Abstract
Background
The challenges of introducing Whole Genome Sequencing (WGS) as NHS standard of care for patients with glioma are reviewed.
Methods
Patients undergoing glioma surgery with WGS sampling were identified retrospectively from WGS reports between 01/01/2022-30/12/2023. Data including demographics, integrated molecular diagnosis, time through critical pathway steps per calendar quarter (Q) and WGS variants were captured from electronic health records.
Results
146 glioma samples were analysed. 91% of patients underwent craniotomy and 1 cm3 tumour sampled for WGS, with median tumour content (MTC) of 69.5% (IQR + /- 30.5). The remainder underwent stereotactic needle biopsy, and one core sampled for WGS, with MTC of 73% (IQR + /- 19%). Median time from tumour sampling to completion of WGS report in Q1:2022 was 255 days (IQR + /- 107.5) versus in Q4:2023 was 137 days (IQR + /- 60.5; p < 0.001). 26/146 (17.8%) patients had molecular variants leading to trial recommendation. 1 patient with glioblastoma and high Tumour Mutational Burden commenced anti-PD1 immunotherapy. 8 patients with glioblastoma had RB1 variants associated with improved progression-free survival.
Conclusions
WGS is feasible for patients undergoing biopsy or craniotomy. NHS infrastructural resources and improvement of WGS technologies are required to improve turnaround time and ensure equitable access for all patients with glioma.
Similar content being viewed by others
Background
Gliomas are the most common intrinsic central nervous system (CNS) tumours. The 2021 WHO Classification of Tumours of the CNS [1] combines histological and molecular information to provide an integrated diagnosis that stratifies gliomas into four grades. WHO grades 1 and 2 are considered low grade glioma. Grade 1 glioma typically represents a well circumscribed, benign and curable glioma following complete surgical resection, with grade 2 diffuse glioma (e.g. oligodendroglioma or astrocytoma) transforming over time to a malignant high-grade glioma (HGG; WHO 3 or 4). Glioblastoma (isocitrate dehydrogenase-wild-type, WHO 4) represents the most common HGG with an estimated 3200 new cases diagnosed in the UK per year [2]. In order to confirm a tissue diagnosis and subsequent treatment plan, an operation is required to provide tumour tissue by either maximal safe surgical resection where appropriate, or image-guided needle biopsy. The integrated molecular diagnosis is typically confirmed by the neuropathologists at the next multi-disciplinary team (MDT) meeting, following serial immunohistochemistry tests, typically with a turnaround time of less than one week. However, if these diagnostic serial tests are non-conclusive, the tissue may be required to be sent for methylation array at Queen Square Laboratories, University College London Hospitals NHS Foundation Trust, with turnaround time of approximately a month. For HGG, further oncological management includes concurrent chemoradiotherapy, followed by adjuvant chemotherapy. Currently, for glioblastoma the alkylating agent Temozolomide remains first line chemotherapy [3], and one of the few effective chemotherapy agents available for this condition. Recurrence is inevitable, however there is no standard of care second line therapy. Treatments in this scenario are dictated by the clinical picture, and may include re-challenge with chemotherapy, repeat resection or debulking surgery and/or irradiation potentially coupled with systemic therapies [4]. Despite decades refining these treatments, median survival time remains just 14.6 months [3], and new therapeutic strategies are desperately needed to improve outcomes.
The NHS Genomic Medicine Service (GMS), initiated in October 2018, aims to provide genomic testing, clinical care, and interpretation for rare diseases and cancer throughout England. This service is structured around a standardised National Genomic Test Directory [5], ensuring uniformity in test methodologies, gene targets, and eligibility criteria in different cancer clinical indications. The service is delivered through a consolidated network of seven NHS England (NHSE) regional Genomic Laboratory Hubs (GLH). Incorporating the evolving knowledge from the 100,000 Genomes Project [6, 7] into the existing molecular testing provision within the NHS, it is the long-term goal of NHSE to accelerate the delivery of molecular testing, including whole genome sequencing (WGS) as part of cancer clinical care [8]. WGS is performed using short read next generation sequencing (NGS) technology and can identify actionable mutations that may influence treatment options, facilitate access to precision medicines through clinical trials and compassionate access, provide germline cancer predisposition genes, and prognostic or predictive biomarkers. The Cancer Programme of the 100,000 Genomes Project reported that in over 90% of glioblastoma cases genomic alterations were present [9]. This study also highlighted the importance of linking WGS and real-world clinical data, including survival analysis to help identify cancer genes that affect prognosis. In the future, precision medicine trials for glioma may require WGS for enrolment and therapeutic stratification.
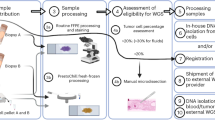
The NHS pathway for WGS is complex (Fig. 1), requiring co-ordination and transport of WGS record of discussion [10] and test order forms [11] together with tissue samples through multiple NHS departments and sites. To practically deliver WGS as NHS standard of care for individuals with brain tumours, numerous challenges need to be over-come. Here, we report our two-year experience in establishing the WGS pathway for patients with glioma, in two neurosurgical centres in the West Midlands. We identify the challenges to deliver WGS, report on the frequency of WGS variants for glioma and aim to provide a framework to help other NHS centres develop the WGS pathway to ensure equitable access across the UK.
Patient are identified at the weekly neuro-oncology multi-disciplinary team meeting (MDT), WGS forms are completed at the weekly neurosurgical clinic. Tissue sampling of paired fresh frozen tumour and blood samples must be co-ordinated with the associated forms at time of surgery, and transported to the pathology laboratory for standard of care serial investigations to achieve neuro-pathology diagnosis. If SOC diagnosis is not achieved tumour samples are sent to Queen Square Laboratories London for methylation array. The diagnostic code and the median tumour content is calculated and WGS request form updated. The tissue and updated forms are transported to the Regional Genomics Laboratory Hub (GLH) for DNA extraction and plating, and then onto Illumina (Cambridge) for sequencing. WGS analysis and variant calling is completed by Genomics England. The data is sent to the GLH for disease-specific analysis, and discussed in the fortnightly Genomics Tumour Advisory Board (GTAB) before a final report is finalised and sent to the treating oncologist. Patients with actionable variants are discussed in the neuro-oncology MDT.
Methods
In 2021, all stakeholders at Queen Elizabeth Hospital Birmingham (QEHB) and Royal University Hospital Stoke (RUHS) neurosurgical units contributed to a WGS Standard Operating Procedure (SOP) for neuro-oncology patients at each unit. There were common themes however the pathways and process are unique to each unit e.g. QEHB pathology serves both units in a hub and spoke model. For an overview of the WGS pathway see Supplementary Information S1.
Patients undergoing a neurosurgical procedure for radiologically-presumed glioma or redo-surgery for glioma progression and fresh frozen tumour sampling for WGS at QEHB and RUHS were identified retrospectively from WGS reports between 01/01/2022-30/12/2023. From 01/01/2022 onwards, patients were prospectively consented for WGS. Twelve patients had fresh frozen tumour previously collected and were retrospectively consented for WGS (One patient from RUHS and eleven from QEHB as part of the Tessa Jowell BRAIN MATRIX platform study [12]. Patients retrospectively consented for WGS were excluded from the timing analysis of the NHS WGS pathway, as some had surgery up to two years prior to WGS consent).
Data including demographics, integrated diagnosis, molecular variants identified in WGS analysis were captured from the electronic health records. The time taken to pass through WGS critical steps were recorded in quarter of a year (Q) time blocks commencing in January 2022. Critical pathway steps were defined as:
-
1)
Surgery-Regional Genomics Hub: time from WGS sampling at neurosurgical procedure, transfer to neuro-pathology hub at QEHB, to sample receipt at the regional GLH. (The Women’s Hospital Birmingham acts as the West Midlands GLH). This reflects the time taken to achieve an integrated molecular diagnosis, documentation of the diagnostic code and calculated median tumour content (MTC) on the WGS paperwork and transfer of the fresh frozen tumour with paired blood sample and updated WGS paperwork to the regional GLH.
-
2)
Regional Genomics Hub-Illumina- GTAB: time for DNA plating, transfer to Illumina Cambridge, variant calling by Genomics England, transfer of initial report back to the GLH and subsequent disease-specific analysis prior to presentation at fortnightly brain tumour GTAB.
-
3)
GTAB-WGS report: time from GTAB and release of completed WGS report to treating oncologist.
Statistical analysis
Statistical analysis and data visualisation was performed using MATLAB R2021b (MathWorks, version 9.11). Most data were not normally distributed (Anderson-Darling test), so median and interquartile range were used as summary statistics, and Wilcoxon rank sum test used for group comparisons.
Results
All patients offered WGS consented for this investigation. Of 152 patients with completed WGS reports (QEHB:RUHS = 127:25), six patients results were excluded. Two were excluded due to limited material for WGS provided at time of surgery. Three were excluded as the histopathological diagnosis was not included for WGS sampling (two lymphoma needle biopsy samples and one sample radio-necrosis with <10% neoplastic cells at redo craniotomy for glioblastoma). In one patient there was insufficient sample for WGS due to technical issues with diagnostic sample and so the WGS samples was instead used for diagnosis.
Demographics
One hundred and forty-six patients had confirmed integrated molecular diagnosis of glioma (65% glioblastoma, Table 1). In approximately 30% of these glioma pathology investigations performed serially were non-conclusive and methylation array at Queen Square Laboratories, London was requested to facilitate an integrated molecular diagnosis. Approximately 65% of patients were male, with a median age of 52.9 ± 14.9 years. All WGS reports were in concordance with the NHS standard of care pathology reports and no changes were made to the integrated diagnosis.
91% of patients underwent craniotomy with at least 1 cm3 sent for WGS. Median tumour content (MTC) in the WGS sample was 69.5% (IQR + /- 30.5). 10% of these were redo surgery for glioma recurrence. 9% of patients had a stereotactic needle biopsy with one needle core (1 cm×0.2 cm) sent for WGS with MTC of 73% (IQR + /- 19%). There was no significant difference on Wilcoxon sign rank test between MTC obtained by craniotomy or core needle biopsy. DNA extraction failed in five patients (three undergoing craniotomy; two undergoing biopsy as MTC < 30%).
Only the pertinent germline variants were reported in thirteen patients because at the time of WGS analysis at the regional GLH prior to GTAB they had a poor performance status or had passed away. Hence one hundred and twenty-eight patients were included in WGS pathway timing and results analysis.
Timing
Over the course of the two-year study, the time taken from tissue sampling at surgery to the final GTAB report improved by nearly five months, from an initial median time of 255 ± 107.5 days down to 137 ± 60.5 days in the final quarter (p < 0.001, Fig. 2). This was driven by progressive incremental improvements in most aspects of the diagnostic pipeline, with the only exception being the time taken for DNA extraction at the plating hub, transfer to Illumina Cambridge, first analysis at GEL and analysis in a disease specific by the clinical scientists at the regional GLH to discussion at the regional GTAB (Fig. 2C).
Germline Findings
Four percent of patients (5/141) had a pertinent germline variant requiring referral to the local clinical geneticist for counselling and further investigation. Germline variants included MSH6 and PMS2 mismatch repair genes associated with Lynch syndrome [13], TP53 variant causing Li-Fraumeni syndrome [14], BRCA2 gene variant associated with breast, ovarian, fallopian tube, primary peritoneal cancer and male breast and prostate cancer [15] and ATM gene variant associated with breast, gastric and colon cancers [16].
Landscape of molecular variants
The frequency or reported molecular variants can be seen in Fig. 3. There was concordance between the glioma IDH status identified by WGS and as per the pathology report. 26 patients (18%) had molecular variants where a trial was recommended. One patient with recurrent glioblastoma, who had exhausted all therapeutic options, was found to have a High Tumour Mutational Burden with GTAB recommendation of anti-PD1 immunotherapy [17]. The patient commenced Nivolumab but passed away with disease progression after completion of three cycles. Eight patients with glioblastoma had RB1 variants associated with improved progression free survival [18].
Discussion
One of the NHS long-term objectives is to accelerate the delivery of genomic medicine, including WGS in clinical cancer care [7]. The NHSE target for WGS analysis and reporting is 42 days (monitored by NHSE). The Central and South NHS genomic medicine service is responsible for serving the West Midlands, Thames Valley and Salisbury/Southampton covering a population of 11 million (the largest catchment population of all GLHs). This was reflected in the West Midlands being the highest recruiter to the 100,000 Genomes Project [8]. Despite this, at a local level there was limited institutional memory, and no extra local resources were made available to roll out WGS for patients with brain tumours. Through regional neuro-oncology MDT training to complete the WGS record of discussion, training and support for a local lead WGS brain tumour specialist nurse, development of local WGS tissue pipelines from theatre to pathology laboratory, establishing regional virtual GTAB meetings and recent appointment of a medical neuro-oncology consultant, we have now established a robust regional hub and spoke infrastructure for this service. This WGS infrastructure can be expanded to support WGS for other cancer types within our region and can also be replicated in other centres to ensure equitable access to WGS for brain tumour nationally, acknowledging that each unit will have its own unique challenges to address.
Since the first patient was recruited in 2021, this service has continued to evolve with optimisation and streamlining of pathways, and with expansion of analytical expertise we have seen a steady improvement in the WGS turnaround time. Improvements included establishing procedures to identify issues with incomplete WGS paperwork, establishing a weekly transfer of samples to the regional GLH. If the serial neuropathology diagnostic investigations are able to provide the diagnosis, the longest time samples on the WGS pathway would need to wait prior to transfer to regional GLH is a week. However, if methylation array is required to achieve a diagnosis, this can take up to six weeks as samples are sent to London and require batching before analysis can proceed. Until a diagnosis is achieved the WGS samples cannot proceed to the next step as a diagnostic code is required to be added to the WGS test order form. Currently, the WGS replace paperwork totals five pages, each requiring patient identification to be added to each sheet. A simplified single page electronic centralised platform would allow visibility of the status of orders, oversight of the test status that, coupled with sample tracking, would significantly reduce duplication and burden of work for the NHS team and allow transparency of data and sample flow.
Confidence in interpretation of the WGS report represented a steep learning curve, but with time and experience it has become progressively easier and quicker to generate a finalised report. Currently for glioma cases with no clinically actionable variants, reports have been signed off, without having to wait for presentation at the fortnightly GTAB. Together this has resulted in shorter discussion times of individual cases at the GTAB meeting, allowing more cases to be discussed, and a quicker time to release the final WGS report. The recent addition of another clinical genomic scientist to the regional GLH team has facilitated further improvement in turnaround time. To date the integrated molecular diagnosis has been complemented by the WGS findings. In the future this may expand the diagnosis as we develop deeper understanding of the influence of molecular variants.
The list of neuro-oncology actionable variants is evolving with the accompanying recommendation of genotype-matched medicines and clinical trials available at the time of reporting. The minimum turnaround for the WGS result in Q4 2023 was 77 days which can be considered clinically actionable, as it corresponds to the cumulative time taken for initial treatment, consisting of recovery time from primary neurosurgery for HGG (one month) followed by treatment with the Stupp regimen [3]. Currently, depending on the pattern of disease progression and performance status, patients with actionable variants and available precision medicine treatments may be offered these as part of a clinical trial or compassionate access. In our study, only one patient with recurrent glioblastoma and high TMB was offered compassionate access Nivolumab, but passed away with disease progression after completion of only three treatment cycles. This is in keeping with other studies that report in recurrent HGG, high TMB alone may not be sufficiently predictive of immune check point inhibitor (ICI) response [19, 20]. Temozolamide induced T-cell exhaustion may be a contributory factor that limits the effectiveness of ICI in the recurrent glioma setting, and requires further investigation [21]. There is a paucity of precision therapies for patients with glioma. It is hoped that with improved academic, pharmaceutical industry collaborations together with novel drug delivery systems and trial designs, that more options will be added to the therapeutic armamentarium.
There is an acknowledgment that initial and repetitive tissue sampling of brain tumours plays an essential role in understanding both resistance mechanisms and vulnerabilities of brain tumours that may be targeted with precision medicine. Recent trends include the recognition of the importance of perioperative and Window of Opportunity (WoO) trials, aimed at tissue sampling before and after experimental therapies to better understand the pharmacokinetics and pharmacodynamics of novel targets [22]. It is crucial that the turnaround time for critical biomarkers such as IDH, MGMT, and others are expedited for diagnosis and also for WGS to aid stratification of patients into clinical trials including WoO and neoadjuvant therapy trials.
We have demonstrated that WGS is achievable both for 1 cm3 samples achieved at craniotomy and also from single needle biopsy cores (1 × 0.2 cm) both achieving well over thirty percent viable tumour cells required for DNA extraction. This opens up potential precision targets for patients who may not be fit for open craniotomy, have deep seated lesions and/or multifocal disease. Performing needle biopsy as a day case can reduce the NHS burden on clinical resources [23] and facilitate access to clinical trials. Understanding the frequency of genomic variants seen in glioma patients will help to better understand the natural distribution of glioma molecular variants providing more accurate power calculations for future trials.
The Tessa Jowell Brain Cancer Mission (TJBCM) recently highlighted that there is significant UK geographical variability in access to genetic testing and precision medicine trials and estimate less than five percent of adult patients with brain tumour are offered WGS [24]. The British Neuro-Oncology Society recently published a Position Statement: Guideline for tissue sampling of brain tumours [25]. The Brainstrust has also published a patient information sheet to assist patients discuss tissue collection with their neurosurgeon preoperatively in centres where this is not yet standard of care [26]. These initiatives aim to facilitate tumour tissue sampling for genomic analysis, research and emerging novel treatments. The TJBCM report [24] also highlights the range of service delivery and logistic challenges including transportation of samples up to five hundred miles during the WGS analysis pathway. Further improvements in NHS infrastructural resources and improvement of WGS technologies are required to improve turnaround time further and ensure equitable access for all patients with glioma.
Emerging technologies and future perspectives
Sequencing technologies are rapidly advancing and are now able to provide real-time genomic analysis including ultra-fast methylation sequencing [27,28,29,30] and dynamic adaptive sampling [9, 31, 32] to achieve a more rapid diagnosis. Since July 2024, we have used ROBIN [29], a research tool based on PromethION nanopore sequencing technology, to provide a methylome classifier in less than two hours, with next day comprehensive molecular profiling including copy number profiles, gene fusions, SNVS of diagnostic relevance, other structural variants and MGMT promotor methylation status in over twenty patients, using local research infrastructure, (unpublished data) through a University of Birmingham-NHS collaboration. Currently, long read sequencing is not validated for NHS use, and research is ongoing to better understand the limitations of these new tools in order to facilitate NHS accredited rapid genomic analysis.
The equitable implementation of novel sequencing technologies in NHS clinical practice is currently limited by several key challenges, including financial constraints, the absence of a robust infrastructure for fresh or snap-frozen tissue sample collection, and a lack of local neuropathology wet-lab facilities for DNA extraction and sequencing. Addressing these barriers is essential for integrating WGS and other advanced genomic technologies into routine practise. Development of a secure NHS cloud-based infrastructure would enable centralised storage and remote access to genomic data, facilitating a remote centralised GTAB. Overtime, an AI-driven genomic interpretation and generation of report, with integration of relevant clinical trials and available precision medicines could enhance efficiency and accuracy of WGS analysis, support clinical decision making and improve accessibility even in resource-limited settings. The NHS platform represents a novel environment in which to grow new models of genomic care provision and application of new genomic technologies.
Conclusion
We have demonstrated feasibility of performing WGS as standard of care for patients with glioma, with improvement of turnaround time due to incremental pathway and infrastructural improvements. New sequencing technologies to provide real time diagnosis using methylation classifiers can guide intra-operative decision making, such as extent of surgical resection and decision for intraoperative tumour cavity chemotherapy insertion, and rapid diagnosis of molecular variants may inform neo-adjuvant trials and expand development and access to novel targeted treatments. There is a need to evaluate and optimise novel sequencing techniques including methylation array so that these tests can receive NHS accreditation and be included in the National Genomic Test Directory [5]. Furthermore, the NHS genomic infrastructure pathways need to be evaluated in terms of efficiency, cost effectiveness, work force planning, service delivery and review of organisational and environmental sustainability to ensure equitable access for genomic sequencing for all patients with brain tumour.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro-Oncol. 2021;23:1231–51.
Brodbelt A, Greenberg D, Winters T, Williams M, Vernon S, Collins VP. Glioblastoma in England: 2007–2011. Eur J Cancer. 2015;51:533–42.
Stupp R, Mason WP, Van Den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. NEJM. 2005;352:987–96.
Ma R, Taphoorn MJ, Plaha P. Advances in the management of glioblastoma. J Neurol Neurosurg Psychiatry. 2021;92:1103–11.
NHS England. National genomic test directory for cancer. Version 13.1. Available from: https://www.england.nhs.uk/publication/national-genomic-test-directories/[Internet] 2025 [Cited 15/10/2025].
Turnbull C, Scott RH, Thomas E, Jones L, Murugaesu N, Pretty FB, et al. The 100 000 Genomes Project: bringing whole genome sequencing to the NHS. BMJ 2018;361:1687.
Turnbull C. Introducing whole-genome sequencing into routine cancer care: the Genomics England 100 000 Genomes Project. Ann Oncol. 2018;29:784–7.
NHS England. Accelerating genomic medicine in the NHS. Version 3. Available from: https://www.england.nhs.uk/long-read/accelerating-genomic-medicine-in-the-nhs/ [Internet] 2022 [Cited 01/07/2024].
Sosinsky A, Ambrose J, Cross W, Turnbull C, Henderson S, Jones L, et al. Insights for precision oncology from the integration of genomic and clinical data of 13,880 tumors from the 100,000 Genomes Cancer Programme. Nat Med. 2024;30:279–89.
NHS England. Record of Discussion Regarding Genomic Testing V4.03. [Internet] 2021 [Cited 01/07/2024] Available from https://www.england.nhs.uk/wp-content/uploads/2021/09/nhs-genomic-medicine-service-record-of-discussion-form.pdf [Internet] 2021 [Cited 01/07/2024].
NHS England. Test order Form-Cancer V1.19. [Internet] 2022 [Cited 01/07/2024] Available from https://www.england.nhs.uk/wp-content/uploads/2021/09/genomic-medicine-dervice-test-order-form-cancer-v1.19.pdf [Internet] 2022 [Cited 01/07/2024].
Tessa Jowell Brain Matrix [Internet] 2019 [Cited 01/07/2024]. Available from: https://www.birmingham.ac.uk/research/crctu/trials/brain-matrix [Internet] 2019 [Cited 01/07/2024].
Abildgaard AB, Nielsen SV, Bernstein I, Stein A, Lindorff-Larsen K, Hartmann-Petersen R. Lynch syndrome, molecular mechanisms and variant classification. Br J Cancer. 2023;128:726–34.
Fortuno C, Feng BJ, Carroll C, Innella G, Kohlmann W, Lázaro C, et al. Brunet J, Feliubadaló L, Iglesias S, Menéndez M, Teulé A. Cancer risks associated with TP53 pathogenic variants: maximum likelihood analysis of extended pedigrees for diagnosis of first cancers beyond the Li-Fraumeni syndrome spectrum. JCO Precision. Oncology. 2024;8:e2300453.
Benusiglio PR, Dardenne A, Fallet V, Cadranel J. Emerging cancer risks in BRCA2 pathogenic germline variant carriers. Eur J Hum Genet. 2023;31:1355–6.
Hall MJ, Bernhisel R, Hughes E, Larson K, Rosenthal ET, Singh NA, et al. Germline pathogenic variants in the ataxia telangiectasia mutated (ATM) gene are associated with high and moderate risks for multiple cancers. Cancer Prevention. Research. 2021;14:433–40.
Goodman A, Kato S, Bazhenova L, Patel S, Frampton G, Miller V, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–608.
Dono A, Ramesh A, Wang E, Shah M, Tandon N, Ballester L, et al. The role of RB1 alteration and 4q12 amplification in IDH-WT glioblastoma. Neuro-Onc Adv. 2021;1:3.
Ahmad H, Fadul CE, Schiff D, Purow B. Checkpoint inhibitor failure in hypermutated and mismatch repair-mutated recurrent high-grade gliomas. Neuro-Oncol Pr. 2019;6:424–7.
McGrail DJ, Pilié PG, Rashid NU, Voorwerk L, Slagter M, Kok M, et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol. 2021;32:661–72.
Badani A, Ozair A, Khasraw M, Woodworth GF, Tiwari P, Ahluwalia MS, et al. Immune checkpoint inhibitors for glioblastoma: emerging science, clinical advances, and future directions. J Neuro-Oncol. 2024;21:1–7.
Hotchkiss KM, Karschnia P, Schreck KC, Geurts M, Cloughesy TF, Huse J, et al. A brave new framework for glioma drug development. Lancet Oncol. 2024;25:e512–9.
Grundy PL, Weidmann C, Bernstein M. Day-case neurosurgery for brain tumours: the early United Kingdom experience. Br J Neurosurg. 2008;22:360–7.
Tessa Jowell Brain Cancer Mission https://www.tessajowellbraincancermission.org/closing-the-gap-report/ [Internet] 2024 [Cited 01/12/2024].
https://www.bnos.org.uk/bnos-guideline-for-tissue-sampling-of-brain-tumours/ British Neuro-Oncology Society Position Statement: Guideline for tissue sampling of brain tumours[Internet] 2023 [Cited 01/12/2024].
Brains Trust https://brainstrust.org.uk/wp-content/uploads/2022/11/brainstrust-know-hows-tissue-collection-2-1-1.pdf [Internet] 2022 [Cited 01/12/2024].
Djirackor L, Halldorsson S, Niehusmann P, Leske H, Capper D, Kuschel LP, et al. Intraoperative DNA methylation classification of brain tumors impacts neurosurgical strategy. Neuro-Oncol Adv. 2021;3:vdab149.
Vermeulen C, Pagès-Gallego M, Kester L, Kranendonk ME, Wesseling P, Verburg N, et al. Ultra-fast deep-learned CNS tumour classification during surgery. Nature. 2023;622:842–9.
Deacon S, Cahyani I, Holmes N, Fox G, Munro R, Wibowo S, et al. ROBIN: A unified nanopore-based sequencing assay integrating real-time, intraoperative methylome classification and next-day comprehensive molecular brain tumour profiling for ultra-rapid tumour diagnostics. Neuro-Oncology. 2025;noaf103.
Brändl B, Steiger M, Kubelt C, Rohrandt C, Zhu Z, Evers M, et al. Rapid brain tumor classification from sparse epigenomic data. Nat Med. 2025;28:1–9.
Patel A, Dogan H, Payne A, Krause E, Sievers P, Schoebe N, et al. Rapid-CNS2: rapid comprehensive adaptive nanopore-sequencing of CNS tumors, a proof-of-concept study. Acta Neuropathologica. 2022;143:609–12.
Weilguny L, De Maio N, Munro R, Manser C, Birney E, Loose M, et al. Dynamic, adaptive sampling during nanopore sequencing using Bayesian experimental design. Nat Biotech. 2023;41:1018–25.
Acknowledgements
Thank you to Professor Andrew Beggs, Department of Cancer and Genomic Sciences, University Birmingham for reviewing the final manuscript.
Funding
University Hospitals Birmingham Charity kindly covered publication costs associated with the manuscript, and funded Sana Manan as Physicians Associate at Queen Elizabeth Hospital Birmingham. Her work as Clinical Research Associate at the University of Birmingham (UoB) was kindly supported by a philanthropic donation by Mr Robert Spier, through the UoB Development and Alumni Relations Office. No other funding has supported this study. The authors received no specific funding for this work.
Author information
Authors and Affiliations
Contributions
VW, EA and CW have all contributed to the conception of the study and study design. VW, VCr, SM, NC, NV, LH, EA, TK and CB collected the clinical data. VW, EA, VCr and SM contributed to co-ordination and execution of the study, analysis and interpretation of the results and manuscript preparation. All authors contributed to and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This study was registered as a qualitative improvement study/audit at each hospital under the local Research and Audit departments and thus no ethical committee approval was needed. All methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
No informed consent for research was required for this work as it was performed as an audit of NHS standard of care practice for brain tumour patients.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wykes, V., Albanese, E., Crispi, V. et al. Introduction of whole genome sequencing as NHS standard of care for glioma patients in two neurosurgical oncology centres: West Midlands. BJC Rep 3, 79 (2025). https://doi.org/10.1038/s44276-025-00168-9
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44276-025-00168-9