Abstract
Wound healing is a complex process involving spatiotemporal patterning of cellular activity across four overlapping phases: hemostasis, inflammation, proliferation, and remodeling, which restore anatomic and functional tissue integrity. Recent advances in cell-based technologies have increased focus on cell populations, heterogeneity, and phenotypes during healing. This manuscript reviews traditional and emerging technologies that advance our understanding of the cellular biology of wounds, from histological methods to high-resolution single-cell, spatial-, and multi-omics.
Similar content being viewed by others
Introduction
Wounds represent a consequential source of morbidity and mortality across the United States, affecting more than 8 million patients and costing upwards of 96 billion dollars in annual Medicare expenditures1. Chronic non-healing wounds, which include pressure ulcers, venous/arterial ulcers, and diabetic foot ulcers (DFUs), affect 10.5 million Medicare beneficiaries in the U.S.2. Chronic wounds are defined by failure to heal within 8–12 weeks and loss of native function and anatomical integrity, leading to impaired mobility and increased risk of prolonged or recurrent hospitalizations due to infections. DFUs are a particularly debilitating type of wound that arise from poorly managed or longstanding diabetes. Up to 34% of patients with diabetes will develop a DFU during their lifetime, with 30% of DFU patients experiencing a recurrence within one year and >90% experiencing recurrence in 10 years2,3. In contrast, acute or complex wounds often progress to hypertrophic scar formation (HTS), a pathological scarring response characterized by excess collagen deposition and fibroblast activity. HTS can lead to functional, cosmetic, and psychological complications, especially when scars form over areas of high mechanical stress or in sensitive areas such as the face. Given the severe morbidity and mortality associated with both acute and chronic wounds, including a 5-year mortality rate of approximately 50% for some chronic wounds, advancements in the field of wound healing are crucial.
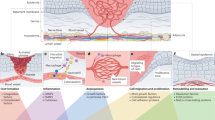
Cutaneous wound healing is a physiological process involving a concerted event of several key cellular events across multiple cell types (e.g., immune cells, fibroblasts, keratinocytes, and endothelial cells) and tissues (Fig. 1).
A–E Schematic demonstrating the stages of wound healing following A tissue injury, B hemostasis, C inflammation, D proliferation, and E remodeling. A Tissue injury (e.g., ischemia, trauma, infection, autoimmune, malignancy) results in a breakdown of physiological tissue integrity, thus initiating innate physiological tissue repair pathways to restore anatomical and physiological function. B Hemostasis (1st stage): Following tissue injury, blood vessels constrict, the coagulation cascade initiates, and activated platelets form a clot to limit bleeding. C Inflammation (2nd stage): Chemoattractant (e.g., chemokines and cytokines) released from platelets and other cells/tissues recruit neutrophils and macrophages to the local wound environment to clear bacteria and debris. These recruited leukocytes, in turn, secrete chemoattractants that result in the recruitment of keratinocytes and fibroblasts. D Proliferation (3rd stage): Proliferation and differentiation of fibroblast cell populations proliferate and differentiate, resulting in collagen synthesis and deposition. In parallel, granulation tissue is formed as the wound microenvironment begins to undergo neovascularization. E Remodeling (4th stage): The wound’s fibrotic extracellular matrix remodels over months to years into mature scar tissue. Figure created in BioRender. Hostler, A. (2025).
The cellular response to injury occurs in four overlapping phases: hemostasis, inflammation, proliferation, and remodeling4. Immediately following tissue injury, the body works to achieve hemostasis, the first stage in wound healing. Platelets are the first to arrive at the site of injury, forming a hemostatic plug, and release numerous growth factors, including platelet-derived growth factor (PDGF), transforming growth factor-beta (TGF-β), and vascular endothelial growth factor (VEGF), which initiate the recruitment of immune cells5,6. The inflammatory phase is in the second stage of wound healing, characterized by homing of immune cells to the wound site. Neutrophils are among the earliest immune cells recruited to the wound site and perform pathogen clearance, releasing reactive oxygen species (ROS) and proteolytic enzymes to neutralize bacteria and digest damaged tissue7. Neutrophils also form neutrophil extracellular traps (NETs), which enhance their antimicrobial capabilities. Macrophages are recruited to the site where they undergo a phenotypic shift to an anti-inflammatory phenotype and secrete VEGF and other anti-inflammatory cytokines to promote the resolution of the inflammatory phase. This event transitions the wound healing process to proliferation, activating collagen-producing fibroblasts and neovascularization. Proliferation dominates between days 5–21 and is marked by fibroblast deposition of extracellular matrix (ECM) components composed predominantly of collagen. During long-term remodeling (>1 year), fibroblasts re-organize and remodel collagen as it matures7.
Each phase involves dynamic interactions among cells, ECM components, and signaling cascades, and dysfunctions in these phases can prolong healing or even lead to chronic and potentially life-threatening complications. For example, excessive neutrophil activity can lead to tissue damage, as observed in chronic wounds8. To better understand the factors behind non-healing wounds, it is critical to investigate the cellular dynamics and mechanisms underlying impaired wound healing. Here, we review the evolution of current techniques to study cells involved in wound healing.
Historically validated techniques in wound healing analysis
Histopathology and immunohistochemistry (IHC)/immunofluorescence (IF)
Histopathological analysis (histology) of human tissue remains the gold standard for cell and wound healing assessment. This method involves the microscopic analysis of tissue morphology to evaluate inflammation, epithelialization, collagen deposition, and tissue remodeling. Common stains include Hematoxylin and Eosin (H&E), which highlight nuclear and cytoplasmic components, respectively; Masson’s Trichrome, which differentiates muscle fibers, collagen, and cytoplasm; and Picrosirius Red, which stains explicitly collagen fibers under polarized light. For example, in our published work using a deep partial-thickness porcine wound model, histological analysis demonstrated significantly accelerated wound closure in wounds treated with a focal adhesion kinase inhibitor (FAKI) hydrogel (Fig. 2).
A–C Representative histological and immunological images of red Duroc pig scars following 25 cm2 deep partial-thickness wounding. Wounds were treated with standard dressings alone (Wound), pullulan-collagen hydrogels (Wound + Hydrogel), or focal adhesion kinase inhibitor (FAKI)-infused hydrogels (Wound + FAKI hydrogel) for 90 days, and scars were allowed to remodel until the postoperative day (POD) 180. A Masson’s Trichrome stained images of healed scars demonstrate more significant collage deposition in Wound and Wound + Hydrogel groups than in Wound + FAKI-hydrogel. Wound + FAKI-hydrogel groups demonstrated a more significant presence of cutaneous glands (black arrows), hair follicles (yellow arrows), and intradermal adipocytes (yellow dashed arrow). Scale bar: 200 µm. B Healed scar images stained with Picrosirius Red (PSR) demonstrating that the pharmacological blockade of FAKI-infused Hydrogels demonstrated a greater basket weave-like collagen fiber network, most like unwounded skin, when compared against Wounds and Wound + Hydrogel. Scale bar: 10 µm. C Immunofluorescent staining for DAPI, nuclear marker, and αSMA, myofibroblast marker, demonstrating reduced αSMA expression in wounds treated with a FAKI-infused hydrogel compared to traditional wound dressings. Scale bar: 200 µm. Images were adapted and reproduced with permission from Chen et al. (2021) Nat. Commun.9.
In these published findings, we observed that scar formation was significantly reduced after 3 months and that scar appearance continued to improve throughout 6 months. Trichrome staining was used to visualize fibrosis and the presence or absence of secondary hair appendages (an indication of regeneration), such as hair follicles or sweat glands, and wounds treated with FAKI hydrogel showed visual hair follicle regrowth with Trichrome Staining at 90 and 180 days after injury. The collagen structural network of FAKI hydrogel-treated wounds closely resembled the structural features of normal unwounded pig skin in which collagen fibrils form a “basket-weave” architecture. Using Picrosirius Red staining, we observed that collagen fibers of untreated control and placebo wounds showed highly aligned scars characterized by elongated bundle structures, demonstrating that FAK plays an important role in collagen and extracellular matrix remodeling following injury9.
While histology provides valuable information about overall tissue structure, it lacks cellular specificity. To overcome this limitation in wound healing research, antigen-specific probing techniques utilizing immunostaining (e.g., immunohistochemistry (IHC) and immunofluorescence (IF)) can be used to visualize specific cellular markers. For example, IF staining of the protein, alpha-smooth muscle actin (αSMA), a marker of myofibroblasts, allowed the evaluation of a molecular feature of scar fibrosis9. IF is also commonly used to identify the infiltration of polarized macrophages by probing for markers such as CD86 for phenotypically “pro-inflammatory” macrophages, or CD163 for phenotypically “anti-inflammatory” macrophages10,11. Neutrophils may also be visualized based on cell surface markers such as ICAM1, CD11a, CD15, CD44, CD45, and CXCR412,13,14. While single-cell protein analysis may be observed with these techniques, the investigator must have previous suspicion to probe for specific markers in each sample. Furthermore, dynamic cellular processes such as active transcription, translation, and secretion of products like growth factors and cytokines cannot be assessed from the visualization of a fixed tissue specimen.
Complementary techniques for collagen analysis
In addition to conventional histology and immunofluorescence methods, several additional techniques have been utilized to evaluate collagen structure and organization during wound healing. These techniques provide complimentary insights into collagen’s biochemical composition, molecular organization, and architecture. Second-harmonic generation (SHG) microscopy is a nonlinear optical imaging technique used to visualize fibrillar collagen without exogenous labeling. Specifically, SHG resolves collagen fiber density, alignment, and remodeling15,16. Other methods such as Fourier transform infrared (FTIR) spectroscopy and Raman spectroscopy provide information on collagen structure and crosslinking at the molecular level. These techniques specifically detect changes to collagen protein secondary structure or functional groups17,18. Small-angle X-ray scattering (SAXS) also allows for the quantitative assessment of collagen fibril orientation and spacing at the nanoscale, characterizing collagen maturation and hierarchical structure19. Together, these techniques offer complementary insights into collagen architecture and extracellular matrix dynamics, enabling a comprehensive understanding of wound healing.
Molecular techniques
Quantitative real-time polymerase chain reaction (qRT-PCR) is commonly used to assess the transcriptional status of select genes and infer changes in cellular transcriptional profiles. Primers are designed to amplify specific wound healing-related genes, such as Transforming growth factor-beta (TGFβ1), matrix metalloproteinases (MMPs) such as MMP1, MMP2, MMP9, and other key factors with expression levels typically normalized to a housekeeping gene such as GAPDH. TGFβ1 is a key gene indicating inflammation and re-epithelialization in wound healing. MMPs are also critical markers for wound healing, as they regulate key cellular processes, such as angiogenesis and tissue remodeling, by targeting various components of the extracellular matrix. If probing a large tissue sample, increased or decreased gene transcription may be a signature of one cell population, but this granularity cannot be assessed using qRT-PCR. Western blot is a typical complement to PCR to quantify proteins of interest in a tissue or cell sample. In line with the pitfalls of IF, genes or proteins of interest must be selected based on previous suspicion or the current body of literature using these techniques. Furthermore, PCR and western blotting are performed on all cells or whole tissue in a sample, which does not differentiate expression between cell types.
Microarray technology, introduced in 1990, is another molecular tool used to identify differentially expressed genes in a cell sample using cDNA probes. cDNA microarray technology is a valuable tool for studying the gene signatures of wound healing over time, as well as comparing hypertrophic and keloid scars20,21. For instance, microarray profiling of human keratinocytes revealed events of wound healing as early as 30 minutes after the injury, up to 15 h, including the expression of c-Fos, c-Jun, and Egr122. However, microarray probes are limited in resolution because they cannot detect unknown gene variants. Additionally, although microarray technology is effective for identifying overall gene signatures, this assay requires pooling mRNA from thousands of cells within a heterogenous population, which reduces the resolution of individual cell populations.
Recently, single-cell resolution has become critical in the study of wound healing, as it aids in identifying rare and dynamic cell types and signatures. Advanced tools, such as microfluidics-based technologies, offer a powerful approach by sorting individual cell populations, elucidating specific cellular markers and offering increased granularity needed to appreciate interactions and mechanisms at play during wound healing.
Microfluidic techniques and scRNA-seq to achieve single-cell resolution to study wounds
Novel single-cell sequencing analytics have identified cellular heterogeneity and enabled the identification of rare and dynamic cell types and transcriptional signatures. At first, microfluidics-based technology allowed researchers to achieve single-cell resolution of cell signatures involved in wound healing. For instance, flow cytometry is a technique used for quantifying cell characteristics such as size, granularity, and the expression of specific surface markers or intracellular components (Fig. 3).
This technique quantifies live cells, but the populations are not sorted or returned to the user. Fluorescence-activated cell sorting (FACS), on the other hand, collects cells based on user-directed criteria, allowing the user to manipulate isolated cell populations (Fig. 3). Both techniques are high throughput and utilize microfluidic technology to manipulate small fluid volumes through microchannels, resulting in single-cell suspension and isolation in a flow stream.
Single-cell RNA sequencing (scRNA-seq) has advanced wound healing research since its debut in 2009 (Fig. 4).
A Droplet-based technology used to isolate single cells from a sample by using capture beads and oil isolation techniques in a microfluidic device. The cells are separated, and a gene expression library is generated; this information is then analyzed through various computational tools. (*) Capture beads, with distinct barcodes on each bead, classify individual cells. The barcode on the capture beads is attached to a unique molecular identifier (UMI), which is attached to the capture sequence of the cell. B The re-clustering of fibroblast subpopulations from wound samples from diabetic or non-diabetic patients. The three clusters (sc1, sc2, and sc3) are transcriptionally unique. Sc1 is a fibrotic and non-regenerating, sc2 is collagen producing, and sc3 is pro-fibrotic cluster27. C Feature plots of known wound healing and fibrosis genes, PTK1, COL1A1, and JUN in fibroblast subpopulations. Figures adapted from Januszyk et al. (2021), Micromachines27.
ScRNA-Seq has transformed gene expression profiling by enabling researchers to analyze individual cell signatures, rather than average gene expression from a bulk tissue or heterogeneous cell sample23. ScRNA-seq uses a droplet-based technology to isolate single cells from a sample containing a large population of heterogeneous cells. Individual cells from a sample are isolated using a microfluidic chip, where single cells are separated and captured by beads with unique barcodes. Each cell’s RNA is then sequenced, generating a library of transcriptional profiles linked to the bead’s barcode. These unique molecular identifiers ultimately provide detailed gene expression data for each cell in the sample24. In the context of wound healing, scRNA-seq data provides valuable insights into heterogeneous genetic signatures expressed during tissue repair, even within the same cell type.
scRNA-seq has previously been utilized to characterize changes in cellular subpopulations over the course of fibrotic wound healing. Specifically, scRNA-seq at early and late time points after full-thickness porcine injuries identified an upregulation of mechanoresponsive macrophage subpopulations at early time points, followed by differentiation of pro-fibrotic fibroblast subpopulations at later time points. Further investigation of heterogeneous fibroblasts identified that normal fibrotic healing triggered upregulation of pro-chondrogenic, ECM-depositing fibroblasts expressing genes such as ACAN or THSB4, while pharmacologic disruption of mechanical signaling with a focal adhesion kinase inhibitor (FAKI) differentiated fibroblasts into a more pro-adipogenic transcriptional profile25.
Conventional preservation methods have proved challenging for the maintenance of cellular membranes and preparing high-quality single-cell suspensions for scRNA-seq analysis. However, recent improvements in cryopreservation techniques now support the successful use of primary cells in scRNA-seq26. Our group has also shown that small volumes of cells obtained from human debridement tissue can be successfully processed for scRNA-seq analysis, even after storage at –80 °C, sample barcoding/pooling, and without FACS27. We demonstrated differences in cell populations within clinical wound samples from diabetic and non-diabetic patients. When comparing whole-transcriptome gene expression data of fibroblast populations from these patient’s foot ulcers, we identified three transcriptionally distinct subgroups (subclusters (sc) sc1, sc2, sc3). These populations were characterized by differentially enriched expression of the known wound healing and fibrosis genes PTK2, COL1A1, and JUN4,28. Notably, diabetic and non-diabetic debridement tissue showed considerable differences in these three clusters, with diabetic cells significantly enriched among collagen-producing and pro-fibrotic subpopulations, consistent with a tissue-wide shift toward a non-healing fibrotic state18. We then identified that sc1 cells showed transcriptional shifts toward either sc2 or sc3 in diabetic debridement tissue, suggesting differentiation into a more fibrotic and non-regenerative phenotype. Theocharidis et al. also described significant transcriptome heterogeneity of fibroblasts collected from patients with healed diabetic foot ulcers compared to non-healers. These healing-enriched fibroblasts demonstrated high expression of extracellular matrix remodeling (MMP1, MMP3) and inflammation-associated (CHI3L1, TNFAIP6) genes27.
A direct hybridization-based transcriptomic technique can be used to measure gene expression from a predefined panel without the need for amplification29. In a study assessing the transcriptome of chronic wounds in diabetic mice, this technique was employed to elucidate metabolic and injury response pathways enriched in chronic wounds30. While this assay is useful in illustrating gene expression using minimal concentrations of RNA and reduces the risk of amplification bias or errors seen in qRT-PCR, it lacks spatial context for the analysis of genes within a specific tissue architecture.
Spatial transcriptomics
While scRNA-seq provides valuable insights into single-cellular gene expression and resolves cellular heterogeneity in a given sample, the technology lacks spatial context. Therefore, spatial transcriptomics offers a more comprehensive view of gene expression within tissue architecture, an important aspect for studying wounds. Spatial transcriptomics technologies can be broadly categorized into two main types: sequencing- and in situ detection-based approaches, with some emerging platforms integrating aspects of both. Sequencing-based methods rely on spatial barcodes and unique molecular identifiers to generate RNA reads that are mapped back to their original tissue coordinates, resulting in a spatially annotated library (Fig. 5).
A–C: A Spatial transcriptomics begins with a unique slide that contains a capture area comprised of 2 × 2 µm spots, with each spot capturing ~1–10 cells. The spots contain primers that have unique molecular identifiers (UMI), a spatial barcode, and a poly(dT)VN. After the genetic reads are picked up at the spots, scRNA data is produced and analyzed, therefore providing single-cell data with a spatial component. B Spatial localization of scRNA-seq-defined fibroblast subpopulations onto tissue slides. C Tissue slides for POD 7 and 14 of mouse excisional wounds. These slides show that Cluster 1 cells migrate from the deep to superficial wound between POD 7 and 14, whereas Cluster 3 cells migrate toward the wound center. Figures adapted from (Foster et al, 2021, PNAS) and (A) created with BioRender. Hostler, A. (2025).
In contrast, in situ approaches detect transcripts directly in tissue samples using multiplexed fluorescence imaging, resulting in subcellular resolution without RNA extraction. Hybrid platforms are emerging, leading to the enhancement of resolution and precision of transcriptomic mapping31.
Our group has previously demonstrated that spatial transcriptomics can localize scRNA-seq-defined cell populations within healing wounds32. Using paired scRNA-seq and spatial transcriptomics data collected from mouse excisional wounds at various time points, we identified four distinct fibroblast subpopulations involved in normal wound healing. Anchor-based label transfer was used to deconvolve each transcriptomic “spot” (consisting of 1–10 cells) into partial memberships from these subpopulations. The inferred cell distribution closely matched the scRNA-seq proportions and manual fibroblast counts, supporting the utility of this approach for spatially resolving fibroblast populations in histological sections.
Theocharidis et al. also used spatial transcriptomics to show that their healing-associated fibroblasts, identified via scRNA-seq, preferentially localize to the wound bed, rather than the edges of the wound, and were not present in non-wounded samples33. In line with these findings, Liu et al. leveraged scRNA-seq and spatial transcriptomics in a unique in vivo human wound healing model, creating a spatiotemporal cell atlas of human wound healing34. Interestingly, their group identified previously underappreciated cytokines (CXCL1 and CXCL5), and keratinocyte ligand-receptor interactions (THBS1, LGALS3, and TNF) involved in the wound microenvironment.
Spatial transcriptomics have been traditionally limited by large spatial regions that combine expression in a region consisting of 5–10 cells. However, these techniques are rapidly evolving with new platforms approaching true single-cell resolution35. Overmiller et al. investigated the role of PITX1, a transcription factor expressed in keratinocytes, in promoting cutaneous wound healing by homing keratinocytes and neutrophils into the wound bed using an advanced single-cell spatial transcriptomics approach36. In contrast to barcoded mRNA capture spots within defined tissue regions, other spatial transcriptomics technologies utilize patterned arrays of DNA nanoballs to capture spatially resolved transcripts at submicron resolution across large tissue areas34,37. This technique enables ultra-high-resolution mapping of entire wounds or multiple healing stages within a single experiment, facilitating detailed analyses of microenvironmental interactions. In addition to experimental approaches to spatial transcriptomics, newer technologies include computational approaches. These methods integrate scRNA-seq data with existing spatial transcriptomics references to infer spatial relationships of cells32.
To further complement transcriptional data, spatial proteomic technologies are now emerging, allowing in situ protein quantification and localization at single-cell resolution. Unlike mRNA, protein levels reflect the functional output of cells and are influenced by post-transcriptional modifications. Developing technologies permit the simultaneous detection of dozens of protein markers across large tissue sections, preserving spatial relationships and tissue architecture. As these methods become more widely available, they will offer additional opportunities to study wound healing in a spatiotemporal manner38,39.
Conclusion and future directions
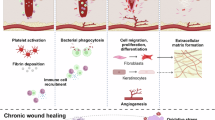
This review underscores the potential of advanced therapeutic approaches targeting specific cellular mechanisms to enhance outcomes in chronic wounds and fibrotic conditions (Fig. 6).
Emerging technologies like single-cell spatial transcriptomics and proteomics offer unprecedented insights into the molecular and cellular landscape of wound healing, highlighting the importance of understanding gene expression profiles and spatial cell interactions. While single-cell RNA sequencing provides valuable gene expression data with excellent resolution, it lacks spatial context—an essential element in wound healing, where cell-cell interactions and localization within tissue niches are fundamental. Therefore, integrating spatial transcriptomics with single-cell resolution and proteomics are crucial for advancing our knowledge of wound healing, enabling more precise identification of cell types, their interactions, and the mechanisms driving tissue regeneration. This enhanced understanding will pave the way for more effective therapeutic strategies for chronic wounds.
The most advanced technologies recently developed for the investigation of wound healing emphasize real-time and spatial visualization of wound progression. Incorporating a temporal dimension into wound healing studies enables researchers to gain a more comprehensive understanding of the dynamic changes underlying these complex biological processes. This concept is exemplified by recent in vivo assays, in which researchers employ 3D cell cultures combined with fluorescent-tagging of specific analytes to visualize both the extent and nature of intercellular communication in real time40. In parallel, advances in temporal transcriptomics have provided new insights into the dynamic molecular landscape of wound healing. For instance, a study by Zhuang et al. constructed a spatiotemporal atlas of human gene expression throughout distinct stages of the wound healing process. By integrating scRNA-seq with spatial transcriptomics (Visium ST-seq) across multiple time points, the study yielded a comprehensive dataset that illuminates the structural organization of the epidermal wound environment and elucidates key regulatory pathways involved in re-epithelialization. Temporal mapping of wound healing dynamics is emerging as a transformative approach in the field, and the integration of single-cell spatial transcriptomics with temporal resolution holds significant promise for improving diagnostic accuracy, prognostic capability, and therapeutic strategies as this methodology continues to evolve.
Further integration of spatial transcriptomic and proteomic methodologies will enable a comprehensive multi-omic view of wound biology, allowing for the correlation of gene and protein expression with cell identity, function, and spatial localization within the tissue microenvironment. Although we have recently performed an initial multi-omic analysis41,42, more work is required integrate our findings within a spatial framework. Together, the integration of temporal and spatial resolution in transcriptomic and proteomic analyses marks a significant leap forward in our ability to study wound healing with unprecedented detail. By capturing the dynamic molecular and cellular landscape of tissue repair, these approaches are reshaping our understanding of wound biology.
Data availability
No datasets were generated or analyzed during the current study.
References
Nussbaum, S. R. et al. An economic evaluation of the impact, cost, and medicare policy implications of chronic nonhealing wounds. Value Health 21, 27–32 (2018).
Sen, C. K. Human wound and its burden: updated 2022 compendium of estimates. Adv. Wound Care 12, 657–670 (2023).
Huang, Z. H. et al. Risk factors for the recurrence of diabetic foot ulcers among diabetic patients: a meta-analysis. Int. Wound J. 16, 1373–1382 (2019).
Gurtner, G. C., Werner, S., Barrandon, Y. & Longaker, M. T. Wound repair and regeneration. Nature 453, 314–321 (2008).
Rodrigues, M., Kosaric, N., Bonham, C. A. & Gurtner, G. C. Wound healing: a cellular perspective. Physiol. Rev. 99, 665–706 (2019).
Barrientos, S., Stojadinovic, O., Golinko, M. S., Brem, H. & Tomic-Canic, M. PERSPECTIVE ARTICLE: growth factors and cytokines in wound healing. Wound Repair Regen. 16, 585–601 (2008).
Singer, A. J. & Clark, R. A. F. Cutaneous wound healing. N. Engl. J. Med. 341, 738–746 (1999).
Wilgus, T. A., Roy, S. & McDaniel, J. C. Neutrophils and wound repair: positive actions and negative reactions. Adv. Wound Care 2, 379–388 (2013).
Chen, K. et al. Disrupting biological sensors of force promotes tissue regeneration in large organisms. Nat. Commun. 12, 5256 (2021).
Lv, X., He, Z., Yang, M., Wang, L. & Fu, S. Analysis of subsets and localization of macrophages in skin lesions and peripheral blood of patients with keloids. Heliyon 10, e24034 (2024).
Adams, S., Wuescher, L. M., Worth, R. & Yildirim-Ayan, E. Mechano-immunomodulation: mechanoresponsive changes in macrophage activity and polarization. Ann. Biomed. Eng. 47, 2213–2231 (2019).
Katayama, Y., Hidalgo, A., Chang, J., Peired, A. & Frenette, P. S. CD44 is a physiological E-selectin ligand on neutrophils. J. Exp. Med. 201, 1183–1189 (2005).
Chen, J. et al. CREB1-driven CXCR4(hi) neutrophils promote skin inflammation in mouse models and human patients. Nat. Commun. 14, 5894 (2023).
Yang, P., Li, Y., Xie, Y. & Liu, Y. Different faces for different places: heterogeneity of neutrophil phenotype and function. J. Immunol. Res. 2019, 8016254 (2019).
Chen, X., Nadiarynkh, O., Plotnikov, S. & Campagnola, P. J. Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat. Protoc. 7, 654–669 (2012).
Campagnola, P. J. & Loew, L. M. Second-harmonic imaging microscopy for visualizing biomolecular arrays in cells, tissues and organisms. Nat. Biotechnol. 21, 1356–1360 (2003).
Al-Kelani, M. & Buthelezi, N. Advancements in medical research: Exploring Fourier Transform Infrared (FTIR) spectroscopy for tissue, cell, and hair sample analysis. Skin Res. Technol. https://doi.org/10.1111/srt.13733 (2024).
Becker, L. et al. Raman microspectroscopy identifies fibrotic tissues in collagen-related disorders via deconvoluted collagen type I spectra. Acta Biomater. 162, 278–291 (2023).
Barreto, I. S. et al. Nanoscale characterization of collagen structural responses to in situ loading in rat Achilles tendons. Matrix Biol. 115, 32–47 (2023).
Chen, W. et al. Analysis of differentially expressed genes in keloids and normal skin with cDNA microarray. J. Surg. Res. 113, 208–216 (2003).
Smith, J. C., Boone, B. E., Opalenik, S. R., Williams, S. M. & Russell, S. B. Gene profiling of keloid fibroblasts shows altered expression in multiple fibrosis-associated pathways. J. Invest. Dermatol. 128, 1298–1310 (2008).
Dayem, M. A. et al. Early gene expression in wounded human keratinocytes revealed by DNA microarray analysis. Comp. Funct. Genomics 4, 47–55 (2003).
Wang, X., He, Y., Zhang, Q., Ren, X. & Zhang, Z. Direct comparative analyses of 10X genomics chromium and Smart-Seq2. Genom. Proteom. Bioinforma. 19, 253–266 (2021).
He, J., Lin, L. & Chen, J. Practical bioinformatics pipelines for single-cell RNA-seq data analysis. Biophys. Rep. 8, 158–169 (2022).
Chen, K. et al. Disrupting mechanotransduction decreases fibrosis and contracture in split-thickness skin grafting. Sci. Transl. Med. 14, eabj9152 (2022).
Guillaumet-Adkins, A. et al. Single-cell transcriptome conservation in cryopreserved cells and tissues. Genome Biol. https://doi.org/10.1186/s13059-017-1171-9 (2017).
Januszyk, M. et al. Characterization of diabetic and non-diabetic foot ulcers using single-cell RNA-sequencing. Micromachines. https://doi.org/10.3390/mi11090815 (2020).
Wong, V. W. et al. Focal adhesion kinase links mechanical force to skin fibrosis via inflammatory signaling. Nat. Med. 18, 148–152 (2011).
Goytain, A. & Ng, T. NanoString nCounter technology: high-throughput RNA validation. Methods Mol. Biol. 2079, 125–139 (2020).
Basu, P., Kim, J. H., Saeed, S. & Martins-Green, M. Using systems biology approaches to identify signalling pathways activated during chronic wound initiation. Wound Repair Regen. 29, 881–898 (2021).
Lim, H. J., Wang, Y., Buzdin, A. & Li, X. A practical guide for choosing an optimal spatial transcriptomics technology from seven major commercially available options. BMC Genomics. https://doi.org/10.1186/s12864-025-11235-3 (2025).
Foster, D. S. et al. Integrated spatial multiomics reveals fibroblast fate during tissue repair. Proc. Natl. Acad. Sci. USA 118, e2110025118 (2021).
Theocharidis, G. et al. Single cell transcriptomic landscape of diabetic foot ulcers. Nat. Commun. https://doi.org/10.1038/s41467-021-27801-8 (2022).
Liu, Z. et al. Spatiotemporal single-cell roadmap of human skin wound healing. Cell Stem Cell 32, 479–498.e478 (2025).
Nagendran, M. et al. 1457 Visium HD enables spatially resolved, single-cell scale resolution mapping of FFPE human breast cancer tissue. J. Immunother. Cancer 11, A1620–A1620 (2023).
Overmiller, A. M. et al. Reprogramming of epidermal keratinocytes by PITX1 transforms the cutaneous cellular landscape and promotes wound healing. JCI Insight. https://doi.org/10.1172/jci.insight.182844 (2024).
Li, S. et al. CellContrast: Reconstructing spatial relationships in single-cell RNA sequencing data via deep contrastive learning. Patterns 5, 101022 (2024).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell 174, 968–981 (2018).
Mascharak, S. et al. Multi-omic analysis reveals divergent molecular events in scarring and regenerative wound healing. Cell Stem Cell 29, 315–327 (2022).
McGhee, A., McGhee, E., Famiglietti, J. E. & Sawyer, W. G. In situ 3D spatiotemporal measurement of soluble biomarkers in organoid culture. In Vitro Model. 1, 309–321 (2022).
Mascharak, S. et al. Multi-omic analysis reveals divergent molecular events in scarring and regenerative wound healing. Cell Stem Cell 29, 315–327.e316 (2022).
Chen, K. et al. Targeting circulating mechanoresponsive monocytes and macrophages to reduce fibrosis. Nat Biomed Eng. https://doi.org/10.1038/s41551-025-01479-5. (2025).
Acknowledgements
We would like to thank Nick Price and Theresa Carlomagno for providing administrative support. Figures 1, 3, 4A, 5A and 6 were created with BioRender.com. Some figure subpanels are adapted from published manuscripts from the authors: Figure 2, images adapted and reproduced with permission Chen et al. (2021) Nature Communications, Figure 4B, C adapted from Januszyk et al. (2021), Micromachines, and Figure 5B, C adapted from Foster et al. (2021), PNAS.
Author information
Authors and Affiliations
Contributions
K.B.F. and E.K.K. designed the prospective parameters, K.B.F. and E.R.G. performed the literature search, wrote the manuscript, K.B.F., E.R.G., and A.H. prepared the figures. K.B.F. reviewed and edited the manuscript. K.C. and G.C.G. conceived the concept, reviewed, and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Berthiaume Fox, K.A., Galvin, E.R., Kness-Knezinskis, E. et al. Decoding wound healing: cellular insights and technological advances. npj Biomed. Innov. 3, 1 (2026). https://doi.org/10.1038/s44385-025-00046-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44385-025-00046-6