Abstract
Background
Head and neck squamous cell carcinomas (HNSCCs) are heterogeneous in terms of origin and aetiology. In addition, there is uncertainty about the genetic evolution from initial diagnosis to recurrence after primary treatments and further disease progression following systemic treatment. Changes in the genetic profile have implications on the selection of appropriate treatments for patients, especially in the era of targeted therapies and immunotherapies.
Methods
We analysed a cohort of nine HNSCC patients with metachronous recurrence. All patients had paired primary and recurrent samples suitable for whole-exome sequencing, while transcriptomic data from seven patients could be analysed (multiple recurrent samples collected at different time points were available for three patients).
Results
At the genomic level, the recurrences shared a fraction of the somatic single nucleotide variants (SNVs) with the index primary tumours, but they also acquired many additional mutations, while losing only a few others. A similar behaviour was also observed when examining the changes of mutational signatures between primary and recurrent samples. Overall, recurrences appeared thus more genetically diverse than the respective primary tumours. The transcriptomic analysis showed that recurrent samples had lower immune cell presence, which was also confirmed by the multiplex immunofluorescence (IF) histology assays performed on the PhenoCycler platform. Several genes related to immune response were significantly downregulated compared to the primary samples.
Conclusions
Our results underline the importance of analysing multiple samples per patient to obtain a more complete picture of the patient’s tumour and advocate a re-biopsy in the event of recurrence and treatment failure, in order to select the most appropriate therapeutic strategy.
Similar content being viewed by others
Introduction
Head and neck squamous cell carcinoma (HNSCC) consists of a group of heterogeneous diseases arising from different subsites including paranasal sinuses, oropharynx, oral cavity, larynx and hypopharynx [1, 2]. Although these cancers tend to be associated with smoking and alcohol intake, in recent years incidence of oropharyngeal cancer linked to human papillomavirus (HPV) infection has increased noticeably [3]. Compared to HPV-negative disease, patients with HPV-positive oropharyngeal squamous cell carcinoma (SCC) have better tumour response to radiotherapy and/or chemotherapy, as well as an improved overall survival [3, 4].
Although the risk is lower for HPV-associated tumours [5], a significant number of HNSCC patients presents with recurrence or metastasis (R/M) following curative treatments and their prognosis is poor (less than one year median overall survival with palliative chemotherapy) [6]. The first-line treatment for HNSCC patients was previously the EXTREME regimen, consisting of platinum and 5-Fluoruracil (5FU) chemotherapy with cetuximab (anti-EGFR antibody) [6]. Nivolumab (anti-PD-1 antibody) was approved as a second-line treatment after failure of platinum-based chemotherapy [7]. The KEYNOTE-048 study showed that pembrolizumab (anti-PD-1 antibody) monotherapy improved overall survival (OS) compared to the EXTREME regimen in the PD-L1 positive populations (in both PD-L1 combined positive score (CPS) ≥ 20 and ≥1% subgroups) [8], leading to its approval as first-line treatment for recurrent or metastatic HNSCC in multiple countries. Furthermore, pembrolizumab with cisplatin and 5FU chemotherapy improved OS compared to the EXTREME regimen in the total population, as well as in the PD-L1 positive population (CPS ≥ 20 or ≥1%) [8].
Unfortunately, only a small number of R/M HNSCC patients respond or benefit from anti-PD-1 antibodies, with HPV-positive patients seemingly benefitting more than HPV-negative patients [7, 8]. The KEYNOTE-028 study has shown that a T cell-inflamed gene expression profile, PD-L1 expression and tumour mutational burden (TMB) can predict the response to pembrolizumab in multiple tumour types [9]. In a pivotal study, it was shown that high nonsynonymous mutational burden, smoking associated mutational signature, high neoantigen burden and DNA repair pathway mutations were associated with efficacy and benefit of anti-PD-1 antibodies [10]. Various articles have investigated the mechanisms conferring resistance to PD-1 blockade, which include neoantigen heterogeneity, upregulation of additional immune checkpoints like LAG3 and TIM3, JAK1/2 mutations and B2M truncating mutations [10,11,12,13,14], although much work is yet to be done.
The genetic alterations underlying recurrence in HNSCC are still poorly understood, hindering the development of effective therapies, including the optimal combination of immunotherapy and other treatments. The main objective of this study is to assess how the genetic and immune landscapes change upon recurrence and under the pressure of treatment in individual patients. This may help to inform the optimal use and sequence of immunotherapy treatment in combination with standard treatment in recurrent HNSCC patients. To achieve this, we performed a comprehensive genomic and transcriptomic analysis of matched primary and recurrent carcinomas from individual patients of a small HNSCC cohort, for whom detailed patient history and treatment information were available (Table 1 and Fig. S1).
Materials and methods
Patient samples and HPV typing
A cohort of nine HNSCC patients (consented through an accelerated programme, Ethics Approval reference 14/YH/1101) from the University Hospitals Birmingham (UHB) NHS trust was included in this study. These patients underwent routine treatments and subsequently presented with metachronous recurrence (Table 1). Eight patients had carcinoma arising from the oral cavity. One additional patient had non-oral cavity SCC (hypopharynx).
HPV status was initially obtained through the immunohistochemical staining of the surrogate marker p16, which was performed as part of the routine standard of care by UHB on the primary tumours of six out of nine patients. The p16 status of patient P08 primary tumour and recurrence was instead assessed by a certified pathologist in the context of this study. To confirm these results and uncover the HPV status of P03 and P06 (not tested with p16), HPV genotyping was also carried out by PCR on DNA from the primary tumour and recurrent samples (if available after whole-exome sequencing), using HPV general primers GP5+ and GP6+ following the protocol outlined in Smeets et al. [15]. An additional genomic control locus within the HPRT1 gene was amplified (forward primer: TTTCTTGACCCAAGTCATCT and reverse primer TCATATAAGCAGCACTGACG). Water was used as negative control and a synthethic HPV p16 fragment as positive control sample. Tested samples and detailed assay results are shown in Table S1, while HPV status is summarized per patient in Table 1.
Primary and recurrent samples were obtained from either biopsy, resection or both (Table 2) and were formalin-fixed paraffin-embedded (FFPE). Only samples from locoregional recurrences were analysed in this study. One patient (P05) presented with three primary tumours: in this study only the primary tumour giving rise to the recurrence was analysed, which was previously described elsewhere [16]. The same sample identifier appearing in the previous publication (i.e. “T2”) was used for this study and the detailed information on this case is not included here since the previous publication covered this case in detail, including the evidence to support the recurrence originated from primary T2 [16]. For three patients, multiple recurrent samples from different time points were available. Tumour area and tumour nuclei of samples included in the analyses were provided by a board-certified pathologist and are shown in Table S1, while detailed patient histories are shown in Fig. S1.
Next-generation sequencing data
DNA was extracted from the primary and recurrent tumour samples from nine HNSCC patients using the Maxwell FFPE DNA kit according to the manufacturer’s instructions. Matched DNA from buffy coats, to be used as germline control, was extracted using the Qiagen Blood and Tissue Kit. Replicate exome capture libraries were prepared from the tumour and normal DNA with the SureSelect Human All Exon V6 kit (Agilent). Libraries were subjected to paired-end sequencing 2 × 50 bp on an Illumina NovaSeq 6000.
RNA extraction from the tumour samples was performed with the FFPE Clear Kit (AmpTech). One library per sample was prepared using 130–814 ng RNA and the Illumina TruSeq RNA Library Prep Kit V2. The libraries were subsequently sequenced paired-end 2 × 50 bp on an Illumina HiSeq 2500.
Bioinformatic analyses
Whole-exome sequencing data
Reads obtained from whole-exome sequencing (WES) of tumour and normal samples were processed as previously described [16]. Briefly, reads were aligned to the hg19 human reference genome with BWA 0.7.10 [17] and duplicates were flagged with MarkDuplicates from the Picard Tools suite 1.110 (http://broadinstitute.github.io/picard/). Somatic single nucleotide variants (SNVs) were identified from the tumour replicate libraries with an established mutation detection process [18], which in addition to a list of SNVs, outputs a separate list of candidate mutation sites.
The overlap of somatic mutations among tumour samples from the same patient was evaluated to quantify the fraction of SNVs unique to primary, recurrences or shared between them, following the procedure outlined in Bresadola et al. [16]. In summary, for each patient we obtained a list of SNVs, by performing the union of the SNV lists of each tumour sample. If a SNV was not present in all tumour samples, we subsequently verified its presence in the list of candidate mutation sites of the remaining tumour samples of the patient. In case the SNV was not found in this list either, we inspected the reads aligning at the mutation position. We required the position to be covered by ≥50 reads: if the variant allele frequency (VAF) was <0.01, the SNV was considered absent in the analyzed tumour sample, while if the VAF was >0.02, the SNV was considered present. If these conditions were not satisfied, the SNV was excluded from the analysis.
For the generation of Fig. 1, in patients with multiple recurrent samples, a SNV was considered shared if it was detected in the primary tumour and in at least one of the recurrent samples.
COSMIC mutational signatures (v2, March 2015) were calculated from the final list of SNVs using the R package YAPSA [19].
RNA-seq data
Transcriptome data was obtained from the tumour samples and processed to determine RPKM gene expression values as previously described [16]. A differential expression analysis between primary and recurrent samples was carried out with the R package DESeq2 1.22.2 [20] to identify genes expressed at significantly different levels in these two sample groups. Besides the group, information about patient provenance of the samples was included in the design formula of DESeq2 as additional covariate. For each patient, only a primary tumour sample and a recurrent sample were included, to prevent patients with multiple recurrent samples to bias the analysis. When more than one recurrent sample was available, the one with higher tumour content was chosen (R1 for P01, R3 for P06 and P09). Patient P02 was not included in the analyses, since RNA-seq data could not be obtained from the recurrent sample, while patient P03 was excluded because of the very low tumour content of the primary sample (Table S1). An enrichment analysis was performed with GOrilla [21] on the set of significant genes, to detect potentially over-represented gene ontology (GO) terms associated with specific biological processes.
RNA-seq data was also analysed with quanTIseq [22], a bioinformatic tool performing deconvolution of the transcriptomic data to estimate the fractions of ten immune cell types in the samples, besides the fraction of other uncharacterised cells. Immune cell types quantified by quanTIseq are B cells, classically activated macrophages (M1), alternatively activated macrophages (M2), monocytes, neutrophils, natural killer (NK) cells, conventional CD4+ T cells, CD8+ T cells, regulatory CD4+ T (Treg) cells and dendritic cells (DC).
Immunohistochemistry and immunofluorescence assays
FFPE tissue sections (3 μm) were stained in the Leica BOND Rx autostainer using a rabbit anti-human PD-L1 antibody (1:400, Cell Signaling, 13684S) and the staining was detected using BOND Polymer Refined Detection Kit (Leica, DS9800). Bright-field images were taken using Leica Aperio AP2 slide scanner. The combined pathology score (CPS) sums up both tumour and immune cell PD-L1 positivity, which is used as a criteria to select patients for pembrolizumab treatment in the UK since March 2020. As the evaluation of PD-L1 positivity was performed by a board-certified pathologist for this retrospective cohort from the pre-approval of pembrolizumab era and not in the context of selecting patients for pembrolizumab, we avoided giving CPS scoring in our study. Instead, we interpreted the PD-L1 stainings as positive or negative based on clear membrane staining of tumor cells as well as immune cells.
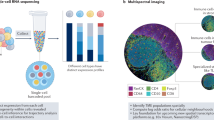
The PhenoCycler®-Fusion system (Akoya Biosciences) was used for spatial phenotyping of the tumour microenvironment. Sample preparation and tissue staining were performed according to the PhenoCycler Fusion user manual (Akoya Biosciences, PhenoImager Fusion SW version: 1.0.5). Briefly, 3 μm FFPE tissue sections were pre-processed by deparaffinization, dewaxing/rehydration, and heat-mediated antigen retrieval (AR9, Akoya Biosciences, AR900125ML). Tissues were then simultaneously stained with the entire barcoded-antibody panel (Table S2). After the staining, a flow cell (Akoya Biosciences, 240205) was placed on top of the slide.
For the reporter plate, unique and spectrally distinct reporters, complementary to the barcodes used in the antibody panel, were organized into groups of three and combined with DAPI (Nuclear Stain, Akoya Biosciences, 7000003). Each mixture was applied to the tissue during separate PhenoCycler cycles. The experimental protocol and reporter plate design were carried out using the PhenoCycler Experiment Designer software. The PhenoCycler run was fully automated and executed by the Controller software. PhenoCycler reporters were delivered to the tissue by the PhenoCycler instrument and detected using the Fusion slide scanner. The repetition of these cycles with different reporters allowed the visualization of the complete antibody panel on the same tissue area.
Histology image analysis and quantification
Image analysis and quantification was performed with Visiopharm Pathology Image Analysis software (Visiopharm). Follow-up analysis of the obtained data was performed using R. Tissue on whole slide scans was separated from the slide background by a low counterstain threshold. Within this tissue annotation, cells positive for CD8, CD20 or FoxP3 were identified by single feature classifiers. Measurements on tissue region and positive cells for area, object coordinates and marker intensity statistics for target markers and supporting markers (Ki67, CD3) were generated and exported.
Exported data was tidied post-hoc by removing false positive objects using supporting markers. CD8 positivity was confirmed by expression of CD3, while CD20 unspecific staining in tumour cells co-localized with unspecific and excessive Ki67 staining. FoxP3 signal was considered only if associated to CD3 positivity. Multiple cells merged together were assigned a weighted cell count taking into account the average size of the respective cell types. This weighted cell count was normalized against the whole tissue area to generate a cell count per area (mm2).
Results
Overview of the cohort genomic profiles
Next-generation sequencing experiments
WES experiments were conducted on all tumour and normal samples included in this study. On average, over samples and replicates, 231.91 million read pairs (sd 35.87) were obtained for the tumour samples and 260.70 million reads pairs (sd 44.45) for the matching normal samples (Table S3). The mean target coverage was 272.42 (sd 40.76) and 287.35 (sd 49.48) for tumour and normal samples, respectively, while the average fold enrichment was 38.07 (sd 2.50) and 35.06 (sd 1.62). The high sequencing depth exceeded the values commonly required in WES studies [23, 24].
The RNA-seq experiment was performed only on the tumour samples and yielded an average of 37.58 million read pairs (sd 8.31). Of these, 95.79% (sd 1.25%) aligned to the human reference genome hg19 and 66.18% (sd 7.77%) to transcript sequences (Table S3).
SNV overlap
The number of detected SNVs varied remarkably across patients (Fig. 1a). The tumours of patients P01 and P04 had more than 300 SNVs, while the lowest number of SNVs was found in patients P02 and P03, whose tumours had less than 50 SNVs when combining the mutations detected in both primary tumour and recurrence. The number of SNVs did not correlate with the number of tumour samples analysed per patient (Spearman’s rho 0.02, p-value 0.96). The mutational burden in primary tumours of smokers (P01, P05 and P09) was on average higher than in those of non-smokers (P03, P07 and P08 - 174 SNVs vs. 90 SNVs, respectively). However, this difference was not statistically significant given the very small sample size and the variability in the number of SNVs (sd 112 and 65, respectively; one-sided Wilcoxon test p-value = 0.35).
The percentage of SNVs shared by primaries and recurrences was also very variable, with a minimum of 18.93% in patient P04 and a maximum of 97.38% in P01 (average was 55.11% with sd 26.49%). Non-synonymous mutations represented 64.86% of SNVs on average across patients (sd 4.65%; Fig. 1b). The overlap among samples was very similar to that observed in the whole mutation sets (Fig. 1a and b), with a mean of 54.99% of non-synonymous SNVs shared between primary and recurrent samples (sd 27.85%).
Overall, recurrences shared a fraction of the SNVs with the respective index primary tumours, but they also acquired many additional mutations, while losing only a few others. On average, 103 (sd 90) SNVs were detected in each primary sample and 165 (sd 97) in each recurrence. This difference is highly significant when taking into account the patient provenance of the samples (paired two-sided t-test p-value = 0.01, performed after verifying homoscedasticity with a Bartlett’s test). Recurrent samples appeared thus to carry more genetic diversity compared to the primaries, although the picture could vary if different regions of the tumours were sequenced.
Mutational signatures
Mutational signatures reflect the processes operating in the tumours and they are useful for understanding the biological mechanisms leading to cancer [25]. We performed mutational signature analysis in the primary and recurrent tumours, but excluded patients P02 and P03 because the low number of mutations detected in these samples (Fig. 1) made the detection of mutational signatures less reliable. Signature 1, which is associated with deamination of 5-methylcytosine, was found in all analysed samples (Fig. 2). This signature is highly associated with ageing and was especially abundant in patient P01. Signature 10 was also detected with variable abundances in all samples analysed. This signature mainly results from altered activity of polymerase epsilon (POLE) and often determines the occurrence of huge numbers of somatic mutations, which however was not the case in these samples. Signature 7 (linked to UV light exposure) was found in all patients, although not in all tumour samples and with a particularly low frequency in P06. Multiple patients showed variable fractions of signatures related to APOBEC activity (signatures 2 and 13), which was previously shown to be associated with HPV activity [26]. The fraction of mutations assigned to these signatures was indeed most abundant in patient P01, the only patient with a confirmed HPV-positive tumour included in our study (Table 1 and S1), although the HPV-negative recurrent sample of patient P04 also showed an important fraction of these signatures. Patients P04, P05, P06 and P09 showed high fractions of signature 16, which is associated with HPV-negativity, as well as with smoking and alcohol consumption [27] (all tumours of these patients were tested or presumed HPV-negative, all were known smokers or ex-smokers). An inverse correlation between CDKN2A expression and frequency of signature 16 was previously reported [27]. Expression of this gene was lower in the tumour samples of these four patients compared to the rest, but the difference was not significant in the small cohort examined in this study (one-sided Wilcoxon test p-value = 0.09, Fig. S2). Interestingly, these patients showed the overall highest increase and change in mutations after recurrence, suggesting that HNSCC tumours driven by tobacco- and/or alcohol-induced mutations might be genetically less stable than tumours driven by HPV and APOBEC activity [27], although the size of the cohort does not allow to generalise confidently this observation.
“No change” indicates that the same signatures were detected in the primary tumours and in the recurrences. “Gain in R sample(s)” indicates that the recurrence(s) acquired additional signatures, not present in the primary tumours. Associations to known mechanisms are reported next to the Signature legend. MMR = mismatch repair, 5meC = 5-Methylcytosine. *Patients P01 and P07 were alive and well at the time of analysis.
In patients P01 and P08, the mutational signatures found in primary and recurrence were very similar across samples, also in terms of proportions, as expected by the high fraction of shared mutations (Fig. 2). In patient P04, the recurrent sample carried the same signatures present in the primary, but the relative proportions differed substantially. We especially observed an increase in signatures 2, 7 and 13 (associated with APOBEC activity and UV damage), while there was a reduction in signatures 6 (associated with DNA mismatch repair) and 16 (tobacco, alcohol). Mutations caused by UV damage could be the result of the location of R2 sample of patient P04 (skin), who also had the overall highest increase in mutational load in the recurrence.
In patients P05, P06, P07 and P09, recurrent samples acquired additional signatures with respect to the primary tumours (Fig. 2), a further indication of recurrences being more genetically diverse than primaries. These included signature 6 (acquired by recurrent samples of P09), signature 7 (P06), as well as signatures 2 (P05 and P06) and 13 (P07).
Analysis of patients with multiple recurrent samples
For three patients (P01, P06 and P09), two or more recurrent samples were sequenced. In this section, we report a detailed investigation of the changes in the genomic profiles of their primary and recurrent samples. The histories of these patients are shown in Fig. S1.
Patient P01
The samples available for this patient originated from the primary (T1) and two recurrent tumours (R1 and R2, Table 2). From WES data, a total of 305 SNVs were detected in primary and recurrences (Table S4). Among these, 297 SNVs were shared among T1, R1 and R2, including non-synonymous mutations in KRAS, TIGIT and NOTCH1 (Table S4). Three SNVs were found in T1, but not in R1 and R2 (Table S5): one of these was non-synonymous and affected PTCHD2, a gene known to be involved in tumorigenesis. Five SNVs were found only in R1 or in both R1 and R2: three of these were non-synonymous and affected CREBZF (only in R1), GALE and PCYT1A (in R1 and R2), all of which are known to play a role in tumorigenesis, cell proliferation and migration of cancer cells (Table S5).
Patient P06
In the primary (T1) and the three recurrent samples from this patient (R1, R2 and R3), 116 SNVs were identified: 63 SNVs were shared among all samples, 47 of which were non-synonymous mutations, including TP53 and ALK mutations (Table S6). Four SNVs were instead detected only in T1 and two of these were non-synonymous: one was found in RAB3GAP1, a gene regulating autophagy, the defects of which are associated with increased tumorigenesis (Table S7); the other SNV was found in CPEB2, which has a tumour suppressor role (Table S7).
All recurrent samples shared 22 SNVs that were not found in the primary tumour T1. Among these, 14 were non-synonymous mutations affecting the genes RUSC1, SELRC1, CAPN1, C14orf135, GNA15, GPATCH1 (two different amino acid substitutions), DYNC1I2, MFNG, CCDC127, HGSNAT, GDA, PRICKLE3 and TTC3P1 (Tables S6 and S7). Many of these genes have unknown significance in cancer, but GDA and GNA15 have been implicated in cancer progression (Table S7).
R1 was obtained from the neck dissection following left neck recurrence (no irradiation), whereas R2 and R3 were obtained after radiotherapy to the left side of the neck (Fig. S1). Five SNVs were found in R2 and R3, but not in R1. One of these was a non-synonymous mutation affecting the gene FADD, which codes for an adaptor protein forming the Death Inducing Signalling Complex (DISC) with the death receptor Fas and caspase-8. The DISC mediates the extrinsic apoptotic program, including apoptosis induced by radiation [28,29,30,31]. While FADD genomic amplifications are frequent in HNSCC [32], FADD somatic mutations are found only in approximately 0.4% of the tumour samples analysed in the TCGA PanCancer Atlas Studies [28].
In R3, which was obtained just before the patient died, we identified 22 unique SNVs. Among these, 17 were non-synonymous and found in genes TAF3, TTLL10, CDON, MAP1A, HERC1, CDH13, PLSCR3, CDC25B, GCKR, ALK, DHX57, ABLIM2, LOC389332, PCDHB18, ZNF467, LYNX1 and RBM12B (Table S6). Most of these genes have been linked to tumorigenesis and cancer progression (Table S7).
Although the primary tumour affected the right side of the neck and the recurrences were detected on the left side, the considerable number of SNVs shared among the samples indicates that the recurrence indeed originated from this primary tumour. This is also corroborated by the mutational signatures, which were very similar between primary and recurrent samples (Fig. 2). The mutational burden was however higher in the recurrences (especially in R3), such that in these samples two additional signatures were identified, as mentioned above.
Patient P09
For this patient, the primary tumour (T1) and two recurrent samples were analysed (R1 and R3; R2 was not used as this resected sample had very low tumour content and matched biopsy R1 - see Table 2). In the three samples, 156 SNVs were detected and 79 were shared among all samples (60 were non-synonymous, including ADAM11 and CLDN18), while 67 SNVs were found only in the recurrences (Table S8). Of the latter subset, 49 SNVs were non-synonymous including PDGFB, which encodes subunit B of the platelet-derived growth factor. Five SNVs were found only in the primary sample and not in the recurrences: three of these were non-synonymous and found in genes PFKM, WWP2 and XG (Table S8). PFKM and WWP2 have tumour suppressive function, while XG regulates cancer cell migration (Table S9). Two synonymous SNVs were found only in R1 and three non-synonymous ones only in R3. These were located in the genes SLCO4A1 (and the uncharacterised LOC100127888), PRR14L and SMARCA2, which have been linked to cancer progression (Table S9).
Transcriptomic analysis of primary and recurrent tumours
We performed a differential expression analysis to uncover genes consistently up- or downregulated in recurrences compared to primary tumours, including only one primary and one recurrent sample per patient. Overall, 243 genes were differentially expressed with |log2 fold change | > 1 and adjusted p-value < 0.05: 54 genes were upregulated and 189 downregulated in recurrent samples compared to primary samples (Fig. 3a). Figure 3b shows the expression in all available samples (not only those included in the differential expression analysis) of the 60 most significant genes (adjusted p-value < 0.005). As expected, the up- and downregulated genes formed two separate clusters in the heatmap. While the sample dendrogram separated overall primaries from recurrences, some exceptions were observed, since the recurrent tumours of patients P01 and P07 clustered together with the primary samples. Interestingly, these two patients were the only ones in the cohort being alive and well at the time of our analyses (Fig. S1). Moreover, the primary of patient P04 was on a distinct branch of the dendrogram, potentially because of its location in a different subsite (hypopharynx) with respect to the other primary tumours (oral cavity).
a Number of significantly up- and downregulated genes in recurrent samples, compared to primary tumours. b Heatmap of the RPKM expression, shown as gene-wise Z-scores, of the genes with the lowest p-values (< 0.005) in all samples of the cohort. Values of -1 and lower are shown with the same light yellow shade, values of 1 and larger with the same dark green shade. Side bar indicates primary tumours (blue) and recurrences (orange). c Gene ontology terms significantly over-represented in the list of downregulated genes. FDR = false discovery rate. * Patients P01 and P07 were alive and well at the time of analysis.
Among the most significant differentially expressed genes shown in Fig. 3b, the upregulated genes included MSLN, FSCN1, PPP1R14B, TMSB10, S100A10, EBNA1BP2, CA9, WNT7A, TGFBI, LAMC2, LRRC42, BBOX1, which have all been found to be over-expressed in tumours and/or involved in tumorigenesis, cell invasion and migration, progression or metastases of various cancers (Table S10a). EGLN3 (PHD3) is a negative regulator of NF-κB pathway and was upregulated in several recurrences of this cohort. Surprisingly, some genes associated with tumour suppression, such as PADI3, PRKCDBP, FEZ1, LCE1B, were found to be upregulated in the recurrent samples. On the other hand, the most significantly downregulated genes included MIR3064, NEU3, GCNT2, LAMC3, LRRC4B, RNF150 and NCALD (Table S10b), which have been either identified as tumour suppressor genes, or their loss of function has been associated with tumour growth or survival. A few genes with oncogenic function were also downregulated in recurrent samples compared to primary tumours, e.g. MIR650, SELE and KCNK5. Other downregulated genes were involved in immune function (BTN3A1, CXCL9, IGJ, POU2AF1, SLC17A9, FCRL5, FHL5; Table S10b) or implicated either in chemoresistance (SIDT1 and GSTM2) or radioresistance (BCAS1). Several genes with unknown role in cancer were also found to be downregulated (Table S10b).
Overrepresented gene ontologies
An enrichment analysis was performed on the set of differentially expressed genes to identify potentially overrepresented GO terms associated with a specific biological process. Among the genes upregulated in the recurrences, no significant overrepresentation was detected. However, in the set of downregulated genes, four GO terms had a false discovery rate (FDR) value < 0.05, namely “regulation of cell adhesion”, “immune response”, “adaptive immune response”, “immune system process” (Fig. 3c and Table S11).
Among the genes associated with these GO terms and downregulated in recurrences, CIITA is important to allow cell interaction with CD4+ T cells [33], PIK3CG promotes immune suppression during inflammation and tumour growth [34], SAMHD1 downregulates innate immune responses [35], ST6GAL1 regulates both T and B cell function [36] and SPN (CD43) regulates CD4+ T cell trafficking [37]. In addition, JAK3 regulates Toll-like receptor-mediated inflammatory cytokine production in innate immune cells [38] and ITK regulates T cell signalling [39]. ZBP1 functions as a central regulator of programmed cell death and inflammatory response [40], while INPP5D serves as an effector for other inhibitory receptors in various immune cell types [41].
Interestingly, the expression of several of these differentially expressed immune-related genes (marked by an asterisk in Table S11) did not decrease in the recurrence of patient P07 (patient history shown in Fig. S1). Their expression was on the contrary higher in R1 than in T1 (examples shown in Fig. S3). These genes contribute to initiation and regulation of the immune response, including T cell activation: CD27 is a co-stimulatory immune checkpoint molecule; CD6 regulates T cell response through the positive regulator SLP-76, which encodes a signal-transducing adaptor protein [42]; INPP5D (SHIP1) regulates the function of various immune cells including T cells, B cells and NK cells [41]; LY9 (SLAMF3) enhances CD4+ T cell sensitivity to IL-2 [43].
Immune cell detection in the tumour samples
In light of the results provided by the GO term enrichment analysis, the presence and proportions of different immune cells in the tumour samples were investigated with quanTIseq [22]. This tool allows the estimation of cell fractions from RNA-seq data, including B cells, classically activated macrophages (M1), alternatively activated macrophages (M2), monocytes, neutrophils, natural killer (NK) cells, conventional CD4+ T cells, CD8+ T cells, regulatory CD4+ T (Treg) cells and dendritic cells (DC).
In spite of patterns varying substantially across patients, immune cell presence was significantly higher in primary tumours than in recurrences (Fig. 4a) when comparing the same samples used in the differential expression analysis: the median percentage of immune cells was 27.95% in primaries (IQR 16.33) and 15.20% in recurrent samples (IQR 5.56; p-value = 0.016 from paired two-sided Wilcoxon test). This was especially evident for patients P04, P05, P06 (three out of the four patients where signature 16 - linked to tobacco and alcohol consumption and HPV-negative tumours - was detected) and P08. This observation was also confirmed by the multiplex immunofluorescence (IF) histology assays performed on the PhenoCycler platform detecting CD8+ T cells, B cells and Treg cells (Fig. 4b, S4–S7). The proportions of CD8+ T cells were generally higher in primary than in recurrent samples in all patients except in P07, who was alive and well at the time of the analyses, and in the R2 sample of P01 (Fig. 4a), a biopsy obtained while the patient was receiving treatment and showing a transient increase in tumour size, before the disease showed a good partial response to treatment. The results were supported by the IF staining (Fig. 4b, S4 and S5). Noticeably, these same recurrent samples of patients P01 and P07 did not show the dramatic contraction in the percentage of B cells which instead was observed in the remaining patients (Fig. 4a). This observation could be seen as bona fide presence of tertiary lymphoid structures (TLS) in these tumours, which have been put forward as good prognostic markers for cancer patients [44]. Indeed, we observed a TLS in the IF assay performed on the R2 sample of patient P01 (Fig. S8). In the case of B cells, some discrepancies between quanTIseq and PhenoCycler results are visible (Figs. S4 and S6). However, some dissimilarities are to be expected, considering that different layers of the FFPE tumour blocks were used for RNA extraction and IF assays. Moreover, quanTIseq estimates are based on gene expression data, while the antibodies used in the PhenoCycler stainings detect the corresponding protein. Similar to B cells, Treg cells were remarkably more abundant in primary tumours than in recurrent samples, determining a microenvironment which from an immunological perspective could have favoured the relapse of the tumour in these patients. As for other immune cell types, patient P07 showed the opposite pattern, i.e. a higher percentage of Treg cells in the recurrence than in the primary sample (Fig. 4a), which however did not prevent the patient from responding well to treatment. These observations were confirmed by the PhenoCycler IF quantifications in the vast majority of the patients (6 out of 7; Fig. 4b, S4 and S7).
a Percentages of immune cells present in the primary and recurrent samples, obtained with quanTIseq. b Phenocycler IF staining of samples T1 and R1 from patient P08 (see Figs. S5–S7 for images of the remaining patients). Whole-slide imaging at single-cell resolution of B cells (CD20) and T cells (CD3, CD8, FoxP3). Insets show representative 20x zoom-in images. Counterstaining: DAPI; scale bar: 250 µm. *Patients P01 and P07 were alive and well at the time of analysis. **Biopsy R2 was taken while patient was receiving treatment and responding to it.
To further characterise the immunoprofile of these tumour samples, we performed immunohistochemistry to detect the immune regulatory marker PD-L1 and evaluated the results for both tumour and immune cells (Table S12; the images of a representative patient are shown in Fig. S9). Eight out of the 18 stained samples were found to be positive when focusing on the tumour, while 14 were positive when examining the immune cells. In four patients (P01, P06, P08 and P09), PD-L1 status (positive vs negative) differed between primary tumour and recurrences, highlighting once more the high level of intertumour heterogeneity that can be observed within a single patient. More importantly, the positive PD-L1 status would have therapeutic implications, rendering the patients eligible for pembrolizumab as first-line treatment according to the standard of care in most countries.
Discussion
In this study, we examined the mutational landscape of matched primary and recurrence pairs from nine patients affected by head and neck squamous cell carcinoma (HNSCC). Overall, we observed that primary tumours and recurrences shared a fraction of the SNVs (55.11% on average; Fig. 1), although the proportion of shared SNVs varied remarkably among patients. This finding is in line with a previous study by Hedberg et al. [45], who performed whole-exome sequencing (WES) on a cohort of 8 HNSCC patients with paired primary and metachronous recurrent tumours. Their analysis highlighted that approximately 60% of SNVs identified in recurrent tumours were transmitted from the index primary tumour, while around 40% arose newly in the recurrences. As emerged from our study, the authors reported a high interpatient variability in the fraction of SNVs shared by primary and recurrent tumours. According to our investigation, recurrent samples had generally a higher tumour mutational burden (TMB) and were more genetically diverse than primaries, as supported by the occurrence of additional mutations and mutational signatures. This observation agrees with what was described by Weber et al. [46], who investigated a cohort of 38 matched HNSCC primary and recurrent samples and detected a higher TMB and an increased fraction of subclonal mutations in the recurrences [46]. This finding might be relevant for its implications on treatment strategies, since a positive correlation between TMB and response to immune checkpoint therapy was previously described [9, 10]. Higher TMB in recurrences was also reported by Schnidrig et al. [47], who hypothesised that the bottleneck linked to the origin of metastases (and local recurrences) might cause the SNVs unique to these samples to acquire a higher clonality in the metastatic site and therefore to be more easily detected by sequencing experiments. Another explanation suggested by the authors involves the effects of treatment on the TMB in recurrences, since it has been shown that both chemotherapy and radiotherapy can induce subclonal mutations [48]. Schnidrig et al. warned therefore against a systematic bias in TMB calculations from primary and recurrent samples [47], which should be evaluated carefully if this metric is used for patient stratification.
In the above-mentioned publication [45], Hedberg et al. [45] found C17orf104 and inositol 1,4,5-trisphosphate receptor, type 3 (ITPR3) to be exclusively mutated in two synchronous metastases, and discoidin domain receptor tyrosine kinase 2 (DDR2) to be exclusively mutated in two metachronous recurrences, but not in the respective primary tumours. None of these three genes were, however, mutated in the HNSCC samples investigated here, neither in the primary nor in the recurrent samples.
Our sample set allowed us to compare gene expression between primaries and recurrences from the same patients and to highlight >200 genes significantly up- or downregulated in one group. Generally, although several genes were differentially expressed between primaries and recurrences, heterogeneous expression levels can still be observed within the groups (Fig. 3b). Besides the pervasive interpatient and intratumour heterogeneity, this might also be explained by the variety of tissues analysed in the study, which made the uncovering of coherent patterns more difficult. Interestingly, the enrichment analysis carried out on the differentially expressed genes revealed that genes annotated with GO terms related to immune responses and processes were significantly overrepresented among the genes downregulated in recurrent samples. This finding might suggest the presence of immune escape mechanisms in the recurrent HNSCC tumours and motivated us to characterise the immune landscapes of the tumour samples. Among the studies previously conducted to characterise this aspect in HNSCC, Mandal et al. [49] examined 280 tumours profiled by The Cancer Genome Atlas (TCGA) and found that both HPV+ and HPV- HNSCC tumours are among the most highly immune-infiltrated cancer types [49]. However, a highly immunosuppressive environment determined by the unbalance between active and suppressive immune cell types was also reported [50]. A study by Chen et al. [51] on RNA-seq data from 522 primary samples from TCGA supports this view: the authors reported that approximately 40% of the HNSCC samples showed enriched inflammatory response, enhanced cytolytic activity and active interferon-γ signalling and could be thus assigned to the “immune class” [51]. However, within this class an “exhausted” subtype was identified, characterised by tumour-promoting signals (e.g. activated stroma, anti-inflammatory M2 macrophages) and by the ability to suppress the patient’s immune response. Such mechanisms could be in place also in our cohort, considering that in approximately half of the samples, tumour cells were PD-L1 positive and might have thus inhibited the activity of the immune system against the disease. When comparing matched primary and recurrent samples, Weber et al. [46] described a lower expression level of the gene signature related to tumour inflammation in the recurrences. In our study, we noticed lower proportions of CD8+ T cells and B cells in recurrent samples than in primary tumours, and generally a lower immune cell presence in recurrences. Interestingly, these patterns were not observed in patients P01 and P07, who were alive and well at the time of analysis. The observed variance could not be attributed to these patients not having received radiotherapy before recurrence, since the recurrent samples from P05 and P08 were also not within the high-dose radiotherapy field. P01 manifested a dramatic response to pembrolizumab and talimogene laherparepvec (T-VEC) immunotherapy (in a clinical trial) and long-term survival. P07 responded very well to radical radiotherapy with concurrent cisplatin chemotherapy following recurrence. This finding agrees with a study assessing how the metastatic immune landscape influences the response to treatment and the outcome of colorectal cancer patients [52]. The authors quantified immune densities in the centre and invasive margin of metastases and detected a positive correlation with improved treatment outcome and patient survival [52]. Also in our study, the cases of P01 and P07 might suggest that a higher immune cell presence in the recurrences was beneficial for those patients, while the lower detection of immune cells in the remaining patients might indicate immune escape, which led to an unfavourable prognosis. However, these reflections need to be validated in a larger cohort and a more detailed analysis of the specific cell types present in the tumour samples would be necessary.
The small size of the cohort here described represents an important limitation of the study: our cohort consists of a mixture of oral cavity and hypopharyngeal SCC patients, among whom eight carried HPV-negative tumors and one had a HPV-positive tumour. While it would be interesting to assess how different sites of origin and HPV status affect the results of genomic profile changes between primary and recurrent tumours, this could not be done since only one hypopharyngeal SCC and one HPV-positive tumour were included in our cohort. Interestingly, the one HPV positive tumour was found to be in patient P01, who presented with an oral cavity SCC (T2N0M0) at diagnosis and underwent a resection of his primary tumour with no adjuvant treatment and subsequently developed locoregional recurrence and distant metastasis (DM). The percentage of HPV positivity in non-oropharynx SCC varies depending on the subsites: according to data from the United States National Cancer Database, 17.7% for hypopharynx, 11% for larynx, 10.6% for oral cavity, in contrast to 62.9% for oropharynx [53]. As expected, HPV positivity was associated with a statistically significant superior overall survival in patients with AJCC stage III to IVB oral cavity SCC (hazard ratio 0.78, p-value = 0.03) compared to non-HPV positive cases [53]. In a retrospective analysis of 1494 HNSCC patients, 23% of HPV positive cases were from non-oropharynx sites, including 8% from oral cavity. In total, 6.4% HPV positive HNSCC developed DM [54]. The median time to distant metastases was 11 months [54], but there was no difference in the time to development of DM between HPV positive HNSCC and the HPV negative/unknown groups. However, a higher rate of DM to the lung was observed in the HPV positive HNSCC group compared to the HPV negative/unknown HNSCC groups [54]. DM to the lung was also observed in patient P01 (Table 1).
Weber et al. [46] described treatment-related changes in transcriptional subtypes between primary and recurrent tumours [46]. Thus, it would have been interesting to further investigate the locoregional recurrence within the high-dose radiotherapy field after radiotherapy or concurrent chemoradiation. In our cohort, one patient underwent primary radiotherapy alone, while the remaining eight patients had primary resection. Only four of them had postoperative radiotherapy (with one having concurrent chemoradiation), the other four patients did not receive adjuvant treatment after resection. Therefore, our cohort consists of a small heterogeneous sample set in term of HPV status, tumour subsites and treatment patterns. This aspect could potentially limit the generalizability of our results. In addition, we were not able to obtain the exact radiotherapy dose and fractionation, as well as surgical margin status for each patient due to this study being a retrospective study using anonymous samples from the biobank. However, the postoperative radiotherapy doses commonly used in the University Hospitals Birmingham NHS Trust correspond to 60 Gy/30 fractions and 64–66 Gy/32-33 fractions in the cases of positive resection margin (with three weekly concurrent cisplatin chemotherapy) and 65 Gy/30 fractions for primary radical radiotherapy. These limitations prevent us from making a firm conclusion in regard to therapy-related selection of resistant clones between primary and recurrent tumours, especially within high-dose radiotherapy field areas.
In our small cohort, all the recurrences occurred within two years after initial treatment, apart from one patient who had recurrence within five years from initial treatment. Recurrences within five years would not normally be classified as secondary cancers, unless they present different histologies (e.g. adenocarcinoma instead of squamous cell carcinoma), or if the pathologists have raised the concern of secondary cancer (due to new cancer having a different histological appearance, e.g. keratinising vs non-keratinising SCC), which was not the case here. Moreover, the substantial overlap of somatic mutations between the primary and the matching recurrence(s) strongly supports the view that these are not secondary cancers. All patients in our cohort had surgery rather than primary radiotherapy at the initial diagnosis, a part from one patient who had radical chemoradiation for recurrence and remained alive and well at the time of data analysis. We could not conclude whether the patients in our cohort had minimal residual disease (MRD) following treatments, since it is not a routine clinical practice in the UK to perform post-treatment PET-CT scan, other imaging examinations or circulating tumour DNA assessment after surgery ± postoperative radio(chemo)therapy at three months.
Despite these limitations, through the analysis of this small cohort of HNSCC patients, we highlighted the complex and remarkable differences occurring in the genomic and transcriptomic landscape of different patients and of matched samples of primary tumours and recurrences. In spite of the high level of heterogeneity among patients, some common patterns could be observed, such as a higher tumour mutational burden characterising recurrent samples with respect to primary tumours. Recurrences also appeared to be genetically more diverse, as shown by the mutational signature analysis. Immune cell presence was generally lower in these samples (with the interesting exceptions of P01 and P07) and several genes related to immune response were expressed at a significantly lower level than in primaries. Overall these results, together with the changes in PD-L1 expression status with therapeutic implications, underline the importance of analysing (when possible) multiple samples per patient to minimize the risk of immune evasion and obtain a more complete picture of the patient’s tumour. The pervasive differences among patients also strongly advocate for personalised therapy approaches for HNSCC patients.
Data Availability
Sequence data supporting the findings of this study has been deposited at the European Genome-Phenome Archive (EGA), which is hosted by the European Bioinformatics Institute (EBI) and the Centre for Genomic Regulation (CRG), under accession numbers EGAS00001004857 (patient P05) and EGAS00001007464 (remaining patients). Further information about EGA can be found on https://ega-archive.org and “The European Genome-phenome Archive of human data consented for biomedical research” (http://www.nature.com/ng/journal/v47/n7/full/ng.3312.html).
References
Haddad RI, Shin DM. Recent advances in head and neck cancer. N Engl J Med. 2008;359:1143–54.
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49.
Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–9.
Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. New Engl J Med. 2010;363:24–35.
Nissi L, Suilamo S, Kyto E, Vaittinen S, Irjala H, Minn H. Recurrence of head and neck squamous cell carcinoma in relation to high-risk treatment volume. Clin Transl Radiat Oncol. 2021;27:139–46.
Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359:1116–27.
Ferris RL, Blumenschein G Jr, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375:1856–67.
Burtness B, Harrington KJ, Greil R, Soulières D, Tahara M, de Castro G Jr, et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. Lancet. 2019;394:1915–28.
Ott PA, Bang YJ, Piha-Paul SA, Razak ARA, Bennouna J, Soria JC, et al. T-cell-inflamed gene-expression profile, programmed death ligand 1 expression, and tumor mutational burden predict efficacy in patients treated with pembrolizumab across 20 cancers: KEYNOTE-028. J Clin Oncol. 2019;37:318–27.
Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8.
Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, et al. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov. 2017;7:188–201.
Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375:819–29.
Restifo NP, Smyth MJ, Snyder A. Acquired resistance to immunotherapy and future challenges. Nat Rev Cancer. 2016;16:121–6.
McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–9.
Smeets SJ, Hesselink AT, Speel EM, Haesevoets A, Snijders PJF, Pawlita M, et al. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer. 2007;121:2465–72.
Bresadola L, Weber D, Ritzel C, Lower M, Bukur V, Akilli-Ozturk O, et al. Comprehensive genomic and transcriptomic analysis of three synchronous primary tumours and a recurrence from a head and neck cancer patient. Int J Mol Sci. 2021;22:7583.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Lower M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–6.
Hubschmann D, Jopp-Saile L, Andresen C, Kramer S, Gu Z, Heilig CE, et al. Analysis of mutational signatures with yet another package for signature analysis. Genes Chromosomes Cancer. 2021;60:314–31.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009;10:48.
Finotello F, Mayer C, Plattner C, Laschober G, Rieder D, Hackl H, et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11:34.
Koboldt DC. Best practices for variant calling in clinical sequencing. Genome Med. 2020;12:91.
Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet. 2014;15:121–32.
Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21.
Riva G, Albano C, Gugliesi F, Pasquero S, Pacheco SFC, Pecorari G, et al. HPV meets APOBEC: new players in head and neck cancer. Int J Mol Sci. 2021;22:1402.
Plath M, Gass J, Hlevnjak M, Li Q, Feng B, Hostench XP, et al. Unraveling most abundant mutational signatures in head and neck cancer. Int J Cancer. 2021;148:115–27.
Marin-Rubio JL, Vela-Martin L, Fernandez-Piqueras J, Villa-Morales M. FADD in cancer: mechanisms of altered expression and function, and clinical implications. Cancers. 2019;11:1462.
Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–41.
Praveen K, Saxena N. Crosstalk between Fas and JNK determines lymphocyte apoptosis after ionizing radiation. Radiat Res. 2013;179:725–36.
Belka C, Marini P, Lepple-Wienhues A, Budach W, Jekle A, Los M, et al. The tyrosine kinase lck is required for CD95-independent caspase-8 activation and apoptosis in response to ionizing radiation. Oncogene. 1999;18:4983–92.
Eytan DF, Snow GE, Carlson S, Derakhshan A, Saleh A, Schiltz S, et al. SMAC mimetic birinapant plus radiation eradicates human head and neck cancers with genomic amplifications of cell death genes FADD and BIRC2. Cancer Res. 2016;76:5442–54.
Bou Nasser Eddine F, Forlani G, Lombardo L, Tedeschi A, Tosi G, Accolla RS. CIITA-driven MHC class II expressing tumor cells can efficiently prime naive CD4(+) TH cells in vivo and vaccinate the host against parental MHC-II-negative tumor cells. Oncoimmunology. 2017;6:e1261777.
Kaneda MM, Messer KS, Ralainirina N, Li H, Leem CJ, Gorjestani S, et al. PI3Kgamma is a molecular switch that controls immune suppression. Nature. 2016;539:437–42.
Chen S, Bonifati S, Qin Z, St Gelais C, Kodigepalli KM, Barrett BS, et al. SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF-kappaB and interferon pathways. Proc Natl Acad Sci USA. 2018;115:E3798–E807.
Oswald DM, Zhou JY, Jones MB, Cobb BA. Disruption of hepatocyte Sialylation drives a T cell-dependent pro-inflammatory immune tone. Glycoconj J. 2020;37:395–407.
Mody PD, Cannon JL, Bandukwala HS, Blaine KM, Schilling AB, Swier K, et al. Signaling through CD43 regulates CD4 T-cell trafficking. Blood. 2007;110:2974–82.
Wang H, Brown J, Gao S, Liang S, Jotwani R, Zhou H, et al. The role of JAK-3 in regulating TLR-mediated inflammatory cytokine production in innate immune cells. J Immunol. 2013;191:1164–74.
Andreotti AH, Schwartzberg PL, Joseph RE, Berg LJ. T-cell signaling regulated by the Tec family kinase, Itk. Cold Spring Harb Perspect Biol. 2010;2:a002287.
Kuriakose T, Kanneganti TD. ZBP1: innate sensor regulating cell death and inflammation. Trends Immunol. 2018;39:123–34.
Pauls SD, Marshall AJ. Regulation of immune cell signaling by SHIP1: a phosphatase, scaffold protein, and potential therapeutic target. Eur J Immunol. 2017;47:932–45.
Hassan NJ, Simmonds SJ, Clarkson NG, Hanrahan S, Puklavec MJ, Bomb M, et al. CD6 regulates T-cell responses through activation-dependent recruitment of the positive regulator SLP-76. Mol Cell Biol. 2006;26:6727–38.
Comte D, Karampetsou MP, Kis-Toth K, Yoshida N, Bradley SJ, Mizui M, et al. Engagement of SLAMF3 enhances CD4+ T-cell sensitivity to IL-2 and favors regulatory T-cell polarization in systemic lupus erythematosus. Proc Natl Acad Sci USA. 2016;113:9321–6.
Sautès-Fridman C, Petitprez F, Calderaro J, Fridman WH. Tertiary lymphoid structures in the era of cancer immunotherapy. Nature Reviews Cancer. 2019;19:307–25.
Hedberg ML, Goh G, Chiosea SI, Bauman JE, Freilino ML, Zeng Y, et al. Genetic landscape of metastatic and recurrent head and neck squamous cell carcinoma. J Clin Invest. 2016;126:1606.
Weber P, Kunstner A, Hess J, Unger K, Marschner S, Idel C, et al. Therapy-related transcriptional subtypes in matched primary and recurrent head and neck cancer. Clin Cancer Res. 2022;28:1038–52.
Schnidrig D, Turajlic S, Litchfield K. Tumour mutational burden: primary versus metastatic tissue creates systematic bias. Immunooncol Technol. 2019;4:8–14.
Pich O, Muinos F, Lolkema MP, Steeghs N, Gonzalez-Perez A, Lopez-Bigas N. The mutational footprints of cancer therapies. Nat Genet. 2019;51:1732–40.
Mandal R, Senbabaoglu Y, Desrichard A, Havel JJ, Dalin MG, Riaz N, et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight. 2016;1:e89829.
Wu AA, Drake V, Huang HS, Chiu S, Zheng L. Reprogramming the tumor microenvironment: tumor-induced immunosuppressive factors paralyze T cells. Oncoimmunology. 2015;4:e1016700.
Chen YP, Wang YQ, Lv JW, Li YQ, Chua MLK, Le QT, et al. Identification and validation of novel microenvironment-based immune molecular subgroups of head and neck squamous cell carcinoma: implications for immunotherapy. Ann Oncol. 2019;30:68–75.
Mlecnik B, Van den Eynde M, Bindea G, Church SE, Vasaturo A, Fredriksen T, et al. Comprehensive intrametastatic immune quantification and major impact of immunoscore on survival. J Natl Cancer Inst. 2018;110:97–108.
Tian S, Switchenko JM, Jhaveri J, Cassidy RJ, Ferris MJ, Press RH, et al. Survival outcomes by high-risk human papillomavirus status in nonoropharyngeal head and neck squamous cell carcinomas: A propensity-scored analysis of the National Cancer Data Base. Canncer. 2019;125:2782–93.
Sacks R, Law JY, Zhu H, Beg MS, Gerber DE, Sumer BD, et al. Unique patterns of distant metastases in HPV-positive head and neck cancer. Oncology. 2020;98:179–85.
Acknowledgements
We thank Karen Chu for providing project management support, together with Ines Schneider and Lisa Bischof. We would also like to thank Matthias Gaida for performing the p16 staining of patient P08 and for providing feedback on the immunohistochemistry images. We are also grateful to Andrea Nubbemeyer for performing the HPV PCR assays.
Author information
Authors and Affiliations
Contributions
U.S. and A.K. designed the study. H.M. and A.K. provided the study material. J.B. and A.G. conducted laboratory analyses. L.B., D.W., C.R., M.L., V.B., O.A.O., C.S., B.S., F.V. and U.S. analysed and interpreted the data. L.B., D.W., O.A.O. and A.K. wrote the manuscript, with contributions and revisions from all authors. The final submitted article was approved by all authors.
Corresponding authors
Ethics declarations
Competing interests
US and VB are employees at BioNTech SE. US is co-founder and shareholder of TRON, co-founder and CEO of BioNTech SE. AK received fees for consulting, advisory, speaker’s roles and/or research funding from PUMA BioTechnology, AstraZeneca, Merck, MSD, Bristol-Myers Squibb, and Avvinity Therapeutics. HM has personal financial interests with AstraZeneca, MSD, GSK, Sanofi Pasteur, Merck, Warwickshire Head Neck Clinic Ltd; institutional financial interests with AstraZeneca, GSK PLC, Sanofi Pasteur, MSD, GSK Biologicals, Silence Therapeutics; leadership roles: Chief of Liteform Trial Steering Committee, Chair of NIMRAD Trial Steering Committee, President of the British Association of Head Neck Oncologists, Trial Steering Group member of the MRC CTU Cancer Trials Steering Committee, Council member of the International Association of Oral Oncology, Director of the Institute for Head Neck Studies and Education, Secretary of the Head Neck Cancer InterGroup. HM is a National Institute for Health Research (NIHR) Senior Investigator. The views expressed in this article are those of the author(s) and not necessarily those of the NIHR, or the Department of Health and Social Care. All other authors have no conflict of interest.
Ethics approval and consent to participate
An informed consent was obtained from all participants in this study. These patients had consented for their samples to be used for translational research and all experimental protocols used in this study were carried out in accordance with the accelerated programme protocol at the University Hospital Birmingham (UHB) NHS trust (Sponsor reference RG14-114, Research Ethics Committee West Midlands, Ethics Approval 14/YH/1101).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bresadola, L., Weber, D., Ritzel, C. et al. Temporal evolution and inter-patient heterogeneity in primary and recurrent head and neck squamous cell carcinoma. BJC Rep 2, 62 (2024). https://doi.org/10.1038/s44276-024-00091-5
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44276-024-00091-5