L. & Mellata, M. Evaluation of live bacterial prophylactics to decrease IncF plasmid transfer and association with intestinal small RNAs. Front. Microbiol. 11, 1–10 (2021).
Stromberg, Z. R., Masonbrink, R. E. & Mellata, M. Transcriptomic analysis of shiga toxin-producing Escherichia coli during initial contact with cattle colonic explants. Microorganisms 8, 1–10 (2020).
Ott, L., Smith, C. & Mellata, M. Dietary zinc supplementation inhibits bacterial plasmid conjugation in vitro by regulating plasmid replication (rep) and transfer (tra) genes. Appl. Environ. Microbiol. https://doi.org/10.1128/AEM.01480-24 (2024).
Carlet, J. The gut is the epicentre of antibiotic resistance. Antimicrob. Resist. Infect. Control 1, 39 (2012).
Hasenstein, J. R., Zhang, G. & Lamont, S. J. Analyses of five gallinacin genes and the Salmonella enterica serovar enteritidis response in poultry. Infect. Immun. 74, 3375–3380 (2006).
Monson, M. S. & Lamont, S. J. Genetic resistance to avian pathogenic Escherichia coli (APEC): current status and opportunities. Avian Pathol. 50, 392–401 (2021).
Gul, H. et al. Genetic resilience in chickens against bacterial, viral and protozoal pathogens. Front. Vet. Sci. 9, 1032983 (2022).
Kaiser, M., Kaufman, J. & Lamont, S. J. Different MHC class I cell surface expression levels in diverse chicken lines, associations with B blood group, and proposed relationship to antigen-binding repertoire. Poult. Sci. 104, 104569 (2025).
Antão, E. M. et al. The chicken as a natural model for extraintestinal infections caused by avian pathogenic Escherichia coli (APEC). Microb. Pathog. 45, 361–369 (2008).
Borodin, А. М. et al. Chickens productivity selection affects immune system genes. Vavilovskii Zhurnal Genet Selektsii 24, 755–760 (2020).
Fleming, D. S. et al. Single nucleotide variant discovery of highly inbred Leghorn and Fayoumi chicken breeds using pooled whole genome resequencing data reveals insights into phenotype differences. BMC Genomics 17, 812 (2016).
Wang, Y. et al. Physiological responses to heat stress in two genetically distinct chicken inbred lines. Poult. Sci. 97, 770–780 (2018).
Zhang, S. et al. Small RNA GadY in Escherichia coli enhances conjugation system of IncP-1 by targeting SdiA. Front. Cell. Infect Microbiol. 14, 1–13 (2024).
Dambal, S., Shah, M., Mihelich, B. & Nonn, L. The microRNA-183 cluster: the family that plays together stays together. Nucleic Acids Res. 43, 7173–7188 (2015).
Li, S. et al. The miR-183 cluster: biogenesis, functions, and cell communication via exosomes in cancer. Cells 12, 1315 (2023).
Han, H., Zhou, S., Chen, G., Lu, Y. & Lin, H. ABAT targeted by miR-183-5p regulates cell functions in liver cancer. Int. J. Biochem. Cell Biol. 141, 106116 (2021).
Qi, C., Liu, L., Wang, J. & Jin, Y. Up-regulation of microRNA-183 reduces FOXO1 expression in gastric cancer patients with Helicobacter pylori infection. Histol. Histopathol. 38, 1349–1357 (2023).
Li, Z. B., Li, Z. Z., Li, L., Chu, H. T. & Jia, M. MiR-21 and miR-183 can simultaneously target SOCS6 and modulate growth and invasion of hepatocellular carcinoma (HCC) cells. Eur. Rev. Med. Pharmacol. Sci. 19, 3208–3217 (2015).
Gupta, N. et al. The miR-183/96/182 cluster regulates sensory innervation, resident myeloid cells and functions of the cornea through cell type-specific target genes. Sci. Rep. 14, 1–23 (2024).
Garcillán-Barcia, M. P. & de la Cruz, F. Why is entry exclusion an essential feature of conjugative plasmids?. Plasmid 60, 1–18 (2008).
Virolle, C., Goldlust, K., Djermoun, S., Bigot, S. & Lesterlin, C. Plasmid transfer by conjugation in gram-negative bacteria: from the cellular to the community level. Genes 11, 1239 (2020).
O’Brien, J., Hayder, H., Zayed, Y. & Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 9, 1–12 (2018).
Felden, B. & Augagneur, Y. Diversity and versatility in small RNA-mediated regulation in bacterial pathogens. Front. Microbiol. 12, 2273 (2021).
MacFarlane, L.-A. & Murphy, P. R. MicroRNA: biogenesis, function and role in cancer. Curr. Genomics 11, 537–561 (2010).
Menendez-Gil, P. & Toledo-Arana, A. Bacterial 3′UTRs: a useful resource in post-transcriptional regulation. Front. Mol. Biosci. 7, 617633 (2020).
Di Ruscio, A. & de Franciscis, V. Minding the gap: unlocking the therapeutic potential of aptamers and making up for lost time. Mol. Ther. Nucleic Acids 29, 384–386 (2022).
Ni, X., Castanares, M., Mukherjee, A. & Lupold, S. E. Nucleic acid aptamers: clinical applications and promising new horizons. Curr. Med. Chem. 18, 4206 (2011).
Nelson, W. C., Howard, M. T., Sherman, J. A. & Matson, S. W. The traY gene product and integration host factor stimulate Escherichia coli DNA helicase I-catalyzed nicking at the F plasmid oriT(*). J. Biol. Chem. 270, 28374–28380 (1995).
Li, Z., Hiasa, H., Kumar, U. & DiGate, R. J. The traE gene of plasmid RP4 encodes a homologue of Escherichia coli DNA Topoisomerase III*. J. Biol. Chem. 272, 19582–19587 (1997).
Achtman, M., Kennedy, N. & Skurray, R. Cell-cell interactions in conjugating Escherichia coli: role of traT protein in surface exclusion. Proc. Natl. Acad. Sci. USA 74, 5104–5108 (1977).
Audette, G. F., Manchak, J., Beatty, P., Klimke, W. A. & Frost, L. S. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology 153, 442–451 (2007).
Rasko, D. A. et al. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190, 6881 (2008).
Zhou, H. & Lamont, S. J. Genetic characterization of biodiversity in highly inbred chicken lines by microsatellite markers. Anim. Genet. 30, 256–264 (1999).
Rehmsmeier, M., Steffen, P., Höchsmann, M. & Giegerich, R. Fast and effective prediction of microRNA/target duplexes. RNA 10, 1507–1517 (2004).
Redweik, G. A. J., Kogut, M. H., Arsenault, R. J., Lyte, M. & Mellata, M. Reserpine improves Enterobacteriaceae resistance in chicken intestine via neuro-immunometabolic signaling and MEK1/2 activation. Commun. Biol 4, 1–11 (2021).
Acknowledgements
We thank Michael G Kaiser for his assistance with heritage birds. This work was supported by project award no. 20236701539078, from the U.S. Department of Agriculture’s National Institute of Food and Agriculture and the USDA Hatch projects IOW05700 and IOW04202 to M.M. and the USDA Animal Health project IOW05620 to S.J.L.
Author information
Authors and Affiliations
Contributions
L.C.O. and M.M. designed experiments; L.C.O. performed *in silico* analyses; C.S. and L.C.O. performed *in vitro* experiments and M.M. helped processing animals; M.M. contributed new reagents/analytic tools; S.J.L. provided heritage birds; C.S. and L.C.O. analyzed data; M.M. supervised the research; and C.S., L.C.O., and M.M. wrote the paper with input from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Skow, C., Ott, L.C., Lamont, S.J. et al. Host genetics regulate bacterial plasmid conjugation differentially in avian ceca explant co-culture through secretory miRNA. npj Antimicrob Resist 3, 95 (2025). https://doi.org/10.1038/s44259-025-00163-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44259-025-00163-7