Abstract
Automatic differential diagnosis (DDx) involves identifying potential conditions that could explain a patient’s symptoms and its accurate interpretation is of substantial significance. While large language models (LLMs) have demonstrated remarkable diagnostic accuracy, their capability to generate high-quality DDx explanations remains underexplored, largely due to the absence of specialized evaluation datasets and the inherent challenges of complex reasoning in LLMs. Therefore, building a tailored dataset and developing novel methods to elicit LLMs for generating precise DDx explanations are worth exploring. We developed the first publicly available DDx dataset, comprising expert-derived explanations for 570 clinical notes, to evaluate DDx explanations. Meanwhile, we proposed a novel framework, Dual-Inf, that could effectively harness LLMs to generate high-quality DDx explanations. To the best of our knowledge, it is the first study to tailor LLMs for DDx explanation and comprehensively evaluate their explainability. Overall, our study bridges a critical gap in DDx explanation, enhancing clinical decision-making.
Similar content being viewed by others
Introduction
Differential diagnosis (DDx), a critical component of clinical care, involves generating a list of potential conditions that could explain a patient’s symptoms1. It facilitates comprehensive case evaluation, identifies critical but subtle conditions, guides diagnostic testing, and optimizes resource utilization. Additionally, DDx fosters patient involvement and trust through improved communication. While numerous automatic DDx systems2,3 have been developed to support decision-making, their black-box nature, particularly in deep learning models, often undermines trust4. To address this, providing interpretative insights alongside diagnostic predictions is essential5. Explainable DDx, which takes patient symptom descriptions as input, generates differential diagnoses, and offers accompanying explanations, is thus highly desirable in clinical practice.
In recent years, large language models (LLMs), such as ChatGPT, trained on extensive corpora, have exhibited remarkable capabilities in various clinical scenarios, including medical question answering (QA)6,7,8,9, clinical text summarization10, and disease diagnosis11,12,13,14,15,16. Motivated by these advancements, some studies have explored LLMs to improve diagnostic accuracy17. For instance, Daniel et al.18 fine-tuned PaLM 2 on medical data and developed an interactive interface to assist clinicians with DDx generation, while Savage et al.19 refined Chain-of-Thought (CoT) prompting20 to harness LLMs’ reasoning capabilities.
Despite these efforts, the potential of LLMs to generate reliable DDx explanations remains largely unexplored, leaving their role in supporting clinical decision-making uncertain. Two key challenges impede progress in this domain. First, the absence of DDx datasets annotated with diagnostic explanations limits model development and evaluation21,22. Second, numerous studies have highlighted LLMs’ inherent difficulties with complex reasoning tasks23,24, such as multi-step logical reasoning25,26 and clinical decision-making27,28. Thus, creating tailored datasets and developing novel methodologies to enable LLMs to synthesize high-quality DDx explanations are worthy of exploration.
In this study, we addressed these challenges by investigating prompting strategies for generating trustworthy DDx explanations. Our contributions are threefold. First, we curated a new dataset of 570 clinical notes across nine specialties, sourced from publicly available medical corpora and annotated by domain experts with differential diagnoses and explanations. To our knowledge, this is the first publicly available structured dataset with DDx explanation annotation21,29, which facilitates automated evaluation and holds substantial potential to advance the field. Second, we proposed Dual-Inf, a customized framework to optimize LLMs’ explanation generation capabilities. The core design lies in enabling LLMs to perform bidirectional inference (i.e., from symptoms to diagnoses and vice versa), leveraging backward verification to boost prediction correctness. Third, we comprehensively evaluated Dual-Inf for explainable DDx, including model explainability and error analysis. The results demonstrated that Dual-Inf achieved superior diagnostic performance while delivering reliable interpretations across various base LLMs (i.e., GPT-4, GPT-4o, Llama3-70B, and BioLlama3-70B). Overall, our findings highlight the effectiveness of Dual-Inf as a promising tool for improving clinical decision-making.
Results
Dataset
We developed Open-XDDx, a well-annotated dataset for explainable DDx, consisting of 570 clinical notes from publicly available medical exercises across nine specialties: cardiovascular, digestive, respiratory, endocrine, nervous, reproductive, circulatory, skin, and orthopedic diseases. Each note includes patient symptoms, differential diagnoses, and expert-derived explanations from the University of Minnesota (Supplementary Appendix 1). The dataset statistics are detailed in Table 1 and Table 2.
Differential diagnosis performance
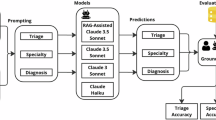
We evaluated differential diagnosis accuracy (Eq. 1) by comparing model predictions to ground-truth diagnoses with prompts (Supplementary Appendix 3). The results with GPT-4 and GPT-4o are depicted in Fig. 1(b), and the results with Llama3-70B and BioLlama3-70B are presented in Supplementary Appendix 4. It showed that Dual-Inf consistently outperformed baselines across nine specialties. Specifically, when built on GPT-4, the overall performance of SC-CoT significantly exceeded CoT (difference of 0.032, 95% CI 0.021–0.043, p = 0.001) and Diagnosis-CoT (difference of 0.019, 95% CI 0.001–0.028, p = 0.004). Dual-Inf further surpassed SC-CoT (0.533 vs. 0.472, difference of 0.061, 95% CI 0.055–0.062, p < 0.001). Concretely, the performance improvement of Dual-Inf over SC-CoT exceeded 16% on cardiovascular and digestive diseases. Similarly, using GPT-4o, Dual-Inf achieved over 0.55 accuracy on nervous, skin, and orthopedic diseases, exceeding the baselines by over 9%. With Llama3-70B and BioLlama3-70B, Dual-Inf outperformed SC-CoT by over 10% in cardiovascular, digestive, and respiratory diseases. The overall performance improvement of Dual-Inf over SC-CoT across the three base LLMs (difference of 0.059, 0.048, and 0.049) was statistically significant (p < 0.001).
a An overview of the Dual-Inference Large Language Model framework (Dual-Inf) for explainable DDx. Dual-Inf consists of four components: (1) a forward-inference module, which is an LLM to generate initial diagnoses from patient symptoms, (2) a backward-inference module, which is an LLM for conducting inverse inference via recalling all the representative symptoms associated with the initial diagnoses, i.e., from diagnoses to symptoms, (3) an examination module, which is another LLM to receive patients’ notes and the output from the two modules for prediction assessment (e.g., completeness examination) and decision making (e.g., filtering out low-confidence diagnoses), and (4) an iterative self-reflection mechanism, which iteratively takes the low-confidence diagnoses as feedback for the forward-inference module to “think twice”. b Differential diagnosis performance built on two base LLMs (GPT-4 and GPT-4o) over nine specialties. The results are averaged over five runs. Standard deviations are also shown.
Interpretation performance
Model explainability was examined through automatic and human assessments. For automatic evaluation, GPT-4o was employed to measure the consistency between ground-truth and predicted interpretations, utilizing prompts detailed in Supplementary Appendix 3. We tested four base LLMs (GPT-4, GPT-4o, Llama3-70B, and BioLlama3-70B). Partial results on GPT-4 are shown in Fig. 2(a), with additional results in Supplementary Appendix 5. In Fig. 2(a), the interpretation accuracy (Eq. 2) of Diagnosis-CoT and SC-CoT was 0.305 and 0.334, surpassing CoT by 0.011 (95% CI 0.004–0.019, p = 0.012) and 0.04 (95% CI 0.038–0.043, p < 0.001), respectively. Dual-Inf achieved even higher accuracy at 0.446, with a 0.112 improvement over SC-CoT (95% CI 0.105–0.118, p < 0.001). Concretely, the performance improvement of Dual-inf over the baselines surpassed 26% in cardiovascular and respiratory diseases. For BERTScore, SentenceBert, and METEOR, Dual-Inf outperformed SC-CoT with comparisons of 0.345 vs. 0.258 (difference of 0.087, 95% CI 0.083–0.090, p < 0.001), 0.427 vs. 0.356 (difference of 0.071, 95% CI 0.067–0.076) and 0.333 vs. 0.251 (difference of 0.082, 95% CI 0.076–0.088). When taking GPT-4o as the base LLM, the interpretation accuracy of Dual-Inf reached 0.488, outperforming CoT and SC-CoT, which scored 0.366 and 0.408, respectively. On the other metrics, Dual-Inf consistently surpassed SC-CoT, with differences of 0.083, 0.064, and 0.08. Similarly, with Llama3-70B and BioLlama3-70B, Dual-Inf exceeded SC-CoT by over 17% across all metrics. In detail, the performance improvement on digestive, respiratory, and endocrine diseases exceeded 25% over the baselines w.r.t interpretation accuracy (Supplementary Appendix 5).
a Interpretation performance w.r.t interpretation accuracy (see Eq. 2) and BERTScore across nine clinical specialties. We implemented the methods with GPT-4. The results were averaged over five runs. Standard deviations were also shown. b Human evaluation results on interpretation. It assessed three aspects: correctness, completeness, and usefulness, with scores ranging from 1 to 5. c Error type analysis on interpretation. We manually examined 100 cases and recorded the count of the error type. Diag-CoT denotes Diagnosis-CoT, and Self-Cont means Self-Contrast. The results were averaged over five runs. The methods are implemented with GPT-4.
The interpretations were also manually examined by clinicians on three qualitative metrics: Correctness, Completeness, and Usefulness (Supplementary Appendix 2). Figure 2b presents the results for 100 randomly selected notes, using GPT-4 as the base LLM. We observed that the Correctness score of Dual-Inf predominantly ranged from 3 to 4, whereas SC-CoT scores mainly fell between 2 and 3. In terms of Completeness score, Dual-Inf achieved 38 scores of 3 and 21 scores of 4, compared to SC-CoT’s 19 and 3, respectively. Regarding the Usefulness score, Dual-Inf had 33 scores of 3 and 25 scores of 4, while SC-CoT had 26 and 10, respectively.
Case study
We further provided case studies to demonstrate the superior explainability of Dual-Inf over the baselines. The example in Fig. 3 showcased that SC-CoT only provided three correct explanations for a differential, i.e., Pneumothorax, while Dual-Inf generated more accurate explanations. Besides, Dual-Inf had one more correct differential, i.e., Hemothorax, with three correct explanations than the baselines. See more examples and detailed illustrations in Supplementary Appendices 7 and 8.
Error analysis on explanation
We analyzed error types in generated explanations by comparing Dual-Inf with the baselines on 100 randomly selected samples with incorrect outputs. Errors were categorized as missing content (missing at least two pieces of evidence), factual errors (medically incorrect), or low relevance (evidence not highly pertinent) based on prior studies30,31. Using GPT-4 as the base LLM (Fig. 2(c)), SC-CoT had 89 cases of missing content versus 76 for Dual-Inf (difference 13.4, 95% CI 11.5–15.2). For factual errors, the baselines achieved similar performance, and the count number comparison between Dual-Inf and SC-CoT was 17 vs. 8.2 (difference 8.8, 95% CI 7.8–9.8). As for low-relevance, Self-Contrast and SC-CoT had fewer errors than the CoT and Diagnosis-CoT, while the comparison between SC-CoT and Dual-Inf was 15.4 vs. 10.8 (difference 4.6, 95% CI 3.9–5.3). All differences were statistically significant (p < 0.001). We further presented the count of errors in each clinical specialty in Supplementary Appendix 6. The results demonstrated that the errors fell into all the specialties, while the nervous and digestive diseases specialty had more errors.
Ablation study
We evaluated the contribution of each component in Dual-Inf through four variants: (1) forward-inference only (FI), (2) FI with excluded backward-inference (FI-EM), (3) FI-EM without self-reflection (FI-EM*), and (4) Dual-Inf without self-reflection (Dual-Inf*). Specifically, we adopted automatic metrics for the evaluation. The results in Supplementary Appendix 12 confirmed that Dual-Inf achieved superior diagnostic accuracy and explainability, highlighting the necessity of all components.
Discussion
Our study demonstrated that Dual-Inf significantly enhanced diagnostic accuracy by filtering low-confidence diagnoses through quality assessment. Specifically, the examination module consolidated outputs from other components to verify correctness, while the self-reflection mechanism enabled the forward-inference module to refine predictions iteratively. To evaluate iterative reflection, we tracked the iteration count for each note in Dual-Inf (Fig. 4a), revealing that most predictions were iteratively revised. For randomly selected ten notes with five iterations, the number of correct diagnoses improved progressively (Fig. 4c), confirming the effectiveness of the iterative reflection mechanism. Besides, we observed that most of the notes’ prediction correctness was boosted or remained stable in the fourth or fifth iteration, demonstrating the necessity of setting the maximum iteration λ to a relatively large value (e.g., 5). Additionally, the distribution of diagnostic accuracy across cases, visualized in Fig. 4b, showed that the median and upper quartile for Dual-Inf (0.495 and 0.746) outperformed SC-CoT (0.434 and 0.652) and Diagnosis-CoT (0.421 and 0.628), with statistically significant improvements (p < 0.001). These findings highlight the efficacy of Dual-Inf in enhancing diagnostic accuracy.
a Data statistics of the iteration number for each note in Dual-Inf. b Distribution visualization of diagnostic accuracy on each note. Diag-CoT denotes Diagnosis-CoT, and Self-Cont means Self-Contrast. c Performance change on Dual-Inf w.r.t diagnosis and explanation after each iteration. We randomly selected ten notes with five iterations. In this figure, the base LLM of the methods is GPT-4. d Distribution visualization of interpretation performance on each note. SC-CoT and Dual-Inf were implemented with GPT-4. The circular points shown as outliers mean that some scores are deviated from the vast majority.
Second, Dual-Inf produced superior DDx explanations. Manual evaluation of 100 cases (Fig. 2b) showed higher scores across all metrics, attributed to bidirectional inferences and iterative prediction refinement. Iterative reflection effectiveness was confirmed through ten notes with five iterations (Fig. 4c) and a case study of intermediate predictions (Supplementary Appendix 7), both demonstrating improved explanations over iterations. Distribution analysis (Fig. 4d) revealed higher median and quartile scores (e.g., BERTScore, METEOR) for Dual-Inf compared to SC-CoT, confirming its ability to generate better explanations across most cases. Notably, although the note snippets were publicly available, the ground-truth of DDx and the corresponding explanations were manually generated by our domain experts. Therefore, the LLMs have not been exposed to the ground-truth, and the evaluation on our dataset is trustworthy.
Third, this study demonstrated that leveraging multiple LLMs mitigates explanation errors in DDx. As shown in Fig. 2c, Self-Contrast and SC-CoT reduced low-relevance errors compared to CoT and Diagnosis-CoT, highlighting the benefit of integrating multiple LLM interpretations to address hallucinations. Dual-Inf further minimized errors across all the types, attributed to its dual-inference scheme: the forward-inference module generated diagnoses, the backward-inference module recalled medical knowledge, and the examination module refined predictions. The self-reflection mechanism further improved explanation quality and reduced hallucinations through iterative refinement. Additionally, the higher error rates observed in the nervous and digestive disease specialties were attributed to their larger sample sizes in the dataset. However, normalizing error counts by sample size revealed comparable error rates across the specialties.
One limitation of this study is that our dataset, encompassing nine clinical specialties, does not fully capture the breadth of real-world scenarios. Besides, the dataset lacks annotations on the priority of each diagnosis within the DDx, as ranking the likelihood of possible diseases presents significant challenges. Furthermore, the backward-inference module’s reliance on internal medical knowledge to generate reference signs and symptoms makes it vulnerable to severe hallucinations or erroneous knowledge, which could impact performance. This issue can be mitigated by implementing Dual-Inf with advanced LLMs32.
In summary, this study established a manually annotated dataset for explainable DDx and designed a tailored framework that effectively harnessed LLMs to generate high-quality explanations. The findings revealed that existing prompting methods exhibited suboptimal performance in generating DDx and explanations, limiting their practical utility in clinical scenarios. Our experiments verified the effectiveness of Dual-Inf for providing accurate DDx, delivering comprehensive explanations, and reducing prediction errors. Furthermore, the released dataset with ground-truth DDx and explanations could facilitate the research field. Future work could expand the dataset to a broader range of clinical specialties or integrate domain knowledge from external databases for superior performance.
Methods
Data acquisition and processing
The data source is publicly available medical exercises collected from medical books33,34 and MedQA USMLE dataset35. There are two key criteria for selecting the clinical notes: (1) the notes must originate from disease diagnosis exercises; (2) they must pertain to one of the nine clinical specialties. We transformed the exercises into free text by preserving the symptom descriptions and removing the multiple-choice options, where applicable. The texts were further preprocessed, including (1) removing duplicate notes, (2) unifying all characters into UTF-8 encoding and removing illegal UTF-8 strings, (3) correcting or removing special characters, and (4) filtering out notes with fewer than 130 characters. Lastly, we collected 570 clinical notes, among which 10 notes were used for prompt development, and 560 notes were preserved for evaluation. The full dataset can be found in Supplementary Appendix 13.
Data annotation
The raw data generally did not have the annotation of DDx, explanation, and clinical specialty. To build a well-annotated dataset, we employed three clinical physicians to curate the dataset manually. Two independent physicians annotated each exercise. When disagreement existed in the annotation, a third physician examined the case and made the final annotation. We checked the inter-annotator agreement (IAA) on DDx, interpretation, and specialty (Supplementary Appendix 1). Additionally, our dataset is well-structured in a standardized format, facilitating automated evaluation.
Model development
How to effectively elicit LLMs’ capability for accurate DDx explanation is challenging. Inspired by the fact that humans usually conduct backward reasoning to validate the correctness of answers when solving reasoning problems36, we proposed performing backward verification (i.e., from diagnosis to symptoms) to examine the predicted diagnoses and elicit correct answers via self-reflection. Accordingly, we developed a customized framework called Dual-Inference Large Language Model (Dual-Inf), shown in Fig. 1a. Specifically, Dual-Inf consisted of four components: (1) a forward-inference module, which was an LLM for initial diagnoses, i.e., from patients’ symptoms to diagnoses, (2) a backward-inference module, which was an LLM for inverse inference via recalling all the representative symptoms of the initial diagnoses, i.e., from diagnoses to symptoms, (3) an examination module, which was another LLM that received patients’ notes and the output from the two modules for prediction assessment and decision making, and (4) an iterative self-reflection mechanism, which iteratively took low-confidence diagnoses as feedback for the forward-inference module to “think twice”.
The pipeline was as follows. First, the forward-inference module analyzed clinical notes to infer initial diagnoses and provide interpretations. Next, the backward-inference module received the initial diagnoses as input and recalled the representative symptoms that the diagnoses generally present, including medical examination and laboratory test results. Given that the recalled symptoms were derived from the LLM’s internal knowledge, they are generally reliable in advanced LLMs30,32 and could serve as a reference for measuring the correctness of the predicted explanations. Afterward, the examination module verified and refined the above results. Specifically, it (i) checked the forward-inference module’s explanations against the recalled knowledge and discarded erroneous ones, (ii) supplemented the interpretations by integrating patient notes with recalled knowledge, (iii) decided whether to accept or filter predictions based on their quality. The underlying idea was that a diagnosis supported by fewer interpretations was deemed less trustworthy. To assess diagnostic confidence, a threshold β was applied: diagnoses with fewer than β supporting interpretations were flagged as low-confidence. Later, the self-reflection mechanism took the low-confidence diagnoses as feedback to prompt the forward-inference module to “think twice.” This iterative process continued up to a maximum limit λ, balancing accuracy with efficiency. Upon reaching this limit, the framework outputted the final results. The prompts for the three modules are detailed in Supplementary Appendix 9. Importantly, the prompts for the forward-inference module were carefully designed to ensure objectivity toward feedback from the examination module, reducing the risk of false negatives undermining correct predictions.
Implementation details
We adopted four baselines: (1) CoT20, a popular prompting method; (2) Diagnosis-CoT19, a customized prompting method for disease diagnosis; (3) Self-Contrast37, an advanced method with multiple prompts and a re-examination mechanism to enhance reasoning; (4) Self-consistency CoT (SC-CoT)38, which assembled multiple reasoning paths to enhance performance. We followed the original papers in the implementation. Specifically, SC-CoT generated five reasoning paths for each note and then selected the most consistent diagnoses and interpretations. The prompts of baselines were shown in Supplementary Appendix 10. As for Dual-Inf, we incorporated CoT into the three LLM-based modules. The maximum iteration number λ was assigned to 5, considering the trade-off between effectiveness and efficiency; the threshold β was set to 3. We further analyzed the impact of the hyper-parameter β on the performance and presented the results in Supplementary Appendix 11. For a fair comparison, all the methods were implemented with the same base LLM, including GPT-4, GPT-4o, Llama3-70B (https://huggingface.co/meta-llama/Meta-Llama-3-70B), and BioLlama3-70B (https://huggingface.co/aaditya/Llama3-OpenBioLLM-70B). For the former two LLMs, we used the API from the OpenAI company (https://platform.openai.com/docs/models), which were “gpt-4-turbo-preview” and “gpt-4o”; for the latter two, we downloaded the models from Huggingface for inference. The temperature parameter was set as 0.1.
Performance evaluation
We conducted automatic evaluation by comparing the ground-truths with the predicted ones. Following related papers27, we used accuracy as the primary metric for assessing diagnostic performance, i.e.,
For interpretation performance, we employed metrics designed to assess the semantic alignment between the reference text and the predicted text, rather than relying solely on string matching. The metrics, including accuracy, BERTScore39, SentenceBert40, and METEOR41, have been widely used in related tasks42,43. Concretely, interpretation accuracy was computed as:
BERTScore39 employs the BERT model44 to determine the semantic similarity between reference and generated text, offering a context-aware evaluation of model performance. SentenceBert40 measures sentence similarity using a BERT model that generates dense vector representations, facilitating efficient and accurate semantic comparisons. METEOR41 assesses the harmonic mean of unigram precision and recall, utilizing stemmed forms and synonym equivalence. The details of automatic and human evaluation are shown in Supplementary Appendix 2.
Data availability
Data is provided in the supplementary information files.
Code availability
The code used for this study is available at https://github.com/betterzhou/Dual-Inf.
References
Adler-Milstein, J., Chen, J. H. & Dhaliwal, G. Next-generation artificial intelligence for diagnosis: from predicting diagnostic labels to “wayfinding. Jama 326, 2467–2468 (2021).
Fansi Tchango, A. et al. Towards trustworthy automatic diagnosis systems by emulating doctors’ reasoning with deep reinforcement learning. Adv. Neural Inf. Process. Syst. 35, 24502–24515 (2022).
Wu, L. et al. Differential diagnosis of secondary hypertension based on deep learning. Artif. Intell. Med. 141, 13 (2023).
Levy, J., Álvarez, D., Del Campo, F. & Behar, J. A. Deep learning for obstructive sleep apnea diagnosis based on single channel oximetry. Nat. Commun. 14, 4881 (2023).
Zhang, Z. et al. Pathologist-level interpretable whole-slide cancer diagnosis with deep learning. Nat. Mach. Intell. 1, 236–245 (2019).
Zakka, C. et al. Almanac—retrieval-augmented language models for clinical medicine. NEJM AI 1, AIoa2300068 (2024).
Wu, S. et al. Benchmarking open-source large language models, GPT-4 and Claude 2 on multiple-choice questions in nephrology. NEJM AI 1, AIdbp2300092 (2024).
Tu, T. et al. Towards generalist biomedical AI. NEJM AI 1, AIoa2300138 (2024).
Singhal, K. et al. Towards Expert-Level Medical Question Answering with Large Language Models. ArXiv, vol. abs/2305.09617, (2023).
Tang, L. et al. Evaluating large language models on medical evidence summarization. NPJ Dig. Med. 6, 158 (2023).
Benary, M. et al. Leveraging Large Language Models for Decision Support in Personalized Oncology. JAMA Netw. Open 6, e2343689–e2343689 (2023).
Kanjee, Z, Crowe, B & Rodman, A Accuracy of a generative artificial intelligence model in a complex diagnostic challenge. Jama 330, 78–80 (2023).
Kwon, T. et al. Large language models are clinical reasoners: Reasoning-aware diagnosis framework with prompt-generated rationales. in Proceedings of the AAAI Conference on Artificial Intelligence. 18417–18425 (2024).
Hua, R. et al. Lingdan: enhancing encoding of traditional Chinese medicine knowledge for clinical reasoning tasks with large language models. J. Am. Med. Informat. Asso. 31, 2019–2029 (2024).
Chen, J. et al. CoD, Towards an Interpretable Medical Agent using Chain of Diagnosis. arXiv preprint arXiv:2407.13301, (2024).
Zhou, S. et al. Large Language Models for Disease Diagnosis: A Scoping Review. ArXiv, vol. abs/2409.00097, (2024).
Tu, T. et al. Towards conversational diagnostic AI. arXiv preprint arXiv:2401.05654, 2024.
McDuff, D. et al. (2023, November 01, 2023). Towards Accurate Differential Diagnosis with Large Language Models. arXiv:2312.00164. Available: https://ui.adsabs.harvard.edu/abs/2023arXiv231200164M.
Savage, T., Nayak, A., Gallo, R., Rangan, E. S. & Chen, J. H. Diagnostic reasoning prompts reveal the potential for large language model interpretability in medicine. NPJ Digit. Med 7, 20 (2023).
Wei, J. et al. Chain-of-thought prompting elicits reasoning in large language models. Adv. neural Inf. Process. Syst. 35, 24824–24837 (2022).
Tchango, A. F., Goel, R., Wen, Z., Martel, J. & Ghosn, J. DDXPlus: A New Dataset For Automatic Medical Diagnosis. in Neural Information Processing Systems, (2022).
Wu, J. et al. Clinical Text Datasets for Medical Artificial Intelligence and Large Language Models — A Systematic Review. NEJM AI 1, AIra2400012 (2024).
Ashwani, S. et al. Cause and effect: Can large language models truly understand causality? in Proceedings of the AAAI Symposium Series, 2–9 (2024).
Chi, H. et al. Unveiling Causal Reasoning in Large Language Models: Reality or Mirage? in The Thirty-eighth Annual Conference on Neural Information Processing Systems.
M. Parmar, et al. “LogicBench: Towards systematic evaluation of logical reasoning ability of large language models. in Proceedings of the 62nd Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), 2024, 13679-13707.
Chen, A. et al. Two Failures of Self-Consistency in the Multi-Step Reasoning of LLMs. Transac. Machine Learning Res. (2024).
Hager, P. et al. Evaluation and mitigation of the limitations of large language models in clinical decision-making. Nat. Med. 30, 2613–2622 (2024).
Williams, C. Y., Miao, B. Y., Kornblith, A. E. & Butte, A. J. Evaluating the use of large language models to provide clinical recommendations in the Emergency Department. Nat. Commun. 15, 8236 (2024).
Macherla, S., Luo, M., Parmar, M. & Baral, C. MDDial: A Multi-turn Differential Diagnosis Dialogue Dataset with Reliability Evaluation. arXiv preprint arXiv:2308.08147, (2023)
Singhal, K. et al. Large language models encode clinical knowledge. Nature 620, 172–180 (2023).
Chen, X. et al. FFA-GPT: an automated pipeline for fundus fluorescein angiography interpretation and question-answer. npj Digital Med. 7, 111 (2024).
Goh, E. et al. Large language model influence on diagnostic reasoning: a randomized clinical trial. JAMA Netw. Open 7, e2440969–e2440969 (2024).
Le, T., Bhushan, V., Sheikh-Ali, M. & Shahin F. A. First Aid for the USMLE Step 2 CS, Third Edition: McGraw-Hill Education, (2009).
Le, T. & Bechis, S.K. First Aid Q&A for the USMLE Step 1, Second Edition: McGraw-Hill Education, (2009).
Jin, D. et al. What Disease Does This Patient Have? A Large-Scale Open Domain Question Answering Dataset from Medical Exams. Appl. Sci. 11, 6421 (2021).
Jiang, W. et al. Forward-backward reasoning in large language models for mathematical verification. in Findings of the Association for Computational Linguistics ACL 2024, 6647–6661 (2024).
Zhang, W. et al. Self-Contrast: Better Reflection Through Inconsistent Solving Perspectives. Annual Meeting of the Association for Computational Linguistics. (2024).
Wang, X. et al. Self-Consistency Improves Chain of Thought Reasoning in Language Models. in The Eleventh International Conference on Learning Representations.
Zhang, T., Kishore, V., Wu, F., Weinberger, K. Q. & Artzi, Y. BERTScore: Evaluating Text Generation with BERT. ArXiv, vol. abs/1904.09675, 2019.
Reimers, N. & Gurevych, I. Sentence-BERT: Sentence Embeddings using Siamese BERT-Networks. Conference on Empirical Methods in Natural Language Processing, (2019).
Banerjee, S. & Lavie, A. METEOR: An automatic metric for MT evaluation with improved correlation with human judgments. in Proceedings of the acl workshop on intrinsic and extrinsic evaluation measures for machine translation and/or summarization, 65–72 (2005).
Abbasian, M. et al. Foundation metrics for evaluating effectiveness of healthcare conversations powered by generative AI. NPJ Digital Med. 7, 82 (2024).
Celikyilmaz, A., Clark, E. & Gao, J. Evaluation of Text Generation: A Survey. ArXiv, vol. abs/2006.14799, (2020).
JDevlin, J., Chang, M.-W., Lee, K. & Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. in North American Chapter of the Association for Computational Linguistics, (2019).
Acknowledgements
This work was supported by the National Institutes of Health’s National Center for Complementary and Integrative Health under grant number R01AT009457, National Institute on Aging under grant number R01AG078154, and National Cancer Institute under grant number R01CA287413. The content is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health. We also acknowledge support from the Center for Learning Health System Sciences, a partnership between the University of Minnesota Medical School and School of Public Health.
Author information
Authors and Affiliations
Contributions
S.Z. and R.Z. conceptualized the study. S.Z. contributed to the literature search and model evaluation. S.Z. and S.D. performed the data collection, prompt design, and model construction. S.Z., R.Z., and G.M. discussed and arranged data annotation and human evaluation. S.Z., M.L., and R.Z. conducted experimental design. S.Z. performed manuscript drafting. R.Z. supervised the study. All authors contributed to the research discussion, manuscript revision, and approval of the manuscript for submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhou, S., Lin, M., Ding, S. et al. Explainable differential diagnosis with dual-inference large language models. npj Health Syst. 2, 12 (2025). https://doi.org/10.1038/s44401-025-00015-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s44401-025-00015-6
This article is cited by
-
Automating expert-level medical reasoning evaluation of large language models
npj Digital Medicine (2025)
-
Uncertainty-aware large language models for explainable disease diagnosis
npj Digital Medicine (2025)
-
Large language models for disease diagnosis: a scoping review
npj Artificial Intelligence (2025)
-
Large language models with retrieval-augmented generation enhance expert modelling of Bayesian network for clinical decision support
International Journal of Computer Assisted Radiology and Surgery (2025)