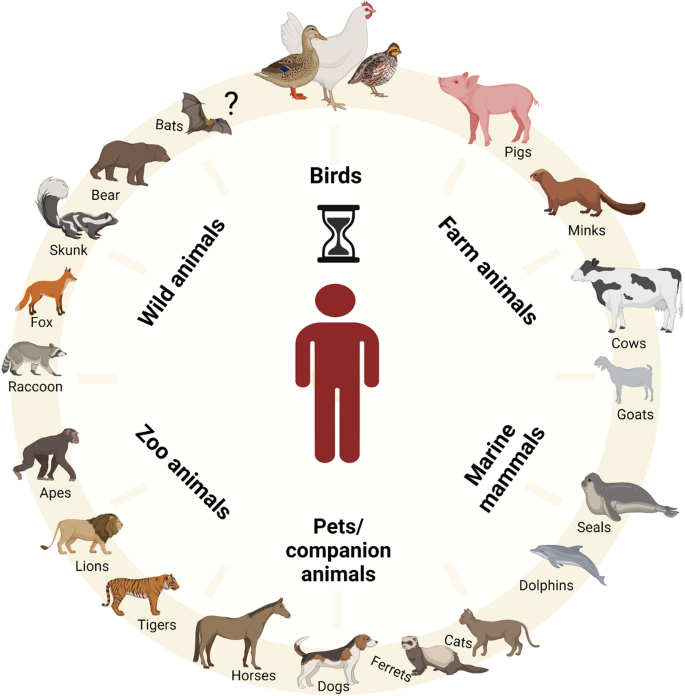
The H5 subtype of highly pathogenic avian influenza viruses represents a significant challenge to animal and human health. H5 clade 2.3.4.4b viruses have experienced an unprecedented global spread, coupled with remarkable genetic plasticity for adaptation in birds and mammals. Although human infections remain very limited, the establishment in wild, marine, and farmed animals, including recently dairy cattle, is of concern. The role of mammalian hosts as intermediaries for zoonotic or even pandemic influenza A viruses should not be underestimated. In order to mitigate the zoonotic risk and be adequately prepared, it is essential to understand and monitor the dynamics of HPAIV H5 at the avian-mammal interface.
Diversity of avian influenza viruses
Avian influenza viruses (AIVs) have their reservoir in wild birds, especially waterfowl1. However, they also continue to cause losses in the poultry industry worldwide, affecting global meat and egg production, threatening wild bird biodiversity and posing zoonotic risks1. The influenza A virus (IAV) genome is segmented into eight distinct, negative-stranded, RNA gene segments. The error-prone nature of the IAV polymerase, which is responsible for viral replication, allows for constant change and adaptation1, which is highly advantageous for viral evolution. When two different IAVs infect a host cell, gene segments can be swapped (i.e., reassortment or antigenic shift) to theoretically generate 254 novel genotypes2. Moreover, stepwise mutations acquired spontaneously during viral replication, may also allow the virus to adapt to new hosts (i.e. antigenic drift). To date, AIVs have been classified into 16 hemagglutinin (HA; H1-H16) and 9 neuraminidase (NA; N1-N9) subtypes based on variation in these surface glycoproteins. The type of HA and NA define the respective subtype (e.g. H5N1) with 144 possible HxNy combinations.
Early history of HPAIV H5N1
The first reports of HPAIV in poultry date back to 1878 by Edoardo Perroncito in Italy3. Over a century later, in 1996, HPAIV of subtype H5N1 was identified in domestic geese in the Guangdong province (Gs/Gd), China4. In 1997, HPAIV H5N1 Gs/Gd was detected in poultry markets in Hong Kong, and 6 out of 18 infected humans died, drawing attention to the zoonotic risk of this virus group. Subsequently, HPAIV H5N1 Gs/Gd was controlled in commercial poultry, but it continued to circulate in the wild bird reservoir. In 2002, Gs/Gd-like viruses jumped again from wild birds to poultry and back to wild birds in the Qinghai Lake region5. The endemicity of this virus in wild birds and poultry, and the continuous exchange between poultry and wild birds resulted in successful evolution and diversification into numerous phylogenetic clades6. Since 2005, HPAIVs H5N1 Gs/Gd have been transmitted in several waves across regions and continents, primarily by migratory wild birds7.
Currently circulating H5N1 of clade 2.3.4.4b
Since 2022, HPAIVs of subtype H5 have become deeply entrenched in poultry and wild birds7,8. The subsequent panzootic clade 2.3.4.4b H5-viruses have a very wide range of avian hosts9 and show numerous spill-over infections to wild carnivores, farmed fur animals such as mink, fox and raccoon dogs, and marine mammals such as sea lions10 (Fig. 1). The unique characteristics of HPAIV H5 clade 2.3.4.4b demand a fundamental rethink of our understanding of these pathogens. Europe is now a new center for HPAIV H5 in wild birds, and reassortment with other European AIV is underway11. HPAIV H5 has been transmitted from Europe to Africa and via Iceland to North and South America. This has resulted in high overall mortality in naïve bird populations and mammals including on mainland Antarctica12,13,14. It is of great concern, that HPAIV H5 has infected well over 150 different bird species and more than 50 mammalian species (Fig. 1)15,16,17. In addition, HPAIV H5N1 of clade 2.3.4.4b has a remarkable genetic elasticity for reassortment with other AIVs resulting in a wide range of highly diverse H5N1, H5N2, H5N3, H5N4, H5N5, H5N6 and H5N8 viruses18,19. Efficient reassortment has resulted in well over 50 different genotypes8, with the HA segment as the stable element. This poses a significant challenge to the study of the epidemiology, pathobiology and genetic characteristics of these viruses20.
Mammals infected with the current AIV H5N1 clade 2.3.4.4b present a significant risk for the emergence of zoonotic viruses. Human infections have been documented in a dairy cow farm worker. Human-adaptation markers have been identified in multiple isolates from mammals, raising concerns about the potential role of these animals as intermediate hosts for the emergence of zoonotic AIV H5N1. The figure was created with BioRender.com.
HPAIV H5N1 in dairy cattle
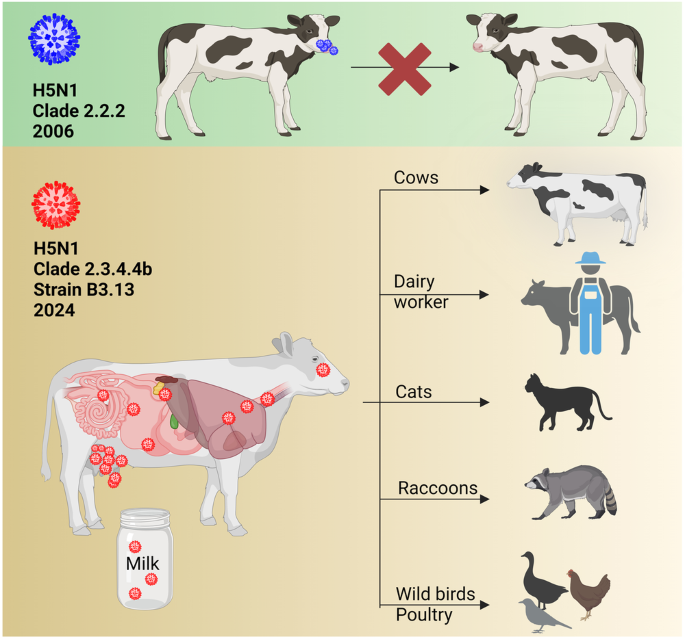
Previous studies at the Friedrich-Loeffler-Institut, Insel Riems, Germany, showed that a 2006 H5N1 isolate from a cat replicated inefficiently in calves following intranasal high-titer infection21. Furthermore, there have been no reports of IAV circulating in cattle, although some spill-over infections of IAV have been described22. Therefore, until recently the role of cattle in the epidemiology of IAV was considered insignificant21. In contrast, the HPAIV H5N1 clade 2.3.4.4b genotype B3.13 efficiently infected dairy cattle in 202423. Infected cattle may be subclinical or clinical and the main clinical signs are a drastic decrease in milk production, changes in milk consistency, decreased feed uptake, dehydration and fever23. The infection is reported to focus on lactating dairy cows and there are very high viral loads in the milk of infected cows. The spread seems to be mainly via the milking process and transport of infected cows to other farms in different states24. Apparently, efficient cow-to-cow transmission has allowed the virus to spread to 63 dairy farms in at least 9 states (status as of 05/24/2024). Sequence analysis and phylodynamics suggest a single introduction several months earlier23,25. Fatal cases in cats after drinking raw milk from infected cows24 and an infection of a dairy farm worker in Texas with a conjunctivitis have been reported26. Another study described the transmission of H5N1 from dairy cows to raccoons and to wild and domestic birds27 (Fig. 2). In addition, H5N1-RNA has been detected in wastewater28, but the source of contamination remains unknown. Importantly, recent research has raised concerns that cattle, once considered a resilient host, may act as a novel IAV host in the future and after further adaptation of H5N129. A recent preprint reports that both avian and human IAV receptors to be present in the upper respiratory tract of cattle and at high densities in bovine udder tissues29. These results were used to consider the risk of cows being a “mixing vessel” as known in pigs and fur animals. However, no other IAVs are currently circulating in cattle. Mixing in cattle can therefore be largely ruled out for the time being15. Cattle are widely infected with another Orthomyxovirus genus, “Influenza D virus (IDV)”. Serological surveillance suggests that IDV can be transmitted to humans, although very rarely compared to AIV30. And IDV uses different receptors than IAV20.
An experimental infection of calves with an early Gs/Gd isolate of H5N1/2006 clade 2.2.2, isolated from a cat in Germany and with the PB2 mutation E627K, demonstrated inefficient replication and a lack of transmission in calves following intranasal high-titer infection21. Conversely, the clade 2.3.4.4b H5N1 B3.13 genotype in the U.S. replicates efficiently in the mammary gland of dairy cows and is capable of transmission to a variety of animals, including wild and domestic birds. The figure was created with BioRender.com.
Seeking answers to many questions
Influenza A viruses are masters of mutation and adaptation. There are undoubtedly serious questions to be answered about the significant change in the epidemiology of HPAIV H5 from Asia to Europe, the dynamics of infection in wild bird reservoirs, the role of climate change in the spread of the virus, the reasons for the genetic plasticity of the current clade 2.3.4.4b viruses to reassort and adapt to new hosts, and the overall low zoonotic potential of H5 viruses from this clade31,32. Vigilance is needed to prevent new waves of infection, especially in mammals, and ultimately to prevent human infection. Dairy cows are the latest and most surprising victims in the list of AIV hosts, but they will not be the last.
Further adaptation of HPAIV H5N1 to cattle must be prevented. Stringent measures such as testing regimes (e.g. via bulk milk diagnostics), strict transport controls, quarantine measures and optimized milking hygiene must be implemented. In addition, much more basic epidemiological data is needed (e.g. seroprevalence on farms, data on non-lactating animals, H5-antibody titers in sera and milk, oro-nasal shedding data). It should also be considered that the most efficient virus shedding cows should be isolated or even euthanized. As a precautionary measure, suitable vaccines should be developed and evaluated, before the virus becomes established in these hosts.
References
Krammer, F. et al. Influenza. Nat. Rev. Dis. Prim. 4, 3 (2018).
Ma, E. J., Hill, N. J., Zabilansky, J., Yuan, K. & Runstadler, J. A. Reticulate evolution is favored in influenza niche switching. Proc. Natl Acad. Sci. USA 113, 5335–5339 (2016).
Perroncito, E. Epizoozia tifoide nei gallinacei. Annali Accad Agri Torino:87–126 (1878).
Sonnberg, S., Webby, R. J. & Webster, R. G. Natural history of highly pathogenic avian influenza H5N1. Virus Res. 178, 63–77 (2013).
Ellis, T. M. et al. Investigation of outbreaks of highly pathogenic H5N1 avian influenza in waterfowl and wild birds in Hong Kong in late 2002. Avian Pathol. 33, 492–505 (2004).
Smith, G. J. & Donis, R. O. Nomenclature updates resulting from the evolution of avian influenza A(H5) virus clades 2.1.3.2a, 2.2.1, and 2.3.4 during 2013-2014. Influenza Other Respir. Viruses 9, 271–276 (2015).
Xie, R. et al. The episodic resurgence of highly pathogenic avian influenza H5 virus. Nature 622, 810–817 (2023).
Fusaro, A. et al. High pathogenic avian influenza A(H5) viruses of clade 2.3.4.4b in Europe—Why trends of virus evolution are more difficult to predict. Virus Evolution 10, veae027 (2024).
Graziosi, G., Lupini, C., Catelli, E. & Carnaccini, S. Highly Pathogenic Avian Influenza (HPAI) H5 Clade 2.3.4.4b Virus Infection in Birds and Mammals. Anim. (Basel) 14, 1372 (2024).
Consortium, E. et al. The role of mammals in Avian Influenza: a review. EFSA Supporting Publ. 21, 8692E (2024).
Fusaro, A. et al. High pathogenic avian influenza A(H5) viruses of clade 2.3.4.4b in Europe-Why trends of virus evolution are more difficult to predict. Virus Evol. 10, veae027 (2024).
Ahrens, A. K. et al. Iceland: an underestimated hub for the spread of high-pathogenicity avian influenza viruses in the North Atlantic. J. Gen. Virol. 105 https://doi.org/10.1099/jgv.0.001985 (2024).
Fusaro, A. et al. Avian influenza overview December 2023-March 2024. Efsa J. 22, e8754 (2024).
Günther, A. et al. Iceland as Stepping Stone for Spread of Highly Pathogenic Avian Influenza Virus between Europe and North America. Emerg. Infect. Dis. 28, 2383–2388 (2022).
Abdelwhab, E. M. & Mettenleiter, T. C. Zoonotic Animal Influenza Virus and Potential Mixing Vessel Hosts. Viruses 15, 980 (2023).
Adlhoch, C. et al. Drivers for a pandemic due to avian influenza and options for One Health mitigation measures. Efsa J. 22, e8735 (2024).
USDA-APHIS (2024) Detections of Highly Pathogenic Avian Influenza in Mammals. Available online at: https://www.aphis.usda.gov/livestock-poultry-disease/avian/avian-influenza/hpai-detections/mammals (Last accessed 15-05-2024).
Verhagen, J. H., Fouchier, R. A. M. & Lewis, N. Highly Pathogenic Avian Influenza Viruses at the Wild-Domestic Bird Interface in Europe: Future Directions for Research and Surveillance. Viruses 13, 212 (2021).
Zeng, J. et al. Spatiotemporal genotype replacement of H5N8 avian influenza viruses contributed to H5N1 emergence in 2021/2022 panzootic. J. Virol. 98, e0140123 (2024).
Uprety, T. et al. Influenza D virus utilizes both 9-O-acetylated N-acetylneuraminic and 9-O-acetylated N-glycolylneuraminic acids as functional entry receptors. J. Virol. 98, e0004224 (2024).
Kalthoff, D., Hoffmann, B., Harder, T., Durban, M. & Beer, M. Experimental infection of cattle with highly pathogenic avian influenza virus (H5N1). Emerg. Infect. Dis. 14, 1132–1134 (2008).
Sreenivasan, C. C., Thomas, M., Kaushik, R. S., Wang, D. & Li, F. Influenza A in bovine species: a narrative literature review. Viruses 11, 561 (2019).
USDA. Confirmed Cases of HPAI in Domestic Livestock https://www.aphis.usda.gov/livestock-poultry-disease/avian/avian-influenza/hpai-detections/livestock (2024). Available online at (Last accessed 15.05.2024).
Burrough, E. R. et al. Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4b Virus Infection in Domestic Dairy Cattle and Cats, United States, 2024. Emerg. Infect Dis. 30 https://doi.org/10.3201/eid3007.240508 (2024).
Virological (2024) Preliminary report on genomic epidemiology of the 2024 H5N1 influenza A virus outbreak in U.S. cattle (Part 1 of 2). Available online at: https://virological.org/t/preliminary-report-on-genomic-epidemiology-of-the-2024-h5n1-influenza-a-virus-outbreak-in-u-s-cattle-part-1-of-2/970/1 (Last accessed 18-05-2024).
Harris, E. CDC: H5N1 bird flu confirmed in person exposed to cattle. JAMA 331, 1615 (2024).
Nguyen, T.-Q. et al. Emergence and interstate spread of highly pathogenic avian influenza A(H5N1) in dairy cattle. Preprint at bioRxiv https://doi.org/10.1101/2024.05.01.591751 (2024).
Tisza, M. J. et al. Virome sequencing identifies H5N1 avian influenza in wastewater from nine cities. Preprint at medRxiv https://doi.org/10.1101/2024.05.10.24307179 (2024).
Kristensen, C., Jensen, H. E., Trebbien, R., Webby, R. J. & Larsen, L. E. The avian and human influenza A virus receptors sialic acid (SA)-α2,3 and SA-α2,6 are widely expressed in the bovine mammary gland. Preprint at bioRxiv https://doi.org/10.1101/2024.05.03.592326 (2024).
Leibler, J. H. et al. Influenza D virus exposure among US cattle workers: A call for surveillance. Zoonoses Public Health 70, 166–170 (2023).
ECDC (2024) European Centre for Disease Prevention and Control. Risk assessment H5 clade 2.3.4.4b viruses. Available online at: https://www.ecdc.europa.eu/en/infectious-disease-topics/z-disease-list/avian-influenza/threats-and-outbreaks/risk-assessment-h5 (Last accessed: 18-05-2024).
WHO. Disease Outbreak News: Avian Influenza A(H5N1) - United States of America https://www.who.int/emergencies/disease-outbreak-news/item/2024-DON512 (2024). Available online at (Last accessed 18-05-2024).
Acknowledgements
The work in this study was partially funded by Deutsche Forschungsgemeinschaft (DFG; grants Nr.: AB567), KAPPA-FLU HORIZON-CL6-2022-FARM2FORK-02-03 (grant agreement No 101084171) and FLU-SWITCH Era-Net ICRAD (grant agreement No 862605).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Abdelwhab, E.M., Beer, M. Panzootic HPAIV H5 and risks to novel mammalian hosts. npj Viruses 2, 22 (2024). https://doi.org/10.1038/s44298-024-00039-z
Published:
Version of record:
DOI: https://doi.org/10.1038/s44298-024-00039-z
This article is cited by
-
The emergence of highly pathogenic avian influenza H5N1 in dairy cattle: implications for public health, animal health, and pandemic preparedness
European Journal of Clinical Microbiology & Infectious Diseases (2025)
-
Expanding wildlife serosurveillance: a study of influenza A virus exposure in Irish carnivores
European Journal of Wildlife Research (2025)
-
H5N1 clade 2.3.4.4b dynamics in experimentally infected calves and cows
Nature (2025)
-
The highly pathogenic H5N1 virus found in U.S. dairy cattle has some characteristics that could enhance infection and transmission among mammals
Lab Animal (2024)
-
Highly Pathogenic Avian Influenza A (H5N1) Virus Outbreak in Ecuador in 2022–2024
Current Infectious Disease Reports (2024)