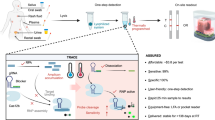
Abstract
Monkeypox (Mpox) has emerged as a critical public health challenge, creating an urgent need for rapid, reliable, field-deployable diagnostic tools for outbreak scenarios. Here, we present Kairo-CONAN, a novel CRISPR-Cas3-based point-of-care diagnostic platform for Mpox, engineered for sustainability and portability. This system uses a disposable hand warmer as a stable heat source and incorporates freeze-dried reagents for ambient temperature stability, enabling device-free, sensitive detection by lateral flow assay strips. Utilizing the unique DNA-targeting and cleavage properties of CRISPR-Cas3, we optimized probe DNA configurations for high specificity and designed clade-specific target CRISPR RNAs (crRNAs). Kairo-CONAN has demonstrated rapid, high-sensitivity, specific detection of Mpox virus (MPXV) DNA across multiple clades, including Clade Ia (Congo), Clade Ib (synthetic DNA), and Clade IIb (Tokyo). Kairo-CONAN addresses logistical and environmental challenges to offer a sustainable, cost-effective, field-adapted solution for infectious disease diagnostics, aligning with the 100 Days Mission framework to enhance global outbreak response efforts.
Similar content being viewed by others
Introduction
The recent global resurgence of monkeypox (Mpox), caused by the orthopoxvirus monkeypox virus (MPXV), has revealed critical public health challenges and underscored the urgent need for rapid, reliable, and accessible diagnostic tools, particularly in outbreak-prone regions1,2,3,4. Traditionally endemic to Central and West Africa, Mpox has spread significantly since 2022, with cases reported across multiple continents5,6. In response, the World Health Organization (WHO) classified Mpox as a Public Health Emergency of International Concern (PHEIC), emphasizing the need for field-deployable diagnostics to effectively monitor and contain outbreaks7. To address this, the G7 initiated the 100 Days Mission (100DM), calling for the development of portable, accurate, and device-free diagnostic tools to enable testing in resource-limited settings (https://ippsecretariat.org/). The current diagnostic methods for Mpox show substantial limitations. PCR-based testing, while highly sensitive and specific, requires a specialized laboratory infrastructure, trained personnel, and a stable power supply, which are often unavailable in remote or emergency settings8,9. Conversely, rapid antigen tests, although portable, lack the sensitivity and specificity required for accurate detection in diverse field environments10,11. These limitations highlight an urgent need for innovative diagnostic platforms for low-resource settings that combine high sensitivity, specificity, portability, and ease of use.
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-based diagnostics have emerged as a promising solution, offering high sensitivity and specificity with rapid, point-of-care potential without relying on complex equipment12,13,14. Some CRISPR systems, including Cas12a and Cas13, are widely used. However, CRISPR-Cas3 offers unique advantages, including the ability to target longer DNA sequences and perform collateral cleavage, enhancing specificity and reducing background noise15. These properties are particularly beneficial for precise viral detection. Our previous work with the CRISPR-Cas3-based CONAN platform16 demonstrated high specificity for nucleic acid detection, inspiring its adaptation for field-based Mpox diagnostics.
In this study, we present Kairo-CONAN, a CRISPR-Cas3-based diagnostic system optimized specifically for Mpox detection. Designed for sustainability and portability, Kairo-CONAN uses a disposable hand warmer (“Kairo” in Japanese) as a stable, device-free heat source17, and incorporates freeze-dried reagents for room-temperature stability15. To maximize detection accuracy, we have developed clade-specific CRISPR RNAs (crRNAs) and optimized the probe DNA sequences, enabling targeted detection across Mpox clades Ia, Ib, and IIb18. Validation of Kairo-CONAN demonstrated robust performance across these clades, highlighting its adaptability and reliability for pandemic preparedness. Kairo-CONAN offers a novel approach to infectious disease diagnostics by combining the specificity of CRISPR-Cas3 with a sustainable and device-free design. This platform aligns with the G7’s 100DM framework by addressing logistical and environmental barriers, supporting rapid deployment in outbreak settings and strengthening global diagnostic preparedness for Mpox and other emerging infectious diseases.
Results
CRISPR-Cas3 mediated diagnostics for monkeypox virus (MPXV)
In the CRISPR-Cas3 system, five distinct proteins (Cas5, Cas6, Cas7, Cas8, and Cas11) and crRNAs containing sequences complementary to the target form a ribonucleoprotein complex called “Cascade” (CRISPR-associated complex for antiviral defense). This Cascade complex is responsible for locating and recognizing target sequences. Subsequently, the Cas3 nuclease protein binds to the Cascade complex to induce DNA cleavage. We optimized the production of functional Cascade complexes by purifying the five Cas components (Cas5, Cas6, Cas7, Cas8, and Cas11) individually using a baculovirus expression system in Spodoptera frugiperda (Sf9) insect cells19. The purified components were then assembled in vitro with synthetic crRNA to form in vitro assembled Cascade complexes (ivCascade)20. This streamlined approach eliminates the need for the purification of recombinant Cascade complexes (rnCascade) from Escherichia coli, as required in previous studies16, significantly simplifying CRISPR-Cas3 diagnostic workflows. Our optimized approach produces stable Cascade components without crRNAs, allowing immediate use simply by mixing these with separately designed and synthesized target-specific crRNAs. This ivCascade production method enables rapid nucleic acid detection, which is particularly advantageous during pandemic scenarios, such as for COVID-19 and Mpox.
To validate the CRISPR-Cas3-based CONAN platform for MPXV detection, target sequences were designed in accordance with the Centers for Disease Control and Prevention (CDC) diagnostic guidelines9 (Fig. 1A). Using these targeting crRNAs, CRISPR-Cas3 collateral cleavage activity specific to MPXV DNA was observed after 10 minutes of incubation at 37 °C (Fig. 1B). Coupling this assay with recombinase polymerase amplification (RPA) enabled detection of MPXV sequences at concentrations as low as 100 aM (approximately dozens of copies), demonstrating the sensitivity and utility of this assay for MPXV detection (Fig. 1C).
A Schematic representation of our CRISPR-Cas3-based CONAN platform for MPXV DNA detection using in vitro assembled Cascade complexes (ivCascade) and Cas3 protein. A crRNA, common #1, was designed to target the same genomic region as the CDC qPCR primer set. B Collateral ssDNA cleavage activity measured sequentially by incubation of EcoCas3-EcoCascade/crRNA complexes with 100 nM of 100-bp dsDNA activator containing a target sequence in the MPXV genome at 37 °C. Changes in fluorescence signal intensity over time are shown in red with dsDNA and black without dsDNA (left figure). CRISPR-Cas3-mediated collateral ssDNA cleavage after targeting MPXV-dsDNA in fragments is shown, represented quantitatively by relative fluorescent units (RFU) after 10 minutes (right graph). Means (n = 3) and standard deviations are presented. Statistical analysis was performed using a two-tailed Student’s t-test. C The CONAN assay with isothermal RPA amplicon products (red) detected several copies of the MPXV activator fragments (10 aM); RFU at 10 min. Means (n = 3), and standard deviations are shown. RPA was performed for 20 minutes on DNA fragments at each concentration and CONAN assays were performed on the RPA products obtained.
Device-free nucleic acid detection using disposable hand warmers (Kairo)
During pandemics of Mpox and other diseases, a lack of approved antigen tests has underscored the need for accurate diagnostics in outbreak regions that have limited infrastructure. To address this, we have developed a device-free nucleic acid detection system that uses a disposable hand warmer (Kairo) composed of activated carbon as a heat source (Fig. 2A). Various models were tested (Fig. S1A and S1B), and a specific Kairo model stabilized at approximately 37 °C within 10 minutes (Fig. S1A), enabling effective nucleic acid amplification. Using a preheated Kairo, RPA reactions achieved comparable performance to those in a 37 °C incubator, with detectable signals within 10 minutes of adding the target sequence (Fig. 2B). The collateral cleavage activity of CRISPR-Cas3 targeting the EMX1 sequence was similarly validated with a Kairo, showing equivalent activity to standard incubation conditions (Fig. 2C). Lateral flow detection further demonstrated the successful integration of a Kairo into a fully device-free workflow (Fig. 2C). In addition, RPA using a Kairo and the CONAN assay using a Kairo also detected Mpox sequences (Fig. 2D).
A Set-up diagram illustrating the use of a hand warmer (Kairo) as an alternative and disposable heat source for the RPA and CONAN assay. B Collateral cleavage activity of EMX1-targeting CRISPR-Cas3 after RPA amplification with incubation using a thermal cycler or a hand warmer. ns: no significance. The template was 10 fM human genomic DNA derived from HEK293T cells. Statistical significance was tested using a two-way ANOVA with Sidak’s post-hoc test. C Collateral cleavage activity and lateral flow detection of CRISPR-Cas3 after the CONAN reaction with hand-warmer incubation for 10 min. RPA products from B were used. #: saturated signals on the CFX Connect device. Positive (red arrow) and negative (black arrow) bands for CONAN. D Kairo-RPA and Kairo-CONAN detecting MPXV oligo DNA. RPA was performed with 10 fM of MPXV_CDC oligo DNA for 40 min and the RPA product was detected by the CONAN reaction for 10 min. All incubations were performed using a Kairo.
We confirmed the applicability of a Kairo for reverse transcription (RT)-RPA, enabling detection of RNA viruses such as SARS-CoV-2 (Fig. S2), and also extended the system to CRISPR-Cas12a diagnostics, with consistent results (Fig. S3). Notably, Kairo-based reactions performed reliably even at 4 °C, demonstrating the feasibility of nucleic acid detection in diverse environments, including cold and off-grid settings (Fig. S4).
Lyophilized CRISPR-Cas3 components for field adaptation
To develop a device-free CRISPR diagnostic tool suitable for field use, we optimized the freeze-drying conditions for the CRISPR-Cas3 components to enable stable storage at room temperature15 (Fig. 3A). Initially, we examined the effect of trehalose, a common cryoprotectant, on the activity of both Cas3 and EMX1-targeting rnCascade16. When either Cas3 or rnCascade was lyophilized with buffers containing different concentrations of trehalose, a buffer containing 10% trehalose maintained the highest CONAN activity (Fig. S5A and S5B). This buffer was then used to lyophilize ivCascade complexes targeting EMX120. These lyophilized complexes retained activity comparable with freshly prepared counterparts after freeze-drying and thawing, confirming the stability of Cascade proteins under these conditions (Fig. 3B). CONAN activity was also observed with lyophilized ivCascade complexes targeting the common region of MPXV (Fig. S6).
A Representative image of lyophilized Cascade proteins targeting EMX1. Proteins were freeze-dried with 10% trehalose and 1× CONAN buffer, then rehydrated with H2O before the reaction. B Collateral cleavage activity of CRISPR-Cas3 with the ivCascade targeting EMX1 before and after lyophilization. ns: no significance. C Collateral cleavage activity of Cascade stored at room temperature for one week, lyophilized and non-lyophilized. D Collateral cleavage activity of CRISPR-Cas3 with Cas3 protein before and after lyophilization. ns: no significance. E Collateral cleavage activity of CRISPR-Cas3 with lyophilized or non-lyophilized Cas3 stored at 4 °C or room temperature for one week. The RFU ratio was calculated as the signal intensity with lyophilized Cas3 or non-lyophilized Cas3 divided by the signal intensity with Cas3 prepared at the time. F Collateral cleavage activity of lyophilized or non-lyophilized Cas3 and Cascade protein mix stored at 4 °C or room temperature for one week. G Collateral cleavage activity of lyophilized or non-lyophilized Cas3 and Cascade protein mix stored at 4 °C or room temperature for 30 days. The RFU ratio was calculated as the signal intensity with lyophilized Cas3 and Cascade protein mix divided by the signal intensity with Cas3 and Cascade protein mix prepared at the time. In B to G, 100 nM of 60-bp EMX1 oligo DNA was used. In B, C, D, E and F, statistical significances were tested using the two-way ANOVA with Sidak’s post-hoc test. In G, one-way ANOVA followed by Sidak’s multiple comparison test was performed. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We further assessed the stability of the lyophilized ivCascade complexes stored at both room temperature and 4 °C for one week, by comparing their activity to non-lyophilized counterparts. Lyophilized complexes exhibited significant cleavage activity under both conditions, often surpassing that of non-lyophilized samples (Fig. 3C). Similarly, Cas3 protein, known for its instability at room temperature and even at 4 °C21, was evaluated under lyophilized conditions. When freeze-dried in a 10% trehalose buffer, Cas3 retained complete activity with no detectable loss compared with pre-lyophilization levels (Fig. 3D). After one week of storage, lyophilized Cas3 maintained over 95% of its initial activity at 4 °C, whereas its activity in buffer solutions stored at the same temperature dropped by more than 70%. At room temperature, lyophilized Cas3 retained approximately 60% of its activity, while Cas3 in buffer lost all activity (Fig. 3E).
To further enhance the field applicability of the assay, we prepared a lyophilized mixture of Cas3-Cascade complexes for single-tube functionality. Stability tests showed similar profiles to those observed with Cas3 protein alone: samples stored at 4 °C maintained activity levels comparable with freshly prepared solutions, while room temperature storage retained approximately 70% of the initial activity (Fig. 3F). Furthermore, the Cas3-Cascade mix retained its activity for as long as 30 days after freeze-drying (Fig. 3G). These results demonstrate that the lyophilized Cas3-Cascade complex is stable under various storage conditions and can be reactivated with water for immediate use, making it suitable for field deployment.
Optimization of the DNA probes for enhanced sensitivity
While the lyophilized, device-free Kairo-CONAN system can be used portably, the signal sensitivity may be affected by environmental factors and the storage conditions. To enhance the CRISPR-Cas3 collateral cleavage activity, we optimized the design of the DNA probes used for single-stranded DNA (ssDNA) detection. Since Cas3 cleaves ssDNA with minimal impact on single-stranded RNA (ssRNA)21, we designed RNA probes with two DNA base substitutions to investigate the cleavage efficiency. Probes with pyrimidine bases (thymine and cytosine) at the 5′ end demonstrated higher cleavage efficiency than probes with purine bases (adenine or guanine), with guanine significantly reducing the cleavage activity (Fig. 4A). Heatmap analysis of the cleavage activity revealed that, although pyrimidine bases at the 5′ end increased cleavage activity, they also induced non-specific cleavage in the absence of target sequences. After screening for optimal signal-to-noise (S/N) ratios, sequences such as CC, CA, and TC were identified as having the highest S/N ratios (Fig. 4B).
A Collateral cleavage activity of EMX1 target sequences using RNA–DNA hybrid probes composed of two deoxynucleotides, and the heat map of the collateral cleavage activity of the two base combinations. B The signal-to-noise ratio in the presence of non-specific cleavage activity without the target. C Evaluation of the collateral cleavage activity with different lengths of adenine and cytosine repeat sequences. D Assessment of the optimal probe sequence with high signal-to-noise ratio by evaluating the collateral cleavage activity of six-base ssDNA probes with various combinations of four deoxyribonucleotides (see also Fig. S7). In A to D, 100 nM of 60-bp oligo DNA was used. E Evaluation of the collateral cleavage activity for MPXV targets with several optimized probes. MPXV 100-bp oligo DNA fragments at 1 nM, 10 nM, and 100 nM were used as templates. For each probe, a calibration curve was created by plotting the target DNA concentration against the RFU values. The relative detectability of each probe was compared by assessing how much lower concentrations of the target could be detected compared to the GAPDH probe. The performance ratio (PR) was defined as the DNA concentration required to reach a given RFU value using the GAPDH probe, divided by the DNA concentration required to reach the same RFU value using the optimised probes. F RPA was performed on 23 fM genomic DNA of the Mpox Zr-599 strain at different incubation times (0, 5, 10, 15, 20, 25, and 30 min) and CONAN activity was measured using improved probes for the RPA products (n = 3). Circle, control probe; triangle, ATATAT probe. #: saturated signals on CFX Connect device.
To determine the optimal probe length, we tested probes featuring AC sequences that were repeated at six-base intervals. While longer probes provided higher signal intensities, they also increased non-specific cleavage, indicating that shorter probes with six-base sequences were more suitable to improve the specificity (Fig. 4C). Further evaluation of the probe sequences revealed that probes with six consecutive bases of single or dual pyrimidines exhibited high specificity. However, thymine-based probes showed lower S/N ratios due to non-specific cleavage, whereas pyrimidine and adenine combinations maintained high activity with reduced non-specific cleavage, achieving improved S/N ratios (Figs. 4D and S7).
This optimization process determined that the ideal probe sequence consists of six bases, combining pyrimidine and adenine bases for enhanced cleavage specificity and S/N ratio (Fig. 4E). The optimized probe enhanced the detectability of low-concentration nucleic acids by up to 2.4-fold compared to the GAPDH (control) probe. Furthermore, in the combination of RPA and CONAN reactions, the optimized probe ATATAT enabled the detection of DNA in shorter times than with the GAPDH probe, of 10 min for RPA and 5 min for CONAN (Fig. 4F).
These optimizations are expected to enhance the overall performance of Kairo-CONAN systems, including those for Mpox detection, by enabling the detection of lower nucleic acid concentrations and reducing the RPA incubation times required.
Kairo-CONAN provides diagnostic methods for the Mpox pandemic response
Following the COVID-19 pandemic, Mpox re-emerged in Central Africa, particularly in the Democratic Republic of Congo, prompting the WHO to classify it as a Public Health Emergency of International Concern (PHEIC) in August 20247. In response, the G7 launched the 100 Days Mission (100DM) initiative to accelerate the deployment of diagnostics, vaccines, and treatments within 100 days of a PHEIC declaration, aiming to ensure rapid and effective medical countermeasures (https://ippsecretariat.org/). Kairo-CONAN, a lyophilized and device-free CRISPR-Cas3 diagnostic platform, has potential as a robust solution for low-resource settings during pandemics. To evaluate its adaptability, we validated clade-specific detection using newly released Clade Ib sequences22,23. Sequences for Mpox Clades Ia, Ib, and IIb were sourced from the CDC and NCBI databases, and target sequences were designed and synthesized as crRNAs8,9,22,23 (Fig. S8). This synthesis process took approximately two weeks, demonstrating the ability of the platform to be deployed rapidly after the release of target sequences.
To evaluate the sequence-specific cleavage activity for each clade, we generated 120-base pair (bp) DNA fragments and validated their clade-specific detection capability. All crRNAs showed robust cleavage signals, confirming the accuracy of the assay, and we were able to select the crRNAs that demonstrated the strongest signals (crRNA #1 for Clade Ia, #3 for Clade Ib, and #1 for Clade IIb; Fig. 5A). We proceeded to validate the Kairo-CONAN platform using genomic DNA from two Mpox strains, Zr-599 (Clade Ia) and TKY220091 (Clade IIb). When testing the detection sensitivity using DNA fragments and qPCR primer sets from the CDC24, we achieved detection sensitivity at the 100 aM level (Fig. 5B). After quantifying the genomic DNA amounts of Zr-599 (Clade Ia) and TKY220091 using qPCR standard curves, we validated the detection specificity and sensitivity of each target-specific crRNA. The common crRNA #1 detected both RPA samples of Clades Ia and IIb MPXV at the aM level. Additionally, the Clade Ia-specific crRNA showed collateral cleavage only with Zr599, the Clade Ib-specific crRNA reacted only with synthetic DNA, and the Clade IIb-specific crRNA showed collateral cleavage only of TKY22009. This demonstrates detection sensitivity in the tens of aM range, comparable with the sensitivity of qPCR (Fig. 5C). For practical field applications, lateral flow strips enabled detection of low viral DNA concentrations in the 150 aM range (Fig. 5D, S9A, S9B, S9C and S9D). Furthermore, Kairo-RPA and Kairo-CONAN could only detect the MPXV genome, even when the human genome was also present (Fig. S10). Finally, the lyophilized Cas3-Cascade mix and optimized probes were used to detect low concentrations of the MPXV genome by Kairo-RPA-Kairo-CONAN (Fig. 5E). We successfully detected 150 aM of the MPXV genome after a 10-min RPA and a 5-min CONAN reaction (Fig. 5F). These results confirm that the Kairo-CONAN assay aligns with the G7’s 100DM framework and represents a valuable diagnostic tool for the response to the Mpox outbreak in resource-limited environments.
A Target-specific collateral cleavage activity of MPXV-targeting CRISPR-Cas3 to MPXV Clade Ia, Ib, and IIb amplicons by the RPA reaction. #: saturated signals on the CFX Connect device. B The limit of detection (LoD) of the CDC qPCR assay amplification of MPXV. Cq, cycle quantification value. C LoD for the combination of the RPA and CONAN assays for the CDC common crRNA and Clade Ia-, Ib- and IIb-specific crRNAs to the genomic DNA from MPXV strains Zr-599 (Clade Ia) and TKY220091 (Clade IIb). For the Clade Ib crRNA, 1 pM of Oligo DNA MPXV Ib was used as a positive control. The RPA reactions were performed using a thermal cycler for 40 min. #: saturated signals on the CFX Connect device. D Clade-specific detection by the Kairo-CONAN-based lateral flow assay on 150 aM of genomic DNA from MPXV strains Zr-599 and TKY220091 (see also Fig. S10). The RPA reaction was performed for 40 min and the CONAN reaction for 10 min, with all incubations performed using a Kairo. The test band intensities on the lateral flow strips were quantified by the gray values using the ImageJ tool. For Clade Ib crRNA, 150 aM of Oligo DNA MPXV Ib was used as the positive control. E Schematic diagram of viral nucleic acid detection by RPA and CONAN using Kairo, lyophilized enzymes and optimized probe. F Lateral flow assay targeting 150 aM genomic DNA from MPXV strain TKY220091 (Clade IIb). The RPA reaction was performed for 10 min and the CONAN reaction for 5 min, and all incubations were performed using a Kairo. A lyophilized Cas3-Cascade protein mix targeting MPXV common #1 and the ATATAT probe were used for CONAN. In the lower panel, test band intensities on the lateral flow strips were quantified by the gray values using the ImageJ tool.
Discussion
The rapid and global reemergence of Mpox has underscored the pressing need for effective, field-deployable diagnostic tools that can operate reliably under outbreak conditions, particularly in regions with limited infrastructure3,4. This study presents Kairo-CONAN, a CRISPR-Cas3-based diagnostic platform optimized for Mpox detection in diverse environments. We also established clade-specific detection for Mpox using crRNAs targeting Clades Ia, Ib, and IIb. By leveraging the unique DNA-targeting properties of CRISPR-Cas3, Kairo-CONAN achieves high sensitivity and specificity, fulfilling key diagnostic criteria in outbreak settings.
The present study achieves portability, which has not been accomplished in previous studies that have combined CRISPR-Cas3 with isothermal nucleic acid amplification. Our work addresses critical limitations by optimizing the reaction conditions and pre-reaction storage under completely electricity-free situations, and by developing novel detection probes that significantly reduce reaction times. The key technical novelties of the Kairo-CONAN method established herein include the complete elimination of electrical equipment by optimizing the conditions for both disposable hand-warmer usage and reagent freeze-drying, and substantial improvements in reaction time, reducing to less than half of that of conventional CONAN methods, by the development of novel detection probes with enhanced signal intensities. These advances enable PCR-level detection sensitivity, with a simplicity comparable with antigen testing. Additionally, we achieved strain-specific detection of viral genomes among several viral strains with similar target sequences.
There have been many reports of CRISPR diagnostics using various CRISPR systems. Unlike Cas13, which recognizes RNA, CRISPR-Cas3 recognizes DNA, similar to Cas12. CRISPR-Cas3 offers unique diagnostic benefits compared with Cas12: it recognizes a different protospacer adjacent motif (PAM) sequence, has a longer target recognition sequence of 27 bases, and exhibits higher specificity, able to distinguish even single nucleotide differences. These features reduce the risks of false positives while enabling specific detection of various viruses with different sequences, as well as drug-resistant strains and variants. Indeed, we have demonstrated that MPXV strains can be easily distinguished without extensive crRNA screening.
While numerous reports have documented field-deployable applications of Cas12 and Cas13, no field-deployable Cas3 systems have been reported. Through optimization under electricity-free conditions and the development of novel probes that significantly reduce reaction times, we expect to achieve point-of-care diagnostic capability equal to, if not superior to, existing methods.
During probe optimization, we found that CRISPR-Cas3 has a sequence preference for ssDNA for collateral cleavage, particularly for the presence of a pyrimidine base in the 5′ position. We propose that Cas3 nuclease activity requires structural changes upon binding to target-recognizing Cascade complexes, allowing ssDNA access to the nuclease domain. Smaller pyrimidine bases may be more readily introduced into the Cas3 cleavage site, resulting in enhanced cleavage activity between pyrimidine bases. On the other hand, the ssDNA cleavage activity of Cas3 nuclease is greatly affected by the concentration of divalent ions21, and thus the observation of this phenomenon in the buffer conditions of the CONAN reaction in this study may vary with changes in ion concentration. Since the crystal structure of E. coli-derived Cas3 remains unsolved, future detailed structural analyses may elucidate the complex molecular mechanisms governing Cas3 ssDNA cleavage.
Here, our device-free design, incorporating a disposable hand warmer (Kairo) as a sustainable heat source17 and freeze-dried reagents for stability15, addresses logistical challenges in low-resource settings. The versatility of Kairo-CONAN has been demonstrated by the detection of RNA viruses, such as SARS-CoV-2, using RT-RPA (Fig. S2). Thus, Kairo-RPA and Kairo-CONAN may become universal virus testing methods, able to detect both RNA and DNA viruses as soon as the nucleic acid sequences are identified. We have also shown that Kairo methods can be applied to DETECTR with Cas12a (Fig. S3). Its compatibility with both Cas12a and Cas13 further broadens its applicability to various nucleic acid targets12,13,14,16,25,26, supporting pandemic preparedness for diverse pathogens.
The ideal is to be able to make diagnoses in all environments. Importantly, the solution temperature is optimally adjusted to around 37 °C. In this study, the usefulness of Kairo-based methods was demonstrated in low-temperature environments and at around 25 °C. The temperature to which a Kairo heats is influenced by the ambient temperature. Our results showed that at low temperatures (~4 °C), Kairo-CONAN was clearly necessary for signal generation. At room temperature (26 °C), Kairo-CONAN provided clear band signals in most experiments, highlighting the importance of defining the environmental factors when developing the package.
Additionally, the lyophilized CRISPR-Cas3 components retained high activity after room-temperature storage for one week and for 30 days (Fig. 3F, G), overcoming the limitations of traditional PCR-based diagnostics that require freezing. However, 4 °C storage provides higher and more stable activity preservation. Since at present we cannot completely maintain activity at room temperature, we currently envision refrigerated storage with room-temperature transport to field sites, although we expect that complete room-temperature storage may become possible through future detailed optimization of the freeze-drying conditions, including additional stabilizers.
We intend to enable people to perform lateral flow assays, allowing interpretation to be performed without special equipment because the simple criterion of the presence or absence of a band is easily understood by general consumers. While here we have used image analysis to present data as numerical values, the presence or absence of the virus can be easily distinguished by comparison with a negative control. During our experiments, we observed that the test strips remained functional even after being left to dry for more than one week, suggesting a low risk of false results. Accordingly, we believe that test results can be easily interpreted even by non-experts. However, while the lateral flow assay sometimes appeared to show a band even when there was no target (Fig. S4A), mechanical performance of the same experiment did not produce false positive signals, as shown in Fig. S4B. In some cases, the quality of the test paper has a large impact on the determination of the presence or absence of bands.
In this study, the procedure involves only simple mixing of solutions and incubation by sandwiching with hand warmers. We are still working to simplify the process, for example by using a single tube instead of the current use of two, and we aim to ultimately produce a kit that can be easily used by anyone, not just experts, that guarantees the stability of the results.
While we were able to validate detection of MPXV DNA samples for Clades Ia and IIb, Clade Ib could not be tested because of the unavailability of appropriate samples22,23. Although we have shown that the MPXV genome can be detected, more practical experiments using clinical samples are required. We contacted various hospital institutions to obtain clinical samples from MPXV patients. Unfortunately, we were unable to secure any clinical samples, and no clear timeline for this is currently available. As an alternative approach to demonstrate clinical applicability, we conducted mock clinical tests using samples containing mixtures of human genomic DNA and MPXV, and showed that only MPXV can be detected. We acknowledge that validation with genuine clinical specimens remains necessary, and plan to perform such validation once samples become available.
In conclusion, Kairo-CONAN represents a significant advance in portable, high-sensitivity Mpox diagnostics, with broader applications for both RNA and DNA viruses. By addressing logistical and environmental challenges, this platform aligns with the G7’s 100 Days Mission, offering a scalable and adaptable solution for outbreak response and pandemic preparedness (https://ippsecretariat.org/). The eco-friendly design, high specificity, and field adaptability position Kairo-CONAN as a transformative tool for global health initiatives aimed at rapid disease detection and containment.
Methods
Protein synthesis
Cas3, a mixture of Cascade components, and recombinant Cascade were synthesized as previously described16,20,21. Briefly, EcoCas3 cDNA was cloned with an octa-histidine tag and a six asparagine-histidine repeat tag into a pFastbac-1 plasmid (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The TEV protease recognition site was also inserted between the tags and EcoCas3 to enable tag removal. All plasmid sequences we used are uploaded as GenBank files.
To produce a mixture of Cascade components, a plasmid was designed in which each Cascade component gene was cloned into pFastbac-1 by incorporating nuclear localization signal (NLS) sequences at the N- and C-termini and a HisTag sequence at either the N- or C-terminus, linked by 2A-peptide. The expression and purification of the mixture of Cascade components in Sf9 was performed by the Bac-to-Bac baculovirus expression system using Sf9 insect cells (Thermo Fisher Scientific Inc., Carlsbad, CA, USA). The plasmid encoding the 2A-tandemed Cascade genes was transformed into DH10bac competent cells. Each purified bacmid was transfected into Sf9 cells, and high titer baculoviruses for each were acquired by repeatedly infecting new cells with baculovirus in the culture supernatant. Acquired baculovirus was added to cultured Sf9 cells at two multiplicities of infection (MOI), then cultured at 28 °C for 24 h. After infection, the culture temperature was lowered to 20 °C for 3 days for protein expression. Sf9 cells were collected and stored at −80 °C until purification. After ultrasonic disruption of the cells, homogenized samples were ultracentrifuged and the supernatant collected, which included the Cas proteins. The Cas proteins in the supernatant were purified using nickel affinity resin (Ni-NTA, Qiagen). Superdex 200 Increase 10/300 GL columns (Cytiva Tokyo, Japan) were used for gel filtration chromatography.
Recombinant Cascade complexes for EMX1 were purified in accordance with previously reported methods21,27,28. Briefly, the recombinant Cascade ribonucleoproteins (RNPs) were expressed in JM109 (DE3) by co-transformation with three plasmids: one plasmid encoding a hexahistidine tag and the HRV 3 C protease recognition site in the N-terminus of Cas11 (plasmid pCDFDuet-1); one plasmid containing the Cascade genes encoding Cas5, Cas6, Cas7, Cas8, and Cas11 (plasmid pRSFDuet-1); and the final plasmid encoding the crRNAs (pACYCDuet-1). The transformed bacteria were cultured in 2× YT medium at 37 °C in a shaking incubator at 130 rpm. After the OD600 reached 0.6–0.8, we added IPTG (final concentration, 0.4 mM) and cultured at 26 °C and 110 rpm for 16 hours. The expressed Cascade-crRNA RNPs were purified by Ni-NTA resin. After removing the hexahistidine tag using HRV 3 C protease, the recombinant Cascade RNPs were further purified by size-exclusion chromatography in 350 mM NaCl, 1 mM DTT, and 20 mM HEPES-Na (pH 7.0), and size-evaluated by SDS-PAGE.
Preparation of DNA and RNA
For the Cascade/Cas3 activator templates, DNA fragments of hEMX1 and MPXV variants (which include a target site) were designed and purchased from Integrated DNA Technologies (IDT) or AZENTA Life Sciences (Tokyo) (Table S1). Monkeypox virus genomic DNA was prepared as previously described29. Two MPXV strains, Zr-599 (Congo Basin strain; clade I) and the 2022 outbreak strain (MPXV/human/Japan/Tokyo/TKY220091/2022; clade IIb; GenBank accession no. LC722946.1), were propagated using VeroE6 cells (kindly gifted by Tokyo Metropolitan Institute of Public Health), and DNA was extracted. The S protein sequence of SARS-CoV-2 Omicron (EPI_ISL_6913953.2) was cloned into the TOPO vector (TOPO TA Cloning Kit) and introduced into E. coli DH5α cells. Plasmid purification was performed using the Pure Link HiPure Plasmid Filter Midiprep kit (Invitrogen K210015). In vitro transcription was performed using SuperScript IV Reverse Transcriptase and RNA obtained for subsequent experiments. The primers used for PCR, qPCR, and RPA are listed in Table S2. The probes were designed with the addition of FAM or HEX and were purchased from IDT or AZENTA (Table S3).
Design and preparation of crRNAs
The Cascade/Cas3 target sites were based on the human EMX1 gene, the N gene from SARS-CoV-2, regions common to MPXV strains, and regions characteristic for each MPXV strain (Table S4). The crRNAs were purchased from Integrated DNA Technologies. Before the CONAN reaction, each crRNA and the Cascade proteins were mixed and incubated at 37 °C for 10 minutes.
Preparation of freeze-dried Cas3 and Cascade protein
Cas3 and/or Cascade protein were mixed in 20 µL freeze-drying buffer containing 10% (w/v) d-trehalose, 60 mM KCl, 10 mM MgCl2, 10 µM CoCl2, 5 mM HEPES-KOH (pH 7.5) and stored at −80 °C before the freeze-drying process. After pre-freezing, freeze-drying was performed for 20–96 h with a freeze-dry system (the Versatile and flexible Freeze Dryer; Tokyo Rikakikai Co., Ltd.).
Recombinase polymerase amplification (RPA)
RPA of the target regions for the CONAN reaction was performed using a TwistAmp Basic kit (TwistDx, Maidenhead, UK). RPA reaction solutions were prepared following the manufacturer’s instructions. Briefly, freeze-dried pellets were dissolved in a solution containing primers and rehydration buffer, template DNA was added, and 280 mM magnesium acetate was added and incubated using a thermal cycler at 37 °C or a Kairo for 5–40 min.
Nucleic acid detection using CONAN
CONAN reactions were performed as previously described16. Briefly, DNA templates containing DNA fragments, PCR products, and RPA products were added to 100 nM Cascade-crRNA complex, 400 nM Cas3, 1 mM ATP, and CRISPR-Cas3 system working buffer (60 mM KCl, 10 mM MgCl2, 10 μM CoCl2, 5 mM HEPES-KOH pH 7.5). Single-stranded DNA probes were added at a final concentration of 1 µM. CONAN reaction solutions were incubated at 37 °C for 10–30 min with a thermal cycler or qPCR system. When using a Kairo, after opening the Kairo bag and shaking it five times, a 200 µL microtube containing the CONAN reaction solution was sandwiched between the Kairo and incubated for 10 min. Changes in the fluorescence values associated with the cleavage of single-stranded DNA probes were measured over time or at endpoints using a CFX Connect (Bio-Rad Laboratories, Hercules, CA).
Detection of MPXV by qPCR
The MPXV genome concentrations were quantified using a SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) and a CFX Connect. The cycling conditions were as follows: 94 °C for 2 min, followed by 40 cycles of 98 °C for 10 s and 60 °C for 15 s. The primers used were as previously reported24 (Table S2).
Detection of CONAN reactions by lateral flow assay
For lateral flow assays, DNA template was added to 50 nM Cascade-crRNA complex, 200 nM Cas3, 1 mM ATP, and CRISPR-Cas3 system working buffer (60 mM KCl, 10 mM MgCl2, 10 µM CoCl2, and 5 mM HEPES-KOH, pH 7.5). After 2 µL of DNA template was added to a total of 17 µL of CONAN reaction solution, 1 µL of biotin-containing single-stranded DNA probes were added at a final concentration of 250 nM. CONAN reaction solutions were incubated with a thermal cycler at 37 °C or a Kairo for 10 min. After adding 80 µL of the Milenia GenLine Dipstick Assay Buffer provided with the Lateral Flow Kit (Millenia GenLine HybriDetect, Milenia Biotec, Gießen, Germany), a lateral flow strip was added to the reaction tube and the appearance of the test band was observed after 3 minutes. The test band intensities on the lateral flow strips were quantified by the gray values using the ImageJ tool.
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism 10 software (GraphPad Software, MA, USA). Image analysis was performed using the ImageJ tool. All data is reported as mean ± standard deviation as described, where applicable. Two-tailed Student’s t-tests were performed in Figs. 1B and S2A. Statistical significance was tested using a two-way ANOVA with Sidak’s post-hoc test in Figs. 2A and 3B, F. A one-way ANOVA followed by Sidak’s multiple comparison test was performed in Fig. 3G. Significance is reported as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Data availability
The research findings presented in this study are supported by data included in the main text and in the Supplementary Information.
References
Mitja, O. et al. Monkeypox. Lancet 401, 60–74 (2023).
Gessain, A., Nakoune, E. & Yazdanpanah, Y. Monkeypox. N Engl J Med 387, 1783–1793 (2022).
Lim, C. K. et al. Mpox diagnostics: Review of current and emerging technologies. J Med Virol 95, e28429 (2023).
Titanji, B. K., Hazra, A. & Zucker, J. Mpox Clinical Presentation, Diagnostic Approaches, and Treatment Strategies: A Review. JAMA https://doi.org/10.1001/jama.2024.21091 (2024).
Olawade, D. B. et al. Strengthening Africa’s response to Mpox (monkeypox): insights from historical outbreaks and the present global spread. 3 https://doi.org/10.1016/j.soh.2024.100085 (2024).
Ilic, I., Zivanovic Macuzic, I. & Ilic, M. Global Outbreak of Human Monkeypox in 2022: Update of Epidemiology. Trop. Med. Infect. Dis. 7 https://doi.org/10.3390/tropicalmed7100264 (2022).
Gostin, L. O., Jha, A. K. & Finch, A. The Mpox Global Health Emergency — A Time for Solidarity and Equity. New England Journal of Medicine 391, 1265–1267 (2024).
Nakhaie, M. et al. Monkeypox virus diagnosis and laboratory testing. Rev Med Virol 33, e2404 (2023).
Minhaj, F. S. et al. Orthopoxvirus Testing Challenges for Persons in Populations at Low Risk or Without Known Epidemiologic Link to Monkeypox - United States, 2022. MMWR Morb Mortal Wkly Rep 71, 1155–1158 (2022).
Davis, I. et al. Development of a specific MPXV antigen detection immunodiagnostic assay. Front Microbiol 14, 1243523 (2023).
Ye, L. et al. Gold-based paper for antigen detection of monkeypox virus. Analyst 148, 985–994 (2023).
Kaminski, M. M., Abudayyeh, O. O., Gootenberg, J. S., Zhang, F. & Collins, J. J. CRISPR-based diagnostics. Nat Biomed Eng 5, 643–656 (2021).
Mohammad, N., Katkam, S. S. & Wei, Q. Recent Advances in CRISPR-Based Biosensors for Point-of-Care Pathogen Detection. CRISPR J 5, 500–516 (2022).
Kostyusheva, A. et al. CRISPR-Cas systems for diagnosing infectious diseases. Methods 203, 431–446 (2022).
Kongkaew, R., Uttamapinant, C. & Patchsung, M. Point-of-care CRISPR-based Diagnostics with Premixed and Freeze-dried Reagents. J. Vis. Exp. https://doi.org/10.3791/66703 (2024).
Yoshimi, K. et al. CRISPR-Cas3-based diagnostics for SARS-CoV-2 and influenza virus. iScience 25, 103830 (2022).
Honma, M. et al. Primary cutaneous anaplastic large cell lymphoma successfully treated with local thermotherapy using pocket hand warmers. J Dermatol 35, 748–750 (2008).
Verma, A. et al. Mpox 2024: New variant, new challenges, and the looming pandemic. Clinical Infection in Practice 24 https://doi.org/10.1016/j.clinpr.2024.100394 (2024).
Hitchman, R. B., Possee, R. D. & King, L. A. Baculovirus expression systems for recombinant protein production in insect cells. Recent Pat Biotechnol 3, 46–54 (2009).
Asano, K. et al. CRISPR Diagnostics for Quantification and Rapid Diagnosis of 2 Myotonic Dystrophy Type 1 Repeat Expansion Disorders. ACS Synthetic Biol. in press https://doi.org/10.1021/acssynbio.4c00265 (2024).
Yoshimi, K. et al. Dynamic mechanisms of CRISPR interference by Escherichia coli CRISPR-Cas3. Nat Commun 13, 4917 (2022).
Vakaniaki, E. H. et al. Sustained human outbreak of a new MPXV clade I lineage in eastern Democratic Republic of the Congo. Nat Med 30, 2791–2795 (2024).
Kinganda-Lusamaki, E. et al. Clade I mpox virus genomic diversity in the Democratic Republic of the Congo, 2018-2024: Predominance of zoonotic transmission. Cell https://doi.org/10.1016/j.cell.2024.10.017 (2024).
Li, Y., Zhao, H., Wilkins, K., Hughes, C. & Damon, I. K. Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA. Journal of Virological Methods 169, 223–227 (2010).
Mustafa, M. I. & Makhawi, A. M. SHERLOCK and DETECTR: CRISPR-Cas Systems as Potential Rapid Diagnostic Tools for Emerging Infectious Diseases. Journal of Clinical Microbiology 59, 00745–00720 (2021).
Zhou, J., Li, Z., Seun Olajide, J. & Wang, G. CRISPR/Cas-based nucleic acid detection strategies: Trends and challenges. Heliyon 10, e26179 (2024).
Hochstrasser, M. L. et al. CasA mediates Cas3-catalyzed target degradation during CRISPR RNA-guided interference. Proc Natl Acad Sci USA 111, 6618–6623 (2014).
Jore, M. M. et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol 18, 529–536 (2011).
Watanabe, Y. et al. Virological characterization of the 2022 outbreak-causing monkeypox virus using human keratinocytes and colon organoids. Journal of Medical Virology 95, e28827 (2023).
Acknowledgements
We thank Tatsuo Serikawa for his valuable insights and contributions to the conceptualization of this study. We are also grateful to Hiromi Taniguchi and Yuko Yamauchi at the University of Tokyo for their technical assistance with the in vitro assays, as well as to Sachiko Yamamoto and Shuku Saji at the RIKEN SPring-8 Center for their support with protein extraction and purification. We thank Catherine Perfect, MA (Cantab), from Edanz (https://jp.edanz.com/ac), for editing a draft of this manuscript. This research was supported in part by JSPS KAKENHI from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT) (grant numbers 19KK0401, 22K19238, 22H02266, 23H00367, 24K02010) and a grant from the Japan Agency for Medical Research and Development (AMED) (grant numbers 23ck0106807, 24bm12230009, 223fa627001, JP24jf0126002, JP243fa627001h0003, JP243fa727002, 23fk0108583, JP24fk0108690, 25fk0310550h0001).
Author information
Authors and Affiliations
Contributions
K.Y. and T.M. conceived and supervised the study, interpreted the data, and drafted the manuscript. R.H. conducted the primary experiments and analyzed the data with assistance from K.A. K.T. was responsible for the preparation and purification of all CRISPR-Cas3 proteins. K.S. and K.J.I. provided insights into study design and assisted in data interpretation. All authors contributed to the manuscript, reviewed it critically, and approved the final version prior to submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hirano, R., Yoshimi, K., Asano, K. et al. Sustainable and portable CRISPR-based diagnostics for high-sensitivity Mpox detection. npj Biosensing 2, 43 (2025). https://doi.org/10.1038/s44328-025-00062-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44328-025-00062-x